Abstract
The contribution of combined sewer overflows (CSO) to the viral contamination of receiving waters was determined. Adenovirus concentrations were determined using the Primary Liver Carcinoma (PLC/PRF/5) cell line and confirmed by Polymerase Chain Reaction (PCR). Norovirus concentration was determined using the Most Probable Number (MPN) and Reverse Transcription-Polymerase Chain Reaction (RT-PCR). Seventy-five water samples were collected during dry weather and 50 samples were collected during wet weather. CSO events significantly increased the concentration of culturable viruses, adenoviruses, and noroviruses in the receiving waters (P < 0.01). During dry weather, 56% of samples were positive for total virus cytopathic effects (CPE), adenoviruses were detected in 41% of the positive cell cultures, and noroviruses in 6% of the concentrates by direct RT-PCR. During wet weather, 100% of the samples were positive by CPE, 84% for adenoviruses, and 40% in the concentrates for norovirus. Our results demonstrate that CSOs can contribute significant viral loading to receiving waters.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The major objective of sewerage systems is to reduce contamination of water bodies by reducing the nutrients and microbial loads, including human pathogens of the discharged treated wastewater (Gerba 2009; Rose et al. 1996). In older large cities, sewage is combined with storm water before treatment. During wet weather, the amount of combined sewage often surpasses the treatment capacity of the wastewater treatment plant (WWTP) and the combined sewer overflow (CSO) is discharged untreated into the water body.
CSOs often result in decreased water quality by increasing the biological oxygen demand as the result of releasing nutrients (Even et al. 2007) and decreasing the microbial quality of the receiving waters (Ellis and YU 1995). In addition, CSOs have been found to increase the concentration of microbial indicators of fecal contamination, such as fecal coliforms, fecal streptococci, Clostridium perfringens, Aeromonas spp., and F-RNA bacteriophages in water (Ferguson et al. 1996). High-rate flocculation and disinfection treatments have the capacity to treat the combined sewage during wet weather conditions, and to reduce the amount of suspended matter and pathogens in the CSO. Disinfection treatment for wet weather sewage flows includes the use of ozone and ultraviolet light (UV) irradiation (Wojtenko et al. 2001a, 2001b).
Adenovirus and norovirus are two of the nine microbiological agents on the United States Environmental Protection Agency’s Contaminant Candidate List (CCL) for regulation in drinking water. Adenovirus is a non-enveloped, double-stranded DNA virus. It has been described as the most resistant enteric virus to UV light disinfection (Thompson et al. 2003; Thurston-Enriquez et al. 2003). Adenoviruses have been detected in surface waters (Fong et al. 2005), source waters (Chapron et al. 2000), and wastewaters (Sedmak et al. 2005).
Norovirus is a non-enveloped, positive strand RNA virus. Norovirus is one of the leading agents responsible for viral gastroenteritis in the United States (Fankhauser et al. 2002). The virus cannot be grown routinely in cell culture; however, reverse transcription polymerase chain reaction (RT-PCR) has proven to be successful for the detection of norovirus in water (Haramoto et al. 2006; Karim and LeChevallier 2004; Katayama et al. 2008). Although, a viral infectivity assay has just been described (Straub et al. 2007), this assay has not been applied to environmental samples.
Information concerning the prevalence and concentration of viral pathogens in CSO is necessary for treatment design and risk assessment during wet weather events. The aim of this research was to extend our previous observations (Rijal et al. 2009, 2011) on pathogens in wet weather events to specifically assess impact of CSO on the concentrations of total culturable viruses (TCV), adenoviruses, and noroviruses on the viral load in receiving waters.
Materials and Methods
Water Sampling and Processing
Water samples were collected in the water ways of three different wastewater treatment plants (WWTP) located in the greater Chicago. In each WWTP waterway, samples were collected in the outfall, upstream, and downstream the outfall (Table 1). The treatment of these WWTPs consisted of activated sludge secondary treatment with no disinfection before discharge. All of the plants discharge high quality secondary treated effluent into a large canal waterway system where recreation is limited to noncontact activity (i.e., boating). Sampling was conducted between the months of June and October covering the recreational season in the CAW for two consecutive years on a monthly basis depending on the weather conditions. Between 120 and 280 l of water were processed using 1-MDS Virosorb filters (CUNO, Meridian, CT), and eluted using beef extract (pH 9.0) and further concentrated by organic flocculation as described USEPA Viradel method (USEPA 2001a). The efficiency of the organic flocculation was determined using poliovirus type 1 (LSc-2ab, obtained from the culture collection of the Department of Virology and Epidemiology at Baylor College of Medicine, Houston, TX) and averaged 60%.
Detection of Culturable Viruses
Concentrated samples were assayed for total culturable viruses using the PLC/PRF/5 cell line as described previously (Rodriguez et al. 2008) and modified from the USEPA Total Culturable Viruses Assay (USEPA 2001b). For confirmation of CPE-positive flask for adenovirus by PCR, CPE-positive flasks were frozen and thawed three times; cell lysates were collected, filtered through a 0.22 μm pore size filter and then stored at −20°C until the confirmatory analysis. CPE-positive flasks from second or third passage were used for the calculation of viral concentration by most probable number (MPN) as described by USEPA (2001b).
Detection of Adenovirus by Nested-PCR (nPCR)
The presence of adenovirus genome was confirmed from cell lysate using an nPCR approach for the detection of human adenovirus from group A–F. The primers for the detection of the hexon capsid protein gene region of the adenovirus genome were obtained from Avellón et al. (2001). Conditions for the first and second round of PCR were described by Rodriguez et al. (2008). Positive flasks for the presence of adenovirus PCR were used to estimate the adenovirus concentration by most probable method as described in previous section, but instead of using CPE positive flasks, PCR positive flask were using for the calculation.
Viral RNA Extraction and RT-PCR for Detection of Norovirus
The samples were extracted in triplicate using a QIAamp viral RNA extraction kit (Qiagen, Valencia, CA) as described by the manufacturer. A sample concentrate of 140 μl was extracted, and the purified RNA was resuspended in 60 μl. The washing steps were repeated twice as recommended by Qiagen technical support to remove inhibitory compounds.
One-step reverse transcription (RT)-PCR was performed for the detection of the norovirus genome polymerase region A using the primers described by Vinjé et al. (2004). The one-step RT-PCR mixture consists of: 1× PCR gold buffer (Applied Biosystems, Roche Molecular Systems Inc, Branchburg, NJ), 3.5 mM of MgCl, 300 μM of each dNTP, 25 units of Murine Leukemia Virus (MuLV) Reverse Transcriptase (Applied Biosystems), 10 units RNase inhibitor (Applied Biosystems), 2.5 mM of Random Hexamers (Applied Biosystems), 0.5 μM of each primer specific for the polymerase region of the norovirus genome (MJV 12 and Reg A), 1.5 units of Amplitaq gold (Applied Biosystems), and 10 μl of sample and water to a total volume of 25 μl. The RT-PCR conditions were the following: 30 min at 42°C, 15 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 50°C, 30 s at 72°C; and a final extension of 10 min at 72°C. For semi-nested PCR, the followed internal primer was used MP 290: 5′-GAY TAC TCY CSI TGG GAY TC-3′ (Maloney 2007). The semi-nested PCR mixture consisted of 1× PCR gold buffer, 2.5 mM of MgCl, 200 μM of each dNTP, 1 μM of internal primer mp 290, and 1 μM of primer Reg A, 1.5 units of Amplitaq gold, 2 μl of RT-PCR product, and water for a total volume of 50 μl. The PCR conditions for the semi-nested PCR were as follows: 5 min at 95°C, 40 cycles of 30 s at 94°C, 30 s at 49°C and 30 s at 72°C, and a final extension of 10 min at 72°C. Samples were assayed in triplicate and diluted 10-, 100-, and 1,000-fold, so that an MPN estimate could be obtained for norovirus genomes in each sample.
Quality Control Assurance
The cell culture facility and incubator were physically separated from the PCR facility. PCR workstations with no airflow and UV light were used to provide clean working areas. One biological hazard type II hood was used for processing the samples. All of the areas for the PCR/RT-PCR analysis were physically separated: one room for sample preparation, one room for gel electrophoresis, and one room for nested PCR. All of the reagents were stored in a separate room from the samples. The workstations were cleaned with 10% bleach solution and exposed to UV light for at least 20 min before and after use. Equipment used in each room was not transported to other areas (i.e., pipettes, tips, and lab coats were exclusively used in only one room). The PCR thermocyclers are located in another room outside of the work area. RNA free water was used as a negative control. All the RT and PCR reagents were mixed in the workstation in the room for reagent preparation.
Statistical Analysis
Viral MPN/100 l was calculated using the MPN general purpose program by Hurley and Roscoe (1983). The differences between samples from sampling locations and the impact of CSOs were analyzed using analysis of variance (ANOVA) and Tukey’s analysis (Minitab 14, Minitab Inc., Cary, NC).
Results
Over the two-year study, 125 samples were collected from the impacted waterways. During dry weather, 75 samples were collected with no combined sewage overflow reported (no-CSO). During the wet weather, 50 samples were collected after reported rain events with CSO discharges. Of the total samples collected, 20 were taken from the outfall, 42 samples were taken upstream (in reference to the outfall), and 63 samples were taken downstream. The lower detection limit of the cell culture assays for the detection of TCV was calculated at between 1 and 0.5 MPN/100 l depending of sample volume concentrated. The lower detection limit for the use of RT-PCR for the detection of norovirus was calculated at between 24 and 58 RT-PCR MPN/100 l.
The results of the samples are presented in Table 2. The CPE-positive PLC/PRF/5 flasks were used to calculate total culturable virus (TCV) numbers, and the adenovirus numbers were calculated using the CPE-positive flasks that were also positive for the confirmation of adenovirus by PCR. Of the 75 samples collected during no-CSO, 42 samples were positive for TCV, 31 were positive for human adenoviruses and five were positive for norovirus. All of the samples collected after CSO demonstrated CPE; adenoviruses were detected in 42 of these samples. Norovirus was detected in 20 of the 50 samples collected during the CSO.
A significant difference (P < 0.01) was noted in the concentration of all viruses between the no-CSO and CSO samples (Table 3). During no-CSO conditions, the average concentration of TCV was 37.5 MPN/100 l at the outfall, 13.4 MPN/100 l downstream, and 8.4 MPN/100 l in the upstream samples. For adenovirus, the average concentrations at the outfall were 20.2 MPN/100 l, 4.3MPN/100 l in the samples collected downstream and 1.2 MPN/100 l in samples collected upstream. During dry weather, no-CSO, there was a significant effect of the sampling location in the concentration of the adenoviruses; no significant difference was observed in the concentration of viruses between the waterways of the three WWTPs (P = 0.007 for sampling location and 0.051 for WWTPs, Table 3). For adenovirus during no-CSO, the concentration was significantly higher in the outfall than in the downstream and upstream samples (Table 4). The average concentration of noroviruses detected in the outfall was 120 RT-PCR MPN/100 l, in downstream samples 2.4 RT-PCR MPN/100 l, and upstream 11.6 RT-PCR MPN/100 l. However, there was not a significant difference between sampling location for the concentrations of noroviruses (Table 3).
For samples collected during wet weather conditions with reported CSOs, the average concentrations of TCV were 593 MPN/100 l, >601 MPN/100 l, and 449 MPN/100 l in the outfall, downstream and upstream, respectively. For adenoviruses, the average concentrations were 182 MPN/100 l, >317 MPN/100 l, and 302 MPN/100 l in the outfall, downstream, and upstream samples, respectively. Owing to the weather conditions, not many samples were collected in the outfall of the WWTP during CSO. The effect of sampling location was not significant for TCV and adenoviruses in the samples collected during CSOs (Table 3). For noroviruses, the concentrations were 418 RT-PCR MPN/100 l, 330 RT-PCR MPN/100 l, and 100 RT-PCR MPN/100 l in the samples collected in the outfall, downstream, and upstream from the WWTP. The difference in norovirus concentration was not significant between sample locations during CSO (Table 3).
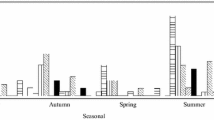
Figure 1 compares the concentration of viruses in samples collected during dry weather and during CSOs (wet weather). The average virus concentrations were higher during reported CSO events, including adenoviruses and noroviruses.
Discussion
The concentrations of total cultivable viruses, infectious adenoviruses and norovirus were monitored in waters receiving the effluent of three wastewater treatment plants for the period of 2 years during dry and wet weather. The concentration of infectious viruses increased more than tenfold after CSO events. We also found that the detection of norovirus by nested RT-PCR increased from 5% positive of the downstream samples in dry weather conditions to 45% of positive downstream samples on wet weather conditions after CSO events. Katayama et al. (2004) monitored the occurrence of noroviruses and infectious enteroviruses in Japan after rain events in the effluent of the WWTP and in the receiving waters of Tokyo Bay. They reported that 26% of the samples collected from the receiving waters were positive for noroviruses and 50% for infectious enteroviruses and these concentrations remained elevated for 4 days after CSO events. CSOs have been receiving attention because of interest in the development of new technologies for treatment of CSO waters before discharges and possible regulations on microbial loads in these waters. Jiang and Chu (2004) have reported increased frequency of detecting adenoviruses by PCR after rainfall events in coastal areas and rivers in Southern California, but these areas receive tertiary disinfected wastewater and the majority of adenoviruses detected were likely noninfectious.
Previously we described the use of the PLC/PRF/5 cell line for the total culturable virus assay of sewage samples. The use of this cell line resulted in an increase of fifty times the culturable viruses detected when compared with the use of the BGM cell line (Rodriguez et al. 2008). Of all culturable viruses detected in that study, 60% of were adenoviruses or enteroviruses as confirmed by PCR analysis of the CPE-positive flasks (Rodriguez et al. 2008). The culturable viral composition in sewage has been described to include echovirus, poliovirus, coxsackieviruses, enterovirus, reovirus, and adenoviruses, but their detection requires the use of several cell lines (Sedmak et al. 2005). Reoviruses, coxsackievirus, poliovirus, and adenoviruses are able to propagate in the PLC/PRF/5 cell line (Grabow et al. 1992, 1999). Therefore, it is probable that the culturable viruses detected with the PLC/PRF/5 cell line comprise a good proportion of viruses usually found in sewage. In the USA, the poliovirus vaccine is administered in a series of injections of inactivated polioviruses; therefore, poliovirus from the oral live attenuated vaccine is not expected to be found in the sewage and the enteroviruses found in sewage are not derived from any vaccination program.
In this study, adenovirus concentration in the outfall of the WWTP ranged from 3.5 to 52.7 MPN/100 l during dry weather conditions and from 118 to 751 MPN/100 l after rainfall events. The concentration of adenovirus in influents (untreated wastewater) of WWTP has been reported to range from 0 to 200 infectious units/l (Sedmak et al. 2005) and 107 genomes/100 l by PCR (He and Jiang 2005). These concentrations are higher than the concentrations found in this study after rainfall events. A dilution of untreated sewage with storm water is expected during CSO events. In spite of the dilution of the sewage with storm water, CSO discharges resulted in increased viral concentration in the receiving waters. Adenoviruses were detected by assaying the cell extract of CPE-positive flasks with PCR. The primers used were specific for the hexon gene of human adenovirus capable of detecting serogroups A–F (Avellón et al. 2001). These groups can cause a wide range of symptoms including but not limited to gastroenteritis, conjunctivitis, and respiratory diseases. Their detection is also important because they are among the most resistant viruses to UV disinfection, which is one of the technologies available for the disinfection of CSO discharges. Human adenovirus has not been detected from any other source other than humans (Jiang 2006), and it has been suggested as a possible candidate for an indicator of the presence of human fecal contamination in water (Fong et al. 2005).
An increase in the number of samples positive for noroviruses was found after CSO events. The primers set used during this study are specific for viral RNA-dependent RNA polymerase (NS7), they have been described to be able to detect a wide range of human noroviruses including GI and GII (Vinjé et al. 2004). The average concentration of norovirus in the waterways went from 29.8 RT-PCR MPN/100 l during dry weather (no-CSO) to 275 RT-PCR MPN/100 l during wet weather (with CSO). The concentration of noroviruses in the sewage can range from 102 to 107 genome copies/l in which GII was more abundant than GI (Haramoto et al. 2006). Wastewater treatment has been demonstrated to reduce by up to 3 logs the norovirus concentration found in raw sewage (as determined by genomes copies); however, norovirus could be easily detected in the WWTP effluents (da Silva et al., 2007). In this study, norovirus concentrations in the secondary treated effluent appear lower than the concentrations of norovirus reported by Haramoto et al. (2006). However, it will be difficult to compare results obtained by real-time RT-PCR with results obtained with nested PCR–MPN because there is no any reference for comparison between both methods. The real-time PCR method is more commonly used and standardized for the quantification of viral genomes in the environment. In addition, this difference in norovirus concentration between both studies may be the result of seasonal changes in the concentration of norovirus in sewage and the incidence of infection within the studied communities (Haramoto et al. 2006; Katayama et al. 2008). Other enteric viruses such as enteroviruses and adenoviruses did not show a clear seasonal variation and their detection is constant throughout the year (Katayama et al. 2008; Choi and Jiang 2005; Sedmak et al. 2005). Finally, the RT-PCR used did not provide any information about the infectivity of the viruses; however, because the wastewater or CSO water was not disinfected, it is assumed that most of the noroviruses detected were infectious.
In conclusion, this study demonstrated that combined sewage overflows and wet weather conditions contribute a significant viral load to the receiving waters. Microbial activity has been described to play an important role in the inactivation of viruses in fresh water (Ward et al. 1986). It will be important to further study the role of microbial activity in the inactivation of viral contamination after CSO events in the receiving waters. Information provided in this study is an important in future decisions for the management of CSO-impacted water bodies.
References
Avellón, A., Pérez, P., Aguilar, J. C., Lejarazu, R., & Echevarría, J. E. (2001). Rapid and sensitive diagnosis of human adenovirus infections by a generic polymerase chain reaction. Journal of Virological Methods, 92, 113–120.
Chapron, C. D., Ballester, N. A., Fontaine, J. H., Frades, C. N., & Margolin, A. B. (2000). Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Applied and Environmental Microbiology, 66, 2520–2525.
Choi, S., & Jiang, S. C. (2005). Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Applied and Environmental Microbiology, 71, 7426–7433.
da Silva, A. K., Saux, J., Parnaudeau, S., Pommepuy, M., Elimelech, M., & Le Guyader, F. S. (2007). Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: Different behaviors of genogroups I and II. Applied and Environmental Microbiology, 73, 7891–7897.
Ellis, J. B., & Yu, W. (1995). Bacteriology of urban runoff—the combined sewer as a bacterial reactor and generator. Water Science and Technology, 31, 303–310.
Even, S., Mouchel, J. M., Servais, P., Flipo, N., Poulin, M., Blanc, S., et al. (2007). Modelling the impacts of combined sewer overflows on the river seine water quality. Science of The Total Environment, 375, 140–151.
Fankhauser, R. L., Monroe, S. S., Noel, J. S., Humphrey, C. D., Bresee, J. S., Parashar, U. D., et al. (2002). Epidemiologic and molecular trends of “Norwalk-Like viruses” associated with outbreaks of gastroenteritis in the United States. Journal of Infectious Diseases, 186, 1–7.
Ferguson, C. M., Coote, B. G., Ashbolt, N. J., & Stevenson, I. M. (1996). Relationships between indicators, pathogens and water quality in an estuarine system. Water Research, 30, 2045–2054.
Fong, T. T., Griffin, D. W., & Lipp, E. K. (2005). Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Applied and Environmental Microbiology, 71, 2070–2078.
Gerba, C. P. (2009). Wastewater treatment and biosolids reuse. In R. M. Maier, I. L. Pepper, & C. P. Gerba (Eds.), Environmental microbiology (2nd ed., pp. 503–530). San Diego: Academic Press.
Grabow, W. O., Puttergill, D. L., & Bosch, A. (1992). Propagation of adenovirus types 40 and 41 in the PLC/PRF/5 primary liver carcinoma cell line. Journal of Virological Methods, 37, 201–207.
Grabow, W., Botma, K. L., Villiers, J. C., Clay, C. G., & Erasmus, B. (1999). Assessment of cell culture and polymerase chain reaction procedures for the detection of poliovirus in wastewater. Bulletin—World Health Organisation, 77, 973–978.
Haramoto, E., Katayama, H., Oguna, K., Yamashita, H., Tarima, A., Nakajima, H., et al. (2006). Seasonal profiles of human norovirues and indicator bacteria in the wastewater treatment plant in Tokyo, Japan. Water Science and Technology, 54, 301–308.
He, J. W., & Jiang, S. (2005). Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Applied and Environmental Microbiology, 71, 2250–2255.
Hurley, M. A., & Roscoe, M. E. (1983). Automated statistical-analysis of microbial enumeration by dilution series. Journal of Applied Bacteriology, 55, 159–164.
Jiang, S. C. (2006). Human adenoviruses in water: occurrence and health implications: A critical review. Environmental Science Technology, 40, 7132–7140.
Jiang, S. C., & Chu, W. (2004). PCR detection of pathogenic viruses in southern Califonia urban rivers. Journal of Applied Microbiology, 97, 17–28.
Karim, M. R., & LeChevallier, M. W. (2004). Detection of noroviruses in water: Current status and future directions. Journal of Water Supply: Research and Technology-AQUA, 53, 359–380.
Katayama, H., Okuma, K., Furumai, H., & Ohgaki, S. (2004). Series of surveys for enteric viruses and indicator organisms in Tokyo Bay after an event of combined sewer overflow. Water Science and Technology, 50, 259–262.
Katayama, H., Haramoto, E., Oguma, K., Yamashita, H., Tajiama, A., Nakajima, H., et al. (2008). One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Research, 42, 1441–1448.
Maloney, A. (2007). Comparison of conventional and taqman real-time RT-PCR assay for the detection of norovirus. MS thesis, University of North Carolina at Chapel Hill.
Rijal, G., Petropoulou, C., Tolson, J. K., DeFlaun, M., Gerba, C., Gore, R., et al. (2009). Dry and wet weather microbial characterization of the Chicago area waterway system. Water Science and Technology, 60, 1847–1855.
Rijal, G., Tolson, J. K., Petropoulou, C., Granato, T., Glymph, T., DeFlaun, M., et al. (2011). Microbial risk assessment for recreational use of the Chicago area waterway system. Journal of Water and Health, 9(1), 169–186.
Rodriguez, R. A., Gundy, P. M., & Gerba, C. P. (2008). Comparison of BGM and PLC/PRC/5 cell lines for total culturable viral assay of treated sewage. Applied and Environmental Microbiology, 74, 2583–2587.
Rose, J. B., Dickson, L. J., Farrah, S. R., & Carnahan, R. P. (1996). Removal of pathogenic and indicator microorganisms by a full-scale water reclamation facility. Water Research, 30, 2785–2797.
Sedmak, G., Bina, D., McDonald, J., & Couillard, L. (2005). Nine-year study of the occurrence of culturable viruses in source water for two drinking water treatment plants and the influent and effluent of a wastewater treatment plant in Milwaukee, Wisconsin (August 1994 Through July 2003). Applied and Environmental Microbiology, 71, 1042–1050.
Straub, T. M., Höner zu Bentrup, K., Orosz-Coghlan, P., Dohnalkova, A., Mayer, B. K., Bartholomew, R. A., et al. (2007). In vitro cell culture infectivity assay for human noroviruses. Emerging Infectious Diseases, 13, 396–403.
Thompson, S. S., Jackson, J. L., Suva-Castillo, M., Yanko, W. A., El Jack, Z., Kuo, J., et al. (2003). Detection of infectious human adenoviruses in tertiary-treated and ultraviolet-disinfected wastewater. Water Environment Research, 75, 163–170.
Thurston-Enriquez, J. A., Haas, C. N., Jacangelo, J., Riley, K., & Gerba, C. P. (2003). Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Applied and Environmental Microbiology, 69, 577–582.
USEPA. (2001a). Concentration and processing of waterborne viruses by positive charge 1MDS cartridge filters and organic flocculation, Chapter 14. In USEPA manual of methods for virology. Washington DC: EPA .Office of research and development.
USEPA. (2001b). Total culturable virus quantal assay, Chapter 15. In USEPA manual of methods for virology. Washington DC: EPA Office of research and development.
Vinjé, J., Hamidjaja, R. A., & Sobsey, M. D. (2004). Development and application of a capsid Vp1 (Region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. Journal of Virological Methods, 116, 109–117.
Ward, R. L., Knowlton, D. R., & Pierce, M. J. (1986). Mechanisms of inactivation of enteric viruses in fresh waters. Applied and Environmental Microbiology, 52, 450–459.
Wojtenko, I., Stinson, M. K., & Field, R. (2001a). Challenges of combined sewer overflow disinfection by ultraviolet light irradiation. Critical Reviews in Environmental Science and Technology, 31, 223–239.
Wojtenko, I., Stinson, M. K., & Field, R. (2001b). Performance of ozone as a disinfectant for combined sewer overflow. Critical Reviews in Environmental Science and Technology, 31, 295–309.
Acknowledgments
Special thanks to Francisco Calderon and Sara Beck for their comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rodríguez, R.A., Gundy, P.M., Rijal, G.K. et al. The Impact of Combined Sewage Overflows on the Viral Contamination of Receiving Waters. Food Environ Virol 4, 34–40 (2012). https://doi.org/10.1007/s12560-011-9076-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-011-9076-3