Abstract
In healthcare services, information extraction is the key to understand any corpus-based knowledge. The process becomes laborious when the annotation is done manually for the availability of a large number of text corpora. Hence, future automated extraction systems will be essential for groups of experts such as doctors and medical practitioners as well as non-experts such as patients, to ensure enhanced clinical decision-making for improving healthcare systems. Such extraction systems can be developed using medical concepts and concept-related features as the part of a structured corpus. The latter can assist in assigning the category and sentiment to each of the medical concepts and their lexical contexts. These categories and sentiment assignments constitute semantic relations of medical concepts, with their context, represented by sentences of the corpus. This paper presents a new domain-based knowledge lexicon coupled with a machine learning approach to extract semantic relations. This is done by assigning category and sentiment of the medical concepts and contexts. The categories considered in this research, are diseases, symptoms, drugs, human_anatomy, and miscellaneous medical terms, whereas sentiments are considered as positive and negative. The proposed assignment systems are developed on the top of WordNet of Medical Event (WME) lexicon. The developed lexicon provides medical concepts and their features, namely Parts-Of-Speech (POS), gloss (descriptive explanation), Similar Sentiment Words (SSW), affinity score, gravity score, polarity score, and sentiment. Several well-known supervised classifiers, including Naïve Bayes, Logistic Regression, and support vector-based Sequential Minimal Optimization (SMO) have been applied to evaluate the developed systems. The proposed approaches have resulted in a concepts clustering application by identifying the semantic relations of concepts. The application provides potential exploitation in several domains, such as medical ontologies and recommendation systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Assigning categories and sentiment to medical concepts as well as contexts is an emerging area under the umbrella of multidisciplinary research in various healthcare services. We generally face challenges in this research area due to a lack of involvement of domain experts and scarcity of domain-specific lexicons. Moreover, the extraction of knowledge-based features and semantic relations of medical concepts is considered as another challenge because, till date, the available medical lexicons do not offer such features as category and sentiment.
Over the past few decades, researchers struggled to design various information extraction systems, such as GENIAFootnote 1 and PennBioIE,Footnote 2 to overcome such challenges. One of their fundamental motivations was to develop structured corpora from unstructured or semi-structured versions. In particular, various statistical and ontology-based approaches were used along with linguistic and machine learning techniques [66]. Such approaches have been employed to extract medical terms or concepts along with their syntactic and semantic features [34].
In the present work, we have developed two systems, one for assigning categories and another for identifying sentiments to medical concepts and their contexts. Both the systems also help to extract the semantic relations among medical concepts. In general, the medical concepts, represented by words or phrases contain attributes with knowledge and information related to medical entities. In order to identify concepts, isolation of stop-words and negation words or phrases are taken into account. The identified concepts are broadly classified as medical and non-medical. For example, abdom-inal_pain and uncontrolled_jerking are presented as medical and non-medical concepts, respectively.
Besides this, each sentence of the corpus is considered as a context in our task. These contexts are classified as medical and non-medical based on the presence of medical concepts. For example, “Abdominal pain is a sign of early pregnancy.” represents a medical context due to the presence of medical concepts such as Abdominal_pain, sign and early_pregnancy. On the other hand, “Green apple is good or red apple.” refers a non-medical context in the absence of any medical concept.
On the extracted concepts and contexts, we have applied our categorization and sentiment identification systems. In case of categorization, the extracted concepts are categorized into five types such as diseases, symptoms, drugs, human_anatomy, and miscellaneous medical terms (unrecognized categories presented as MMT in rest of the paper). For instance, the medical concept “abdominal_pain” indicates disease category. These categories were proposed by medical practitioners and experts based on their observations and evaluations on the primary occurrences of the extracted concepts and contexts in our corpora. The categories of individual concepts help to assign the overall category to their corresponding context. In addition to this, eleven categories of medical contexts were identified in a pair-wise manner such as disease-symptom, disease-drug etc.
For example, the context, “Ranitidinedrug belongs to a class_of_drugsMMT which is primarily known as H2_blockersdrug.” in overall is categorized as drug-MMT. Such categories of medical concepts as well as contexts assist in identifying the knowledge-based relations among various concepts within a context.
Similarly, to make provision for acquiring the sense-based information with respect to both concepts and contexts, we have built a sentiment identification system [6, 7, 9]. We have considered only positive and negative sentiments. For example, “amnesia” is a negative medical concept, whereas the context “Pregnancy is a beautiful experience”, is presented with a positive sentiment.
We have observed that the sentiment identification results are not uniform for different categories of concepts. For example, the medical concepts of category such as human_anatomy mostly contain neutral sentiment, whereas symptom category carries either positive or negative sentiment.
In order to develop these systems, we have used our previously built medical lexicon viz. WordNet of Medical Event (WME) which helps to extract medical concepts from contexts. Moreover, the lexicon supports in assigning the linguistic and sentiment features of medical concepts [67]. To tackle this, two versions of WME, namely WME 1.0 (WME version 1) [46] and WME 2.0 (WME version 2) [47], were prepared by us.
WME 1.0 lexicon contains 6415 number of medical concepts and their linguistic features such as POS, gloss with polarity score and sentiment. This version is unable to provide the knowledge-based information and semantic relation of the concepts. Therefore, we have designed an enriched version of WME (WME 2.0) with 10186 number of concepts and their knowledge-based features such as affinity score, gravity score and similar sentiment words (SSW).
Thereafter, we have adopted a hybrid approach by employing the existing linguistic features of WME 2.0 into a machine learning framework along with additional features like uni-gram, bi-gram, and negation [48]. The uni-gram and bi-gram features help to identify the categories of medical concepts, whereas negation feature was adopted to identify the underlying sentiment of medical contexts [3, 30, 49].
In order to develop the categorization system, we have applied two supervised classifiers viz. Naïve Bayes and Logistic Regression [32]. These classifiers achieved an average F-Measures of 0.81 and 0.86 for assigning categories to medical concepts and contexts, respectively.
The sentiment identification system has been developed using Naïve Bayes and support vector oriented Sequential Minimal Optimization (SMO) classifiers along with the presence of WME 2.0. These classifiers achieved an average F-Measures of 0.91 and 0.81 for identifying sentiment of medical concepts and contexts, respectively.
The category and sentiment identification systems also help to recognize the intensity of a context at the time of communication. Thus, the proposed systems can provide support to build a medical annotation system [73] which could guide to prepare structured corpus from an unstructured or semi-structured corpus. Besides, the structured corpus also assists in designing domain-specific applications like medical question answering, summarization, and recommendation systems for enhancing the quality of treatment in healthcare services [12, 13, 22, 36].
Finally, in the paper, we have also reported how the semantic relations between category and sentiment of medical concepts and contexts are used for designing various domain-specific applications.
Overview
Category and sentiment identification of the medical concept is a contributory research in the domain of Biomedical Natural Language Processing (Bio-NLP), which helps to extract the conceptual information from medical corpora. The conceptual information assists in representing structured corpora from a large number of the daily produced unstructured and semi-structured corpus. Researchers have applied ontology and lexicon such as SenticNet, SentiWordNet, and GENIA for identifying the conceptual information, in the absence of domain experts. Unfortunately, these resources are unable to provide an adequate output in the domain of Bio-NLP due to the paucity of medical concepts and multidisciplinary nature of the corpus [60].
Besides, machine learning approaches are used for identifying linguistic, statistical, and semantical features to overcome the challenge. To the process, the features identification task is presented as a contributory task in this domain. In the present paper, we have used our previously developed domain-specific lexicon, namely WME 2.0 to categorize and identify sentiment from medical concepts as well as contexts. The POS, SSW, affinity score, gravity score, polarity score, and sentiment features of medical concepts and machine learning classifiers are combined and presented as a hybrid approach. The hybrid approach applies for assigning and identifying the category and sentiment of concepts and contexts, which refer the semantic relations for the medical corpus.
Moreover, the extracted category and sentiment of medical concepts and contexts assist in designing domain-specific applications such as medical concept clusters. To develop the categorization and sentiment identification system, the overall structure of the paper is as follows: “Baseline System” illustrates the system’s baseline preparation; “Category Assignment Process” and “Sentiment Identification Process” explain the medical category assignment process and the medical sentiment identification process, respectively; “Evaluation” describes the evaluation of the proposed framework; “Semantic Relation Extraction using Sentiment and Category” illustrates semantic relation extraction using sentiment and category; “Related Work” proposes related work; finally, “Conclusion and Future Scope” sets out conclusion and future scope.
Baseline System
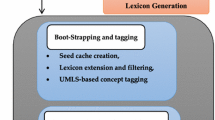
In Bio-NLP, a domain knowledge lexicon is essential for extracting conceptual information from medical corpora [4, 17]. To this end, we have borrowed the knowledge from WordNet of Medical Event (WME 2.0), a domain-specific lexicon [47]. WME 2.0 has both structural and linguistic features such as POS, gloss and conceptual features such as affinity score, category, gravity score, polarity score, sentiment, and SSW. These features assist in categorizing and assigning sentiment of medical concepts as well as contexts. Currently, two different versions of WME are available; one WME 1.0 and another WME 2.0, according to the versatility of medical concepts and the presence of their features.
WME 1.0
The initial version of WME (WME 1.0) was developed with 6415 numbers of medical concepts and their features such as POS, gloss, polarity, and sentiment [46]. The resource was prepared with initial 2479 number of medical concepts, which were acquired from the trial and training datasets of SemEval-2015 Task-6.Footnote 3 These concepts were applied on WordNet, a conventional resource, and a pre-processed English Medical DictionaryFootnote 4 to expand the lexicon, and find POS and gloss for the concepts. Polarity and sentiment for the medical concepts were introduced by the following sentiment lexicons like SentiWordNet,Footnote 5 SenticNet,Footnote 6 Bing Liu subjective list,Footnote 7 and Taboada’s adjective listFootnote 8 [14, 26, 69]. For example, the medical concept “abdominal_cavity” assigns -0.5 polarity score and negative sentiment under WME 1.0, as shown in Fig. 1.
The next version of WME (WME 2.0) was built to enhance the number of medical concepts and their extended semantic features [47, 54].
WME 2.0
The current version of WME, WME 2.0, is able to extract more statistical and semantic features with 10186 number of medical concepts [47]. WME 2.0 added affinity score, gravity score, SSW and additional category features with the earlier features of WME 1.0, namely POS, gloss, polarity score, and sentiment. These features enhance WME 2.0 resource to emulate human thought as a recommendation of medical advice, serving a potential foundation of a higher-order cognitive model under Natural Language Processing. For instance, the medical concept “maltreatment” recognize SSW “abuse, misuse, mismanage, and overlook” with negative sentiment in WME 2.0 lexicon.
Affinity score indicates a sentiment linking between medical concepts and their SSW with a probability from 0 indicating no relation and 1 suggesting a strong relation. Besides, gravity score recognizes the sentiment relevance between concepts and their glosses. The gravity score ranges from -1 to 1 including 0. While -1 suggests no relation, 0 describes neutral situations of either concept or gloss without any assigned sentiment, and 1 indicates strong relations either positive or negative, which helps to identify a proper gloss for concepts.
On the other hand, category extracts the subjective information of concepts and contexts such as diseases, drugs, treatments, human_anatomy, and MMT. The assigned categories of the medical concepts of WME 2.0 lexicon was validated by the manual annotators, who are medical practitioners. The annotators provide statistics applied through Cohen’s kappa coefficientFootnote 9 based agreement analysis. The analysis produced 0.78 satisfactory agreement score for validating the category of the concepts.
In the present paper, this category feature plays an important role in categorizing the medical contexts. In addition, WME 2.0 lexicon is used as a baseline (refer Fig. 2) to build the categorization and sentiment identification system for the medical concepts as well as the contexts, which are explained in “Category Assignment Process” and “Sentiment Identification Process” respectively.
Category Assignment Process
Category refers to the broadest fundamental classes of concepts and contexts that present the explicit knowledge-based information [62]. Such information helps to convert structured corpora as a large amount unstructured or semi-structured textual corpora produced by the medical practitioners. In the present paper, we have adopted WME 2.0 lexicon based five categories which assigns the category to the medical concepts. Therefore, we have used these five basic categories of concepts to identify eleven types of categories for the medical contexts. The concept and context categorization processes are described in the following subsections.
Concept Categories
Medical concepts and their category feature assist in understanding the medical concepts in the absence of domain experts such as doctors and medical practitioners. In order to recognize these categories of concepts, we proposed an automated categorization system. The linguistic and semantic features viz. POS, SSW, and sentiment of the concepts are employed into supervised machine learning classifiers that are combined and presented as a hybrid approach for building an overall system. To evaluate the proposed system, we have applied our previously built WME 2.0 lexicon as a baseline, as mentioned in “Baseline System”, with its medical concepts and features.
Therefore, we first identify the medical concepts from the contexts using nltk packageFootnote 10 based tokenization, stemming, and lemmatization method with the help of WME 2.0 lexicon. The lexicon provides the support to assign POS, SSW, and sentiment features to the extracted medical concepts. Also, these features of extracted concepts have been processed through Naïve Bayes and Logistic Regression supervised classifiers for identifying the categories. The categories are diseases, drugs, treatments, human_anatomy, and MMT (miscellaneous terms) that are similar to the categories of WME 2.0 concepts. For example, medical concepts “Simvastatin ZOCOR 20-mg tablet” and “mouth” are assigned with drug and human_anatomy categories by the system.
To evaluate the categorization system, we have collected the datasets from SemEval-2015 Task-6Footnote 11 and MedicineNetFootnote 12 resources due to the unavailability of the labeled corpus. The SemEval 2015 Task-6 resource was acquired in a completely different way from the development dataset of our baseline system. Thereafter, the dataset was split into two-parts as training and test dataset to validate the proposed system. The training dataset has been labeled with medical concepts and their POS, category, SSW, and sentiment features in the presence of medical practitioners.
On the other hand, the assigned five categories of concepts assist in assigning the categories of the contexts, as described in the following “Context Categories”.
Context Categories
Context category is essential to understand the subjective and conceptual information such as disease, treatment, and emotional condition from the corpus. To identify the category of medical contexts, we have used each of the extracted medical concepts and their category from the context. For example, the medical concepts and their categories like “passage, air, supply, and oxygen” as MMT and “lungs and body” as Human_anatomy assist in assigning the overall category Human_anatomy-MMT for the following medical context.
“The passage of air into and out of the lungs to supply the body with oxygen.”
Hence, we have applied the following algorithm to identify the category of medical contexts in the presence of concept categorization system.
- Step 1: :
-
Initially we assign the category of medical concepts of the context using concept categorization system. Each of the medical concepts and its category is presented as MC in a context
- Step 2: :
-
Consider the consecutive medical concepts and their category from the context, which is presented as Partial Context Category (PCC)
- Step 2.1: :
-
If the consecutive pair of concept categories are same then PCC is:
$$ \text{\textit{PCC}} = MC_{1} \cap MC_{2} $$(1) - Step 2.2: :
-
else PCC is:
$$ \text{\textit{PCC}} = MC_{1} \cup MC_{2} $$(2)
where MC1 and MC2 indicate two consecutive medical concepts and their category in a context.
- Step 3: :
-
Extracted Partial Context Category (PCC) helps to recognize the overall Context Category (CC) as:
$$ \text{\textit{CC}} = PCC_{1} \cap PCC_{2} $$(3)where PCC1 and PCC2 refer the partial context category of the context. The following example shows the extracted categories of medical contexts and their medical concepts.
For example, “We have earlier found that in Jurkat_cells__MMT activation of protein_kinase_C_(PKC)__MMT enhances the cyclic_adenosine_monophosphate__Disease accumulation induced by adenosine_receptor_stimulation__MMT.” medical context is able to assign the category MMT-Disease using our proposed system. To this process, we first extract the partial context categories (PCC) such as “MMT-MMT” and “disease-MMT”, which help to assign the overall context category (CC) as “MMT-disease” for the mentioned context.
Sentiment Identification Process
In recent years, sentiment analysis has become increasingly popular for processing social media data on online communities, blogs, wikis, microblogging platforms, and other online collaborative media [8]. Sentiment analysis is a branch of affective computing research [55] that aims to classify text (but sometimes also audio and video [57]) into either positive or negative (but sometimes also neutral [19]). Most of the literature is on English language but recently an increasing number of publications is tackling the multilinguality issue [41].
Sentiment analysis techniques can be broadly categorized into symbolic and sub-symbolic approaches. The former include the use of lexicons [2], ontologies [28], and semantic networks [16]; the latter consist of supervised [52], semi-supervised [31] and unsupervised [40] machine learning techniques that perform sentiment classification based on word co-occurrence frequencies. There are also a few hybrid approaches [10] that leverage both symbolic and sub-symbolic techniques for polarity detection.
While most works approach it as a simple categorization problem, sentiment analysis is actually a suitcase research problem [15] that requires tackling many NLP tasks, including named entity recognition [42], word polarity disambiguation [75], personality recognition [44], sarcasm detection [56], and aspect extraction [43].
Sentiment analysis has raised growing interest both within the scientific community, leading to many exciting open challenges, as well as in the business world, due to the remarkable benefits to be had from financial [76] and political [23] forecasting, user profiling [45] and community detection [18], manufacturing and supply chain applications [77], human communication comprehension [80] and dialogue systems [79], etc.
Sentiment information also assists in building a structured corpora from unstructured corpora in the domain of Bio-NLP. The researchers have applied various sentiment ontologies and lexicons like SenticNet and SentiWordNet to recognize sentiment of the concepts. Unfortunately, these lexicons are unable to offer an adequate output due to a lack of involvement domain experts and less occurrence of medical concepts. Hence, we have employed a domain-specific lexicon, namely WME 2.0, as a baseline to build sentiment identification system for the medical concepts and contexts which are described in the following subsections.
Concept Sentiment
Medical concepts and their related sentiments help to identify the conceptual knowledge of the corpus such as the situation of the patient, outcome of the prescription, study of diagnosis reports etc. The conceptual knowledge represented as positive and negative sentiment, which intern judges the impact of the medical condition of patients and effects of the treatment [20, 74]. For instance, the medical concept colon_cancer appears as negative sentiment, and shows the behavior of the concept.
In order to develop the sentiment identification system of the medical concept, we employed WME 2.0 lexicon as a baseline (as referred in “Baseline System”). The lexicon provides affinity score, polarity score, and SSW features of medical concepts. Two supervised classifiers viz Naïve Bayes and support vector oriented Sequential Minimal Optimization (SMO) are applied together as a hybrid system to extract sentiment of the medical concepts as positive and/or negative. For example, medical concepts “acute_brain_disorder” and “impregnate” are identified as negative and positive sentiment by the proposed system.
The identified sentiments of the concepts have been validated by the medical practitioners who generated a labeled training dataset. The training dataset contains the sentiment and semantic features like polarity score, sentiment and SSW of the concepts. The rest of the extracted concepts and their sentiments are represented as a test dataset, which is applied through supervised machine learning classifiers, for measuring the F-Measure of the proposed system. Both training and test datasets are collected from the previously mentioned SemEval 2015 Task-6 and MedicineNet resources.
Context Sentiment
Context sentiment identification presents an important research to resolve the semantic ambiguity of the corpus, which is mainly generated due to the short length of the contexts.
For example, the medical context “Drugs to treat pain.” is considered as semantic ambiguity for the medical concept, pain. The medical concept pain carrying more than one meaning, such as, “suffering” or “hurt”, can change the conceptual information obviously based on the situation.
Moreover, context sentiment also reflects the opinion of the doctors or physicians about the health status of patients for better treatment [11, 21, 29]. To observe these challenges, we have used WME 2.0 lexicon with mentioned concepts (as referred in “Concept Sentiment”) for identifying sentiment of the overall medical context. The proposed system is developed in two phases, namely pre-processing and learning.
The pre-processing phase helps to extract key concepts from the contexts using data extraction, cleansing, and formatting as well as assign the conceptual features for the extracted medical concepts. Thereafter, the learning phase combines the conceptual features with machine learning approaches which present a hybrid approach for building sentiment identification system of the context. The learning phase produces context sentiment, based on the polarity scores of overall concepts, presented in context [61]. Besides, we have also taken care of negation words like “no”, “not”, “never”, and “neither” to recognize the correct sentiment of the medical contexts [24, 27, 30].
Hence, we have designed the following algorithm to assign sentiment to the medical contexts with the help of sentiment identification system developed for the medical concepts.
- Step 1: :
-
Initially, we assign the polarity score (Polarityc) and sentiment of each concept (medical and non-medical) of the context. The proposed concept sentiment identification system and various sentiment lexicons such as SenticNet and SentiWordNet are applied to assign the polarity score of these concepts.
- Step 2: :
-
Identify the negation words or phrases to assign the correct sentiment of the context.
- Step 3: :
-
Calculate the overall polarity score of the context using following equation,
$$ Polarity_{context} = \sum\limits_{n = 1}^{k} Polarity_{c} $$(4)where Polaritycontext indicates the overall polarity score of the context and Polarityc refers individual polarity score of each concept in a context.
- Step 4: :
-
Therefore, based on the comparison of overall polarity score of the context, we have assigned the sentiment of the context as positive and negative.
Figure 3 is an example with flow diagram, on how we identify negative sentiment for the medical contexts. Similarly, this approach has been applied to identify positive sentiment for the medical contexts.
Evaluation
A domain-specific lexicon is crucial when it is embedded with linguistic, structural, and statistical features to extract conceptual information from different sources of medical corpora such as discharge summaries, prescription, and reports in the domain of Bio-NLP. In this paper, we have presented category such as diseases, treatments, and symptoms and sentiment as positive and negative for the medical concepts as well as contexts, which assist in identifying subjective information from the corpus. The category and sentiment features help to develop the semantic relation based medical concept clusters [1]. Moreover, our aim is to build an intelligent cognitive system by combining a domain knowledge lexicon, namely WME 2.0, by including category and sentiment to improve the quality of the services in healthcare [59].
In order to evaluate the proposed categorization and sentiment identification systems, we have collected medical concepts and contexts both from SemEval-2015 Task-6Footnote 13 and MedicineNetFootnote 14 resources due to the unavailability of labeled datasets. In addition, we have considered another set of data from SemEval-2015 Task-6, which is not used in developing the baseline system (WME 2.0). Overall, the collected data is presented as training and test dataset in the research. Table 1 shows the distribution of raw and the unique number of collected concepts and contexts from these resources.
We have also applied machine learning approaches in the presence of WME 2.0 lexicon features to validate the extracted categories as well as sentiments for both medical concepts and contexts.
Validation of Category Assignment System
Concept Category
To evaluate the proposed categorization system for the medical concepts, we have prepared a training dataset under the observation of medical practitioners from the collected data. The training dataset contains 9332 number of medical concepts, which are arbitrarily collected from SemEval-2015 Task-6 and MedicineNet resources such as 4212 and 5120 number of the medical concepts respectively. The medical practitioners have individually verified the training data and labeled 2246, 1204, 2108, 428, and 3346 medical concepts as diseases, symptoms, drugs, human_anatomy, and MMT categories. The remaining medical concepts, 5574 and 4714 from SemEval-2015 Task-6 and MedicineNet resources are presented individually as a test dataset. The test dataset has been processed through the proposed categorization system of the concept to assign the categories. Table 2 shows the distribution of training and test datasets.
Thereafter, the training and test datasets are applied through Naïve Bayes and Logistic Regression supervised classifiers in the presence of WME 2.0 lexicon driven features as POS, SSW, and sentiment of the concept. To select these classifiers, we have observed that, the Naïve Bayes classifier helps to combine the newly added training dataset with the existing training dataset. Here Logistic Regression assists in presenting the observations as a form of convenient probability scores over other well-known classifiers. Each of the classifiers provide an average F-Measure of 0.81 with four different models, namely use training set, supplied test set, cross-validation folds 10, and percentage split %66 as shown in Table 3.
To observe the importance of baseline system, we have conducted a statistical comparison between the proposed system and WME 2.0, acquired categories of the concepts, which refereed to Table 4. Besides, we also analyzed the number of linguistic and structural features between SemEval-2015 Task-6 and MedicineNet resource provided unique concepts and compared with WME 2.0. The features are the count of uni-gram, bi-gram, and tri-gram [33]. Table 5 shows the distribution of these linguistic features for SemEval-2015 Task-6, MedicineNet, and WME 2.0.
Context Category
The category assignment system for the medical contexts has been evaluated by Naïve Bayes and Logistic Regression supervised classifiers, with the collected medical contexts as mentioned in Table 1. These contexts are distributed in training and test datasets as 4774 and 9042 number of contexts from SemEval-2015 Task-6 and MedicineNet resources, individually, as shown in Table 2.
Afterward, both datasets were processed with mentioned classifiers in the presence of the following features. The features are the number of medical concepts, number of words in the context, uni-gram, bi-gram count of the context, and concept categories. Finally, the classifiers offer F-Measure of 0.86 for the categorization system of contexts as shown in Table 6.
The proposed category assignment system is able to assign eleven types of categories for the medical contexts. Table 7 presents the distribution of the number of contexts and their assigned categories. These categories assist in discovering subjective information of the contexts related to the domain knowledge. The domain knowledge helps to build a medical annotation system for producing the structured corpus from unstructured corpora. Figure 4 describes the steps of category assignment process of the contexts.
Validation of Sentiment Identification System
Concept Sentiment
To evaluate the sentiment identification system for the concepts, we have manually prepared 5000 labeled medical concepts as a training dataset, which was collected from two different resources previously mentioned, as shown in Table 1. The labeled training dataset classifies as positive and negative sentiment with 1892 and 3108 number of medical concepts individually by the medical practitioners. Besides, the rest of the acquired data is presented as a test dataset to validate the system. Table 8 shows the arbitrary distribution of the resources such as training and test dataset.
Thereafter, the medical concepts of training and test datasets are labeled with POS, gloss, polarity score, and SSW conceptual features and applied through Naïve Bayes and support vector based Sequential Minimal Optimization (SMO) supervised classifiers. The Naïve Bayes classifier helps to enhance the training dataset if new training data is received, where SMO assists in handling a very large training dataset with faster computation process.
Two classifiers obtain an average F-Measure of 0.91 with four different models, namely use training set, supplied test set, cross-validation folds 10, and percentage split %66 for the sentiment identification of medical concepts. Tables 9 and 10 present the F-Measure calculation and a statistical comparison of extracted sentiment for medical concepts between the mentioned two resources and baseline system (WME 2.0).
Context Sentiment
The pre-processing and learning, two-phase-based sentiment identification system of medical contexts, are validated through the supervised Naïve Bayes and SMO classifiers. Hence, we have prepared the training and test datasets from SemEval-2015 Task-6 and MedicineNet resources. These resources obtained 4900 and rest of 8916 number of unique medical contexts, which are presented as a training and test dataset individually, as shown in Table 8.
The training dataset labeled as 2032 and 2868 number of the positive and negative sentiments by the medical practitioners. The test dataset has been applied through proposed sentiment identification system of medical contexts.
Thereafter, the training and test dataset are applied with Naïve Bayes and SMO classifiers, which provide an average F-Measure of 0.81 for identifying sentiment of the contexts. Table 11 shows the F-Measure for sentiment identification system of the contexts with four models of each classifiers.
The proposed system identified positive and negative sentiment of medical contexts, which helped to extract conceptual information from corpora. Table 12 shows the distribution of contexts and their assigned sentiments. Finally, Fig. 5 describes the steps of sentiment identification process for the medical contexts.
Semantic Relation Extraction using Sentiment and Category
Semantic relation between medical concepts or contexts refers as an information retrieval system that helps to identify the subjective meaning and conceptual features from the unstructured corpus. The relation assists in extracting similar meaning oriented links between concepts, which is considered as an automatic relation extraction system with visualization effects. To identify the semantic relation from the corpus, category and sentiment of the medical concept are play an important role in understanding the natural linking between them, in the absence of domain experts [82].
The category and sentiment are both essential to consider due to the absence of uniform characteristic between categories and sentiments. The positive and negative sentiments do not depend on the different categories of concepts such as diseases, symptoms, and drugs etc. If a medical concept is presented as symptom then we can not assure the sentiment of concept as positive or negative. Similarly, in case of drug and human_anatomy category, major cases consider neutral sentiment instead of positive and negative.
To overcome the mentioned challenges and to identify the semantic relation between medical concepts, category feature is essential along with sentiment feature of the concept. Besides, the overall sentiment and category of the context help to understand the meaning of the corpus.
The category and sentiment features of medical concepts as well as contexts combined applications, support the expert and non-expert groups of people to enhance their understanding of the medical concepts and their subjective relations. Figure 6 presents a sample application as semantic clusters of medical concepts, which have been developed using the proposed categorization and sentiment identification systems.
Related Work
Semantic relation extraction from the medical corpus is an essential task in the biologically inspired natural language processing (NLP) domain [53]. The corpus categories and sentiments can be used to represent semantic relations, for identifying conceptual knowledge from the corpus. The category conveys the semantic information, where sentiment shows the opinion or fact of medical concepts and contexts.
In recent years, researchers have introduced several lexicons to reduce the gap between cognitive human and machine language processing, by identifying medical concepts and their semantic features. To design these lexicons, they have combined domain-specific knowledge with linguistic, statistical, and machine learning driven approaches. The linguistic and statistical approaches help to identify concept specific features such as negation, uni-gram, and bi-gram and POS, gloss, and similar sentiment words. The linguistic features of the concepts assist in extracting knowledge-based rules to determine the sentiment of the medical corpus [24, 51, 68].
Smith and Fellbaum [63] developed a Medical WordNet (MEN) with two sub-networks, namely Medical FactNet (MFN) and Medical BeliefNet (MBN), to evaluate consumer health reports. The formal architecture of Princeton WordNet used for MEN [37]. In addition, while MFN aims to serve non-expert groups to extract and present a better understanding of basic medical information, MBN identifies the fraction of beliefs on medical phenomena. Their primary motivation was to develop a network of medical information retrieval system with visualization effects. These lexicon-oriented networks are not able to provide adequate accuracy output due to a lack of conceptual knowledge involvement from the domain experts.
In order to overcome the aforesaid problem, researchers have introduced machine learning-based approaches with supervised techniques [64]. Various supervised classifiers viz. Standard and Multinomial Naïve Bayes and Support Vector Machine (SVM), have been used with uni-gram, bi-gram, Parts-Of-Speech (POS), and negation features. To improve the accuracy of sentiment extraction from the clinical corpus, the researchers utilized the hybrid framework that combines both linguistics and machine learning approaches [5, 58, 72].
Sohn et. al. [65] built an emotion identification system from suicide notes using the hybrid approach. The suicide notes provided by Informatics for Integrating Biology and the Bedside (I2B2) challenge organizers. Machine learning and rule-based combined approach were applied to the training dataset of the suicide notes. The approach provided micro-average score of 0.5640 as the F-Measure. Birks et. al. [4] employed RIPPER (Repeated Incremental Pruning to Produce Error Reduction), multinomial Naïve Bayes classifier, and manual pattern matching rules combined with hybrid approach for identifying emotions from the sentences. To this process, we have developed a domain-specific lexicon viz. WordNet of Medical Events (WME) for extracting medical concepts and their related POS, gloss, SSW, affinity score, gravity score, polarity score, and sentiment features [47].
Besides, to extract the semantic relations between the medical concepts, researchers have introduced several linguistic and conceptual features such as POS and gloss, and category of the concepts [60]. Kambhatla [35] used a linguistic feature-based method as the feature vector to identify the semantic relation. Embarek and Ferret [25] developed an alignment algorithm to extract the relations, like treats, signs, and cures from the medical entities (concepts) by constructing automatic patterns. Yetisgen-Yildiz et. al., [78] applied AMT technique to extract named-entities from clinical trial descriptions.
Moreover, the researchers have also proposed several well-known open source bio-medical annotation tools, like GENIA, PennBioIE, and GENETAG [38, 39, 70]. These tools assist in identifying the semantic relation of the concepts. To this process, they used heuristic rule-based approaches as the combination of the medical ontologies and standard information extraction techniques [50, 71]. Zhang et. al. [81] amalgamated tree and string kernels under machine learning approach to improve the accuracy of the semantic relation extraction system.
In the present research, we have presented two conceptual features (namely, category and sentiment) extraction systems of medical concepts as well as contexts. The categorization system helps to assign five and 11 different types of categories for the concepts (e.g., symptoms) and the contexts (e.g., symptom-drug) respectively. On the other hand, the sentiment identification system assists in extracting positive and negative sentiment for both medical concepts and contexts. In order to design and validate both of the systems, we have combined WME 2.0 lexicon driven features and various well-known supervised classifiers as a hybrid approach.
Finally, we have amalgamated the extracted categories and sentiments of the medical concepts to build the semantic relation application, namely semantic clusters. The application provides support to the expert and non-expert groups for understanding the natural linking between medical concepts. Moreover, the category, sentiment, and semantic relation of the concepts will help to develop summarization and recommendation systems in Bio-NLP domain [22].
Conclusion and Future Scope
This work is a first attempt at building an annotation system through identification of semantic relations based on sentiments and categories of medical concepts and contexts. Furthermore, the work shows how a domain knowledge lexicon, namely WME 2.0 can facilitate the development of novel applications. The basic motivation behind this research is to support the expert and non-expert groups of people to enhance their understanding of key information from a text corpus.
We proposed the categories as disease and disease-drug and sentiment as positive and negative for the medical concepts and contexts. To design these systems, we have initially identified the features of medical concepts as well as contexts using baseline WME 2.0 lexicon. The output of these systems assists in designing a domain-specific application as a semantic relation extraction system between medical concepts.
To evaluate these systems, we have utilized widely applied supervised classifiers such as Naïve Bayes, Logistic Regression, and support vector-based Sequential Minimal Optimization (SMO). These classifiers have provided an average F-Measure of 0.81, 0.86 and 0.91, 0.81, for the categorization and sentiment identification system of concepts and contexts individually.
In the future, we will try to enhance the semantic relations identification system with automatically identified patterns from contexts. These patterns and features can assist in extracting similar relations of concepts from contexts. The extracted relation could appropriately classify relations as a treatment (e.g., medication) and a disease (e.g., problem). Such relations could enable construction of recommendation systems with summarized outputs to support expert and non-expert groups in their respective applications.
Notes
References
Abacha AB, Zweigenbaum P. A hybrid approach for the extraction of semantic relations from medline abstracts. In: International conference on intelligent text processing and computational linguistics, pp 139–150. Springer. 2011.
Bandhakavi A, Wiratunga N, Massie S, Deepak P. Lexicon generation for emotion analysis of text. IEEE Intell Syst 2017;32(1):102–108.
Basili R, Pazienza MT, Vindigni M. Corpus-driven unsupervised learning of verb subcategorization frames. In: Congress of the Italian Association for Artificial Intelligence, pp 159–170. Springer. 1997.
Birks Y, McKendree J, Watt I. Emotional intelligence and perceived stress in healthcare students: a multi-institutional, multi-professional survey. BMC Med Educ 2009;9(1):1.
Boytcheva S, Strupchanska A, Paskaleva E, Tcharaktchiev D, Str DG. Some aspects of negation processing in electronic health records. In: Proceedings of international workshop language and speech infrastructure for information access in the balkan countries, pp 1–8. Citeseer. 2005.
Cambria E. An introduction to concept-level sentiment analysis. In: Mexican international conference on artificial intelligence, pp 478–483. Springer. 2013.
Cambria E. Affective computing and sentiment analysis. IEEE Intell Syst 2016;31(2):102–107.
Cambria E, Das D, Bandyopadhyay S, Feraco A. A practical guide to sentiment analysis. Switzerland: Springer, Cham; 2017.
Cambria E, Jie F, Bisio F, Poria S. Affectivespace 2: Enabling affective intuition for concept-level sentiment analysis. In: AAAI, pp 508–514. 2015
Cambria E, Hussain A. Sentic computing: A Common-Sense-Based Framework for Concept-Level Sentiment Analysis. Switzerland: Springer, Cham; 2015.
Cambria E, Hussain A, Durrani T, Havasi C, Eckl C, Munro J. Sentic computing for patient centered application. In: IEEE ICSP, pp 1279–1282. 2010.
Cambria E, Hussain A, Durrani T, Havasi C, Eckl C, Munro J. Sentic computing for patient centered applications. In: IEEE 10th International Conference on Signal Processing Proceedings, pp 1279–1282. IEEE. 2010.
Cambria E, Hussain A, Eckl C. Bridging the gap between structured and unstructured healthcare data through semantics and sentics. In: ACM WebSci. 2011.
Cambria E, Poria S, Bajpai R, Schuller B. SenticNet 4: A semantic resource for sentiment analysis based on conceptual primitives. In: COLING, pp 2666–2677. 2016.
Cambria E, Poria S, Gelbukh A, Thelwall M. Sentiment analysis is a big suitcase. IEEE Intell Syst 2017;32(6):74–80.
Cambria E, Poria S, Hazarika D, Kwok K. SenticNet 5: Discovering conceptual primitives for sentiment analysis by means of context embeddings. In: AAAI. 2018.
Cambria E, Schuller B, Xia Y, Havasi C. New avenues in opinion mining and sentiment analysis. IEEE Intell Syst 2013;28(2):15–21.
Cavallari S, Zheng V, Cai H, Chang K, Cambria E. Learning community embedding with community detection and node embedding on graphs. In: CIKM, pp 377–386. 2017.
Chaturvedi I, Ragusa E, Gastaldo P, Zunino R, Cambria E. Bayesian network based extreme learning machine for subjectivity detection. Journal of The Franklin Institute. 2018. https://doi.org/10.1016/j.jfranklin.2017.06.007.
Denecke K, Deng Y. Sentiment analysis in medical settings. Artif Intell Med 2015;64(1):17–27.
Deng Y, Stoehr M, Denecke K. Retrieving attitudes: Sentiment analysis from clinical narratives. In: MedIR@ SIGIR, pp 12–15. 2014.
Dey M, Mondal A, Das D. Ntcir-12 mobileclick: Sense-based ranking and summarization of english queries. In: NTCIR-12 Conference. 2016.
Ebrahimi M, Hossein A, Sheth A. Challenges of sentiment analysis for dynamic events. IEEE Intell Syst 2017;32(5):70–75.
Elkin PL, Brown SH, Bauer BA, Husser CS, Carruth W, Bergstrom LR, Wahner-Roedler DL. A controlled trial of automated classification of negation from clinical notes. BMC Med Inform Decis Mak 2005;5(1): 13.
Embarek M, Ferret O. Learning patterns for building resources about semantic relations in the medical domain. In: LREC. 2008.
Esuli A, Sebastiani F. Sentiwordnet: A publicly available lexical resource for opinion mining. In: Proceedings of LREC, vol 6, pp 417–422. Citeseer. 2006.
Goldin I, Chapman WW. Learning to detect negation with ‘not’in medical texts. In: Proc workshop on text analysis and search for bioinformatics, ACM SIGIR. 2003.
Grassi M, Cambria E, Hussain A, Piazza F. Sentic web: A new paradigm for managing social media affective information. Cogn Comput 2011;3(3):480–489.
Greaves F, Ramirez-Cano D, Millett C, Darzi A, Donaldson L. Use of sentiment analysis for capturing patient experience from free-text comments posted online. J Med Internet Res 2013;15(11):e239.
Huang Y, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assoc 2007;14(3):304–311.
Hussain A, Cambria E. Semi-supervised learning for big social data analysis. Neurocomputing 2018;275: 1662–1673.
Jacob SG, Geetha Ramani R. Discovery of knowledge patterns in clinical data through data mining algorithms: multi-class categorization of breast tissue data. Int J Comput Appl 2011;32(7):46–53.
Jang H, Shin H. Effective use of linguistic features for sentiment analysis of korean. In: PACLIC, pp 173–182. 2010.
Jiang M, Chen Y, Liu M, Trent Rosenbloom S, Mani S, Denny JC, Hua X. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc 2011;18(5):601–606.
Kambhatla N. 2004. Combining lexical, syntactic, and semantic features with maximum entropy models for extracting relations. In: Proceedings of the ACL 2004 on Interactive poster and demonstration sessions, pp 22. Association for Computational Linguistics.
Katz JE, Rice RE. Public views of mobile medical devices and services: A us national survey of consumer sentiments towards rfid healthcare technology. Int J Med Inform 2009;78(2):104– 114.
Kilgarriff A, Fellbaum C. Wordnet: An electronic lexical database. 2000.
Kim J-D, Ohta T, Tateisi Y, Tsujii J. Genia corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 2003;19(1):i180—i182.
Kulick S, Bies A, Liberman M, Mandel M, McDonald R, Palmer M, Schein A, Ungar L, Winters S, White P. Integrated annotation for biomedical information extraction. In: Proceedings of the human language technology conference and the annual meeting of the North American chapter of the association for computational linguistics (HLT/NAACL), pp 61–68. 2004.
Li Y, Pan Q, Yang T, Wang SH, Tang JL, Cambria E. Learning word representations for sentiment analysis. Cogn Comput 2017;9(6):843–851.
Lo SL, Cambria E, Chiong R, Cornforth D. Multilingual sentiment analysis: From formal to informal and scarce resource languages. Artif Intell Rev 2017;48(4):499–527.
Ma Y, Cambria E, Sa G. Label embedding for zero-shot fine-grained named entity typing. In: COLING, pp 171–180. 2016.
Ma Y, Peng H, Cambria E. Targeted aspect-based sentiment analysis via embedding commonsense knowledge into an attentive LSTM. In: AAAI. 2018.
Majumder N, Poria S, Gelbukh A, Cambria E. Deep learning-based document modeling for personality detection from text. IEEE Intell Syst 2017;32(2):74–79.
Mihalcea R, Garimella A. What men say, what women hear: Finding gender-specific meaning shades. IEEE Intell Syst 2016;31(4):62–67.
Mondal A, Chaturvedi I, Das D, Bajpai R, Bandyopadhyay S. Lexical resource for medical events: A polarity based approach. In: 2015 IEEE International Conference on Data Mining Workshop (ICDMW), pp 1302–1309. IEEE. 2015.
Mondal A, Das D, Cambria E, Bandyopadhyay S. Wme: Sense, polarity and affinity based concept resource for medical events. In: Proceedings of the 8th global wordnet conference, pp 242–246. 2016.
Mondal A, Satapathy R, Das D, Bandyopadhyay S. A hybrid approach based sentiment extraction from medical context. In: 4th workshop on sentiment analysis where ai meets psychology (SAAIP 2016), IJCAI 2016 Workshop, July 10, Hilton, New York City, USA. 2016.
Morante R, Liekens A, Daelemans W. Learning the scope of negation in biomedical texts. In: Proceedings of the conference on empirical methods in natural language processing, pp 715–724. Association for Computational Linguistics. 2008.
Na J-C, Kyaing WYM, Khoo CSG, Foo S, Chang Y-K, Theng Y-L. Sentiment classification of drug reviews using a rule-based linguistic approach. In: International conference on asian digital libraries, pp 189–198. Springer. 2012.
Niu Y, Zhu X, Li J, Hirst G. Analysis of polarity information in medical text. In: AMIA. 2005.
Oneto L, Bisio F, Cambria E, Anguita D. Statistical learning theory and ELM for big social data analysis. IEEE Comput Intell Mag 2016;11(3):45–55.
Patel CO, Cimino JJ. Using semantic and structural properties of the unified medical language system to discover potential terminological relationships. J Am Med Inform Assoc 2009;16(3):346–353.
Pedersen T, Pakhomov SVS, Patwardhan S, Chute CG. Measures of semantic similarity and relatedness in the biomedical domain. J Biomed Inform 2007;40(3):288–299.
Poria S, Cambria E, Bajpai R, Hussain A. A review of affective computing: From unimodal analysis to multimodal fusion. Inf Fus 2017;37:98–125.
Poria S, Cambria E, Hazarika D, Vij P. A deeper look into sarcastic tweets using deep convolutional neural networks. In: COLING, pp 1601–1612. 2016.
Poria S, Cambria E, Hazarika D, Mazumder N, Zadeh A, Morency L-P. Context-dependent sentiment analysis in user-generated videos. In: ACL, pp 873–883. 2017.
Prabowo R, Thelwall M. Sentiment analysis: A combined approach. J Inf 2009;3(2):143–157.
Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc : JAMIA 2011;18(5):594–600.
Rosario B, Hearst MA. Classifying semantic relations in bioscience texts. In: Proceedings of the 42nd annual meeting on association for computational linguistics, pp 430. Association for Computational Linguistics. 2004.
Sarker A, Mollá-Aliod D, Paris C, et al. Outcome polarity identification of medical papers, pp 105–114. 2011.
Shukla RS, Yadav KS, Rizvi STA, Haseen F. An efficient mining of biomedical data from hypertext documents via nlp. In: Proceedings of the 3rd international conference on frontiers of intelligent computing: Theory and applications (FICTA) 2014, pp 651–658. Springer. 2015.
Smith B, Fellbaum C. Medical wordnet: a new methodology for the construction and validation of information resources for consumer health. In: Proceedings of the 20th international conference on Computational Linguistics, pp 371. Association for computational linguistics. 2004.
Smith P, Lee M. Cross-discourse development of supervised sentiment analysis in the clinical domain. In: Proceedings of the 3rd Workshop in Computational Approaches to Subjectivity and Sentiment Analysis, pp 79–83. Association for Computational Linguistics. 2012.
Sohn S, Torii M, Li D, Wagholikar K, Wu S, Liu H. A hybrid approach to sentiment sentence classification in suicide notes. Biomedical Inf Insights 2012;5(Suppl. 1):43.
Spasic I, Ananiadou S, McNaught J, Kumar A. Text mining and ontologies in biomedicine: making sense of raw text. Brief Bioinform 2005;6(3):239–251.
Swaminathan R, Sharma A, Yang H. Opinion mining for biomedical text data: Feature space design and feature selection. In: The 9th international workshop on data mining in bioinformatics, BIOKDD. 2010.
Szarvas G, Vincze V, Farkas R, Csirik J. The bioscope corpus: annotation for negation, uncertainty and their scope in biomedical texts. In: Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing, pp 38–45. Association for Computational Linguistics. 2008.
Taboada M, Brooke J, Tofiloski M, Voll K, Stede M. Lexicon-based methods for sentiment analysis. Comput Linguist 2011;37(2):267–307.
Tanabe L, Xie N, Thom LH, Matten W, Wilbur JW. Genetag: a tagged corpus for gene/protein named entity recognition. BMC Bioinf 2005;6(1):1.
Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18(5):552–556.
Román JV, Pérez SC, Serrano SL, Carlos J, Cristóbal G. Hybrid approach combining machine learning and a rule-based expert system for text categorization. In: Proceedings of the 24th international Florida artificial intelligence research society conference. AAAI. 2011.
Wilbur JW, Rzhetsky A, Shatkay H. New directions in biomedical text annotation: definitions, guidelines and corpus construction. BMC Bioinf 2006;7(1):1.
Xia L, Gentile AL, Munro J, Iria J. Improving patient opinion mining through multi-step classification. In: International conference on text, speech and dialogue, pp 70–76. Springer. 2009.
Xia Y, Cambria E, Hussain A, Zhao H. Word polarity disambiguation using bayesian model and opinion-level features. Cogn Comput 2015;7(3):369–380.
Xing F, Cambria E, Welsch R. 2018. Natural language based financial forecasting: A survey. Artificial Intelligence Review. https://doi.org/10.1007/s10462-017-9588-9.
Chi X, Cambria E, Tan PS. 2017. Adaptive two-stage feature selection for sentiment classification. In: IEEE SMC, pp 1238–1243.
Yetisgen-Yildiz M, Solti I, Xia F, Halgrim SR. Preliminary experience with amazon’s mechanical turk for annotating medical named entities. In: Proceedings of the NAACL HLT, 2010 Workshop on creating speech and language data with amazon’s mechanical turk, pp 180–183. Association for computational linguistics. 2010.
Young T, Cambria E, Chaturvedi I, Zhou H, Biswas S, Huang M. Augmenting end-to-end dialog systems with commonsense knowledge. In: AAAI. 2018.
Zadeh A, Liang PP, Poria S, Vij P, Cambria E, Morency L-P. Multi-attention recurrent network for human communication comprehension. In: AAAI. 2018.
Zhang M, Zhang J, Su J, Zhou G. A composite kernel to extract relations between entities with both flat and structured features. In: Proceedings of the 21st international conference on computational linguistics and the 44th annual meeting of the association for computational linguistics, pp 825–832. Association for Computational Linguistics. 2006.
Zheng H-T, Kang B-Y, Kim H-G. Exploiting noun phrases and semantic relationships for text document clustering. Inf Sci 2009;179(13):2249–2262. Special Section on High Order Fuzzy Sets.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed Consent
Informed consent was not required as no human or animals were involved.
Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Rights and permissions
About this article
Cite this article
Mondal, A., Cambria, E., Das, D. et al. Relation Extraction of Medical Concepts Using Categorization and Sentiment Analysis. Cogn Comput 10, 670–685 (2018). https://doi.org/10.1007/s12559-018-9567-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-018-9567-8