Abstract
Cytochrome c is known as an electron-carrying protein in the respiratory chain of mitochondria. Over the last 20 years, however, alternative functions of this very versatile protein have become the focus of research interests. Upon binding to anionic lipids such as cardiolipin, the protein acquires peroxidase activity. Multiple lines of evidence suggest that this requires a conformational change of the protein which involves partial unfolding of its tertiary structure. This review summarizes the current state of knowledge of how cytochrome c interacts with cardiolipin-containing surfaces and how this affects its structure and function. In this context, we delineate partially conflicting results regarding the affinity of cytochrome c binding to cardiolipin-containing liposomes of different size and its influence on the structure of the protein and the morphology of the membrane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
At first glance, writing a review focusing on cytochrome c (Fig. 1) seems to be a superfluous task, because this elementary heme protein has been researched now for nearly 75 years (Edman 1979; Alvarez-Paggi et al. 2017). The number or research papers on cytochrome c is enormous and it is still increasing every year. In view of the fact that cytochrome c seems to be a comparatively simple protein in structural and functional terms, skeptics might suspect that any results reported nowadays merely constitute incremental progress and that the broad field of biophysical chemistry has more pressing issues to worry about than answering any open questions regarding cytochrome c. Is there anything new under the sun?
Cartoon representation of horse heart cytochrome c with a focus on the active site at the heme group (PDB 1AKK) (Banci et al. 1997). The central iron complex of the heme group is indicated in brown. The zoomed in image visualizes the covalent attachment of the heme to the CxxC motif flanked by C14 and C17. The axial ligands H18 and M80 are labeled as well for the reader’s orientation. The figure was taken from Soffer (2013)
A closer look at the history of research on cytochrome c reveals that the interest of researchers has been refocused several times. The discovery of cytochrome c by Keilin identified it as an electron transfer protein in the respiratory chain of parasites (Hartree 1973). After the pioneering work of Theorell and Åkesson (1941), who identified five different protonation state of oxidized cytochrome c (Fe3+, cyto), it took quite some time before the availability of the crystal structures of the different oxidation states of the protein (Tanaka et al. 1975; Takano and Dickerson 1981; Bushnell et al. 1990; Louie and Brayer 1990; Berghuis and Brayer 1992; Sanishvili et al. 1995; Numata et al. 2015) facilitated in-depth studies of the electron transfer processes by which the protein shuttles electrons from the CoQH2-cytochrome c reductase to the cytochrome c oxidase complex(Smith et al. 1977; Speck et al. 1979; König et al. 1980; Trumpower 1990; Therien et al. 1991; Trumpower and Gennis 1994; Hunte et al. 2002). In the 90s, cytochrome c became an ideal laboratory for protein folding/unfolding studies from which researcher learnt how changes of pH, ionic strength, temperature, and denaturant concentration could produce different unfolded states of the same molecule (Bai et al. 1993; Elöve et al. 1994; Colón et al. 1996; Pinheiro et al. 1997; Takahashi et al. 1997; Yeh et al. 1997; Englander et al. 1998). The foldon model of Englander and coworkers (Englander et al. 1998), which is now being frequently applied to other proteins (Bhardwaj et al. 2008; Englander and Mayne 2017), also emerged from studies on cytochrome c folding. The structural characterization of the protonation states of oxidized cytochrome c by optical, resonance Raman, and NMR spectroscopy revealed multiple non-native conformations in which the natural M80 ligand is replaced by a lysine residue, a hydroxl ion, or a water molecule (Cohen and Pielak 1994; Döpner et al. 1998; Rossel et al. 1998; Assfalg et al. 2003; Alessi et al. 2010). Based on these results, the question arose to what extent structural changes from the native, folded state into non-native, partially unfolded states bestow new functions on the protein. Textbook wisdom stipulates that a protein can only function in its fully folded state. However, research conducted over the last 20 years revealed additional peculiar functionalities of the protein that seem to require that the protein adopts a non-native conformation. The most prominent among them is the ability to acquire peroxidase activity, which facilities the protein’s release from inner mitochondrial space and its subsequent involvement in mitochondrial apoptosis (Jiang and Wang 2004). Alternatively, in the presence of H2O2, cytochrome c released from mitochondria into the cytoplasm can facilitate the aggregation of α-synuclein (Hashimoto et al. 1999), a process implicated in Parkinson disease. Moreover, the protein can function as a scavenger of superoxide, an important reactive oxygen species (ROS) (Atlante et al. 2000; Paradies et al. 2000). Reduction of cytochrome c by O2− in the intermembrane space interferes with its role as electron acceptor in the respiratory chain. Finally, it is noteworthy that multiple lines of evidence suggest that the protein undergoes structural changes upon its interaction with, e.g., cytochrome c oxidase and cytochrome c peroxidase (c.f. the review article of Hannibal et al. for a summary, detailed references will be given below) (Hannibal et al. 2016).
As indicated, the protein has to change the ligation state of its functional heme group and most likely also the spin state of the heme iron in order to perform some of these functions. This notion particularly applies to its peroxidase activity, because the native protein with its heme iron in a hexacoordinate low-spin state with histidine and methionine as axial ligands is ill equipped for this purpose owing to the low accessibility of the active site to the solvent and the inability of M80 to accept a proton. However, since the heme coordination with methionine is pivotal for maintaining the high redox potential of the protein (Battistuzzi et al. 2002), such changes would eliminate its primary function, namely its electron transfer capability. It is unclear how the protein is balancing these rather essential yet competing tasks.
Cardiolipin (CL) plays a major role in regulating the binding of cytochrome c to and its function on the inner membrane of mitochondria owing to its two phosphate head groups that can strongly interact electrostatically with cytochrome c. The protein has an abundance of positively charged groups on its surface (Sanishvili et al. 1995). Work aimed at exploring the determinants of the peroxidase activity of cytochrome c has mostly utilized cardiolipin-containing liposomes as model systems for the inner membrane of mitochondria. CL is generally mixed with neutral, zwitterionic phospholipids to mimic the mixtures of the inner mitochondrial membrane (Fig. 2) (cf. (Hannibal et al. 2016) and references cited therein). Some researchers have focused on binding studies (Rytöman et al. 1992; Rytöman and Kinnunen 1994; Heimburg and Marsh 1995; Gorbenko et al. 2006; Sinibaldi et al. 2008; Pandiscia and Schweitzer-Stenner 2015a, b), others on exploring the structure of the protein with spectroscopic means (Heimburg et al. 1991; Oellerich et al. 2004; Sinibaldi et al. 2010; Hanske et al. 2012; Capdevilla et al. 2015a; Mandal et al. 2015). Work of the Kagan group has revealed great insight with regard to the external conditions that favor peroxidase activity (Belikova et al. 2006). There is also a limited number of studies from the groups of Kagan and Nantes that compare cytochrome c binding to liposomes and the inner mitochondrial membrane (Belikova et al. 2006; Kawai et al. 2009). Fluorescence microscopy studies probing the influence of cytochrome c binding to CL-containing giant unilamellar vesicles by Groves and coworkers revealed valuable information about theoretically predicted lipid demixing (Heimburg et al. 1999) involving the formation of CL-rich domains (Beales et al. 2011; Bergstrom et al. 2013).
While there is a rather voluminous literature documenting these latest research activities on cytochrome c, a clear and unambiguous picture of different modes of cytochrome c binding to anionic phospholipid membranes and their respective biological significance has still to emerge. As we will show in this article, the results of binding studies are in fact partially contradictory, which is not always being appreciated by researchers in the field. Studies examining structural changes of membrane-bound proteins and morphological changes on membrane surface are not frequently related to binding studies. There is currently no clear understanding about the exact structural requirements for cytochrome c to function as a peroxidase. Finally, the specifics of hydrophobic interactions in general and the role of a proposed lipid insertion into the hydrophobic interior in particular have still to be clarified. It is thinkable that the latter is a prerequisite for CL-oxidation to occur (Kagan et al. 2005; Abe et al. 2011).
This review has two main goals. First, we will compare the results of different binding studies and highlight the differences with regard to experimental protocols and obtained results. In this context, we will discuss different thermodynamic models that have been used to describe the binding of (mostly though not exclusively oxidized) cytochrome c to anionic membrane surfaces. We will discuss how conformational transitions, protein aggregation, phase transitions of the membrane (lipid-lipid demixing), and protein insertion can affect the binding isotherms. Second, structural studies will be reviewed and related to the results of binding studies. In this context, the review also draws the attention of the reader to studies of cytochrome c function. This section will highlight the conditions at which the studies of electron transfer and peroxidase activity have been conducted and how the latter might be related to binding processes and structural changes. Finally, resolved and, more importantly, unresolved issues on which researchers might want to focus in the future will be highlighted. Needless to say, that even a rather comprehensive review cannot cover all interesting topics in depth, certainly not in the field of cytochrome c–surface interactions. We will pay not much attention to how the properties of membranes are affected by the presence of cardiolipin, which is a subject of a very nice review by Lewis and McElhaney (2009). Neither will we discuss how the lipid chemistry of cardiolipin (e.g., the existence of double bonds) affects its interaction with cytochrome c (Kagan et al. 2005; Abe et al. 2011; Ruíz-Ramírez et al. 2015). Interactions of cytochrome c with non-biological surface will be mentioned only when it fits into the context. If the reader wants to inform him/herself about other non-classical functions of this protein and about its role in the apoptopic process, he/she should consult the review of Hüttermann et al. (2011). A recently published very comprehensive review by Alvarez-Paggi et al. informs the reader about the structural and functional properties of a large variety of cytochrome c derivatives (Alvarez-Paggi et al. 2017).
Analysis and comparison of binding studies
In this section, we focus on studies of cytochrome c to anionic lipid surfaces which have been carried out over the last 25 years. A particular emphasis will be put on experimental work accompanied by theoretical analyses. Before going into details, it has to be emphasized that investigations of cytochrome c binding to charged lipids had actually started in the 70s. Results of these studies suggest that cytochrome c has a high affinity for binding to anionic lipids, particularly cardiolipin, that this binding is predominantly electrostatic in nature, that it might involve penetration into the membrane, and that it causes conformational changes of the protein (De Kruijff and Cullis 1980; Heimburg et al. 1991; Spooner and Watts 1992). Of particular interest is a paper by de Kruijff and Cullis which provides evidence for the formation of an inverted hexagonal HII phase on the inner site of cardiolipin vesicles upon cytochrome c binding (De Kruijff and Cullis 1980).
The binding studies of the Kinnunen group
Over a period of 20 years, the study of cytochrome c binding to anionic lipids by Kinnunen and associates has greatly influenced the field. The basic model that emerged from this work still frames all current discussion of cytochrome-lipid interactions. Therefore, these works deserve to be discussed in great detail in this review.
Let us start with basic aspects of their experimental protocol (Rytöman et al. 1992; Rytöman and Kinnunen 1994; Rytömaa and Kinnunen 1995). They investigated different types of anionic lipids (e.g., cardiolipin and phosphatidylglycerol) with zwitterionic lipids and small fractions of fluorescent-labeled phosphocholine lipids (PPHPC:1-palmitoyl-2-[6-(pyren-1-y]hexanoyl-sn-glycero-3-phosphocholine, PPDPC: 1-palmitoyl-2-[6-(pyren-1-yl)]decanoyl-sn-glycero-3-phosphocholine) by recording the fluorescence of the latter as a function of the added cytochrome c concentration. Owing to fluorescence resonance energy transfer (FRET) from these fluorophores to the heme group, their fluorescence is quenched. Thus, binding isotherms can be obtained via fluorescence titration. The results of a very representative set of experiments are shown in Figs. 3 and 4 (Rytöman et al. 1992). The displayed data sets reflect the fluorescence quenching of the donor due to oxidized cytochrome c (cyto) binding to the surface of large unilamellar micelles (LUVs) composed of 10 mol% cardiolipin (CL), 5 mol% PPHPC, and 15 mol% egg phosphatidylglycerol (PG) for different pH values. CL was taken from bovine heart. Obviously, the fluorescence quenching and the apparent binding affinity observed at pH 7 were significantly lower than the corresponding values observed at more acidic pH (4, 5, and 6, Fig. 3). To check whether the binding to cyto might be electrostatic in nature, Rytömaa et al. measured the fluorescence intensity of the above cyto–vesicle mixtures as a function of NaCl concentration (Fig. 4) (Rytöman et al. 1992). At pH 5, 6, and 7, the addition of salt lead to a recovery of fluorescence, which indicates a reduction of cyto binding. However, complete fluorescence recovery (i.e., total elimination of cyto binding) was achieved only at pH 7. The behavior at pH 4 was found to be totally different; here, the data indicate further fluorescence quenching. Similar results were obtained with MgCl2 as salt, which emerged as an even stronger binding inhibitor. The addition of ATP, ADP, and AMP led to a similar pH-dependent reduction of fluorescence quenching.
Quenching of the pyrene monomer emission (RFI) from PPHPC induced by the binding of cyto to egg PC vesicles (LUVs) containing 5 mol% PPHPC and 10 mol% cardiolipin. The total lipid concentration was 25 μM. Measurements were repeated at different pH, namely 4.0 (open circles), 5.0 (solid circles), 6.0 (open triangles), or 7.0 (solid triangles). The figure was taken form ref. (Rytöman et al. 1992)
Reversal of the quenching of the pyrene monomer emission (RFI) from PPHPC induced by the binding of cyto to egg PC vesicles (LUVs) containing 5 mol% PPHPC and 10 mol% cardiolipin measured as a function of NaCl concentration. The total lipid concentration was 25 μM. Measurements were repeated at different pH, namely 4.0 (open circles), 5.0 (solid circles), 6.0 (open triangles), or 7.0 (solid triangles). The figure was taken form ref. (Rytöman et al. 1992)
The results of the above described experiments led Kinnunen and coworkers to propose that the charge density on the surface of vesicles is a key determinant of cytochrome c binding (Rytöman and Kinnunen 1994). To test this hypothesis further, they investigated the binding of cyto to LUVs with different cardiolipin content. For this experiment, the authors used fluorescence-labeled CL as donor (1-palmitoyl-2[10-(pyren-1-yl)]decanyl-sn-glycerol-3-phosphoglycerol). PG was again used as anionic lipid. Figure 5 shows (a) that apparent binding affinity and fluorescence quenching level increases with cardiolipin content and (b) that binding inhibition by ATP decreases with increasing cardiolipin content and that it is practically absent for CL fractions above 50%. A similar observation was made for NaCl-induced inhibition. The observed influence of the increasing fraction of anionic lipids on cyto binding is not cardiolipin specific, since similar results were obtained with PG. In all these experiments, the concentration of alkyl chains was kept constant. The obtained results seem to corroborate surface charge density as an important determinant of cyto binding.
(Left) Quenching of the pyrene monomer emission (RFI) from pyrCL induced by the binding of cyto to egg PC vesicles (LUVs) containing 17.5 (open circles), 25 (filled circles), 33 (open triangles), 49 (filled triangles), 67 (open squares), and 100 mol% (filled squares) CL (including pyrCL). (Right) Corresponding changes of fluorescence as a function of added ATP concentration at the highest cytochrome concentration used for the data displayed on the left-hand side. Taken from ref. (Rytöman and Kinnunen 1994) with permission
Another important quite puzzling experiment on cyto binding to CL-containing liposomes has been carried out by Rytömaa and Kinnunen (1995). First, they allowed 1.2∙10−7 M cyto to react with a 1.06∙10−6 M mixture of CL (17.6 mol%), PC, and 1 mol% PPDPC (Fig. 6). Subsequently, they added liposomes without any fluorescence label to the mixture. The CL-content of the non-labeled liposomes was varied (17.6, 33.3, and 100). Interestingly, the addition of non-labeled liposomes led only to a minimal recovery of fluorescence emission, which seem to indicate that cyto binding was not reversible. Some fluorescence increase (binding inhibition) was obtained if the unlabeled liposomes were added to cytochrome c prior to the labeled ones. A similar experiment where CL was replaced with PG led to a very slow fluorescence recovery.
(Left) Quenching of the pyrene monomer emission (RFI) from PPGPC induced by the binding of cyto to egg PC vesicles (LUVs) containing 17.6 mol% cardiolipin. The vesicle mixture contained different mixtures of pyrene labeled and unlabeled liposomes. The corresponding molar ratios of labeled to unlabeled liposomes were 1:1 (open square), 1:5 (open circle), 1:10 (open triangle, pointing up), and 1:20 (open triangle, pointing down). (Right) Change of fluorescence quenching due to the addition of unlabeled liposomes with 17.6 (filled square and +), 33.3 (filled circle), and 100 (filled rhombus). The upward arrow indicates the gain in fluorescence due to the addition of 150 mM NaCl. The lipid concentration indicated on the abscissa refers to the concentration of phosphate groups. The + data points were obtained for a cytochrome c concentration of 123 nm, at which non-labeled liposomes with 17.6 mol% CL were added. The figure was taken form ref. (Rytömaa and Kinnunen 1995)
The situation was observed to be different if a fraction of PG was substituted by lyso-PG. The elimination of an alkyl chain decreased the level of fluorescence quenching while it had no discernible effect on the apparent binding affinity. Interestingly, the addition of non-labeled liposome caused substantial fluorescence recovery/binding inhibition, very much in contrast to what was observed for intact PG and CL containing liposomes.
How do all these observations fit together? Rytömaa and Kinnunen derived the following picture from their combined observations (Rytöman et al. 1992; Rytöman and Kinnunen 1994; Rytömaa and Kinnunen 1995). At neutral and slightly acidic pH, cyto binding to CL is electrostatic in nature and occurs via the so-called A-site, which was later identified with a patch of positively charged lysine groups (Fig. 7). Upon decreasing the pH, one of the phosphate groups becomes protonated thus reducing the possibility of electrostatic binding. Now another mechanism, the so-called C-site binding, takes over. It involves hydrogen bonding between the protonated phosphate group and the N52 side chain of the protein. C-site binding is therefore not amenable to inhibition by the addition of salt and nucleotides. In line with some reports in the literature, the authors explained the high apparent pK value of the phosphate group with the presence of a second negatively charged group and a higher effective hydronium ion concentration on the surface of their liposomes (in other words, they assumed the existence of a Stern-layer formed by hydronium ions). In their earlier papers, Rytömaa et al. (1992; Rytöman and Kinnunen 1994) invoked the possibility that binding to CL involves the penetration into the liposome interior where the protein could induce the inverted hexagonal phase of the innermembrane reported by De Kruijff and Cullins (1980). However, at a later stage, they replaced this idea by the so-called extended lipid insertion model, which predicts that one of the CL alkyl chains moves into the hydrophobic cavity of the bound protein (Rytömaa and Kinnunen 1995; Tuominen et al. 2002). This lipid insertion was proposed to occur subsequently to both, A- and C-site binding. Naturally, such a peculiar lipid/protein interaction would doubtless add a very hydrophobic component to the binding process and significantly enhance the binding affinity. This model was later corroborated by the observation that brominated lipid derivatives of CL quenched the fluorescence of cytochrome c in which iron was substituted by Zn2+ (Tuominen et al. 2002).
Several attempts have been made to arrive at a quantitative analysis of fluorescence titrations of cyto to fluorescence-labeled membranes. Here, we focus on the work of Gorbenko et al. (Domanov et al. 2005; Gorbenko et al. 2006; Trusova et al. 2010), since it offers the most comprehensive theoretical approach which we discuss in some detail in the following. The authors described the binding process itself in terms of a scaled particle theory approach of Minton (1995, 2000), which considers the dependence of the effective equilibrium constant on the liposomes’ surface occupancy by the protein. The somewhat convoluted binding isotherm was written as follows:
where i labels the binding site to which the protein binds with an apparent binding constant Ki. In the Minton theory, i actually denotes the conformation of proteins on the surface rather than binding site. ρi is the number density of proteins bound to site i. [cyto]f is the free cytochrome c concentration in the bulk. The activity coefficient for the ith-site is given by:
ai and si are area and circumference of the ith binding site. 〈ρ〉 is the average number density of bound proteins, and 〈ρa〉 is the average product of number density and binding area.
In their paper, Gorbenko et al. considered only one type of binding site/protein conformation. In such a case, the activity coefficient solely depends on the CL-fraction and on the number of already bound proteins. As shown by Minton (2000), the activity term accounts for a reduced binding at very high protein concentration, or alternatively at low lipid/protein ratios.
In order to account for the assumed electrostatic character of the investigated binding processes, Gorbenko et al. invoked the theory of Heimburg and Marsh (1995), who formulated the electrostatic component of the binding constant as follows:
where T is the absolute temperature, kB is the Boltzmann constant, and ΔFel is the gain in electrostatic free energy upon cytochrome c binding which is of course a function of the number of adsorbed proteins. ΔFel was calculated as follows:
where s denotes the surface and p the protein. Fsel is the electronic free energy of ta surface area with σprot ligands bound, Fsel (0) is the electronic free energy of the surface without ligands, and Fpel is the electronic energy of the protein in solution. The electrostatic free energy of cytochrome c in solution was calculated using classical theories describing uniform spherical spheres. The authors claimed that they used the classical Gouy-Chapman theory to calculate the free electrostatic energy of the utilized liposomes with and without adsorbed proteins, but unfortunately they do not clearly state the conditions for which their solution is valid.Footnote 1 Since they explicitly considered the neutralization of surface charges by cations of added salt, their approach goes beyond the Heimburg-Marsh theory, which solely considers the reduction of the double-layer potential caused by the accumulation of cations close to the membrane surface. Fluorescence quenching via FRET from membrane donors to heme acceptors were modeled using the classical Förster theory as formulated for surface attached proteins (Dale et al. 1979). The results of their analysis suggest that there are indeed two modes of cytochrome c binding to CL-containing liposomes and that the transition between the two modes depends on the protonation state of the respective phosphate groups. The effective binding affinities of the two protonation states are quite different. As shown in Fig. 8, the equilibrium constant for liposomes containing 40 mol% CL with double-protonated CL-phosphate groups varies between 4.5∙105 M−1 at low cytochrome c concentration and slightly acidic pH (6.35) and 104 M−1 at high protein concentration (ca. 2.7 μM) and neutral pH (7.55). For vesicles with single-protonated phosphate groups, the binding constant varies between 4∙107 and 7.9∙105 M−1. These values indicate a much stronger binding capacity of the semi-protonated state, which would imply the involvement of non-electrostatic interactions. Interestingly, the authors found the situation to be different for liposomes with very low CL-content.
Dependence of effective binding constants on protein concentration derived by Gorbenko et al. from an analysis of fluorescence quenching/binding isotherms data reflecting the binding of cyto to vesicles with 40 mol% CL and a total lipid concentration of 60 μM. The ionic strength for the experiment was 5 mM. Taken from ref. (Gorbenko et al. 2006) with permission
Though rather comprehensive, the studies of Kinnunen and coworkers have left several issues unresolved. The theory of Gorbenko et al. treats cyto adsorption with an equilibrium model. This contradicts the finding that cyto binding to CL and PG containing liposomes is not reversible if one dilutes a solution with fluorescence-labeled liposomes with unlabeled ones. The latter finding is itself at variance with the observation that cyto binding can be inhibited by adding NaCl and/or adenine nucleotides. These two very conflicting observations cannot be reconciled by assuming a two-step mechanism, i.e., electrostatic binding followed by, e.g., the insertion of an extended lipid chain into the cytochrome c cavity. If the latter step is irreversible on a measurable time scale, the addition of NaCl cannot make the binding process reversible, irrespective of whether it is added prior or after the addition of cytochrome c. The fluorescence titration curves reported in several papers of the Kinnunen group are clearly indicative of an equilibrium binding process; in the case of irreversible binding, the initial slope would be linear until a saturation level is reached. If the binding was really irreversible, the hyperbolic and sigmoidal response curves of, e.g., Trusova et al. (2010) would indicate a reduction of fluorescence quenching with increasing binding. Finally, it must be noted that their A-site/C-site model is based on the assumption that one of the two phosphate groups of cardiolipin has a comparatively high pK value which allow for its protonation below pH 6 (Olafsson and Sparr 2013). This notion has been discussed controversial in the literature for some time, but has recently been debunked by three mutually corroborating experimental studies. First, an IR-study of CL-containing liposomes by Malyshka et al. used the marker bands for protonated and deprotonated phosphate groups to show that the protonation of the respective phosphate groups only occurs at pH values below 4 (Malyshka et al. 2014). Their results were later confirmed by a more comprehensive study of Sathappa and Alder via measurements of zeta potentials(Sathappa and Alder 2016) and surface charge densities and by a 31P NMR study of Kooijman et al.(2017).
It is obvious that A-site binding must involve positively charged lysine groups on the proteins surface. Site-directed mutagenesis studies of Sinibaldi et al. provided some strong indication for the involvement of K72, K72, and K79 (Sinibaldi et al. 2013). Interestingly, these are also residues which function as sixth ligand in the different state IV isomers of cyto in solution (Döpner et al. 1998; Blouin et al. 2001).
The lipid insertion model is accepted by many researchers in the field, because there are several lines of indirect experimental evidence in its favor (Abe et al. 1978; Tuominen et al. 2002; Kalanxhi and Wallace 2007; Sinibaldi et al. 2010). However, several open questions remain. We will return to this issue below when we discuss possible protein conformations on anionic surfaces.
Modeling of electrostatic binding of cytochrome c to anionic liposomes
In the last section, we briefly described how a theory mostly based on the electrostatic binding model of Heimburg and Marsh was used to describe the binding of cyt0 to CL-containing liposomes under a variety of conditions (different CL-content of liposomes, different pH, ionic strength, etc.) (Heimburg and Marsh 1995). Here, we briefly review their own attempt to analyze the binding of cyto to DOPG at neutral pH. Proteins were added to a suspension of lipids which formed multilayers only after their addition. The concentration of bound proteins was deduced from the bulk concentration determined by ultracentrifugation. The authors measured binding isotherms for a large number of solutions with ionic strengths varying between 0.21 and 104 mM. In their theoretical approach, they considered the ensemble of adsorbed proteins as a van der Waals gas for which they formulate the following binding isotherm:
withFootnote 2
where ν is the number of adsorbed proteins bound to a surface area nΔA (ΔA is the surface area covered by a single ligand). K0 is the binding constant of an ideal highly diluted protein gas. Kagg is a measure of the interaction energy between proteins in close proximity in units of RT, and σprot denotes the number of bound ligands per liposome.
Equations (3) and (4) reveal the general concept of the electrostatic binding theory. Obviously, the use of a general applicable formalism would require a solution of the Poisson equation for a double layer, which is mathematically impossible, but analytical solutions have been developed for the low and the high potential limit (with regard to the respective formalism for the electrostatic free energy, the reader is referred to the paper of Jähnig (1976)). Heimburg and Marsh used his high potential limit formalism which for their system reads as follows:
where
α is the number of lipid charges involved in protein binding, Z is the number of charges on the ligand, σel is the charge density on the membrane surface, and cion is the concentration of cations in solution. Heimburg and Marsh obtained the following equation for the free electrostatic energy associated with cytochrome c binding:
with
and
where ε0 is the vacuum permittivity.
Two aspects of the analysis of the binding data obtained by Heimburg and Marsh are noteworthy. Figure 9 shows the fits to their binding isotherms. The data clearly suggest that the addition of NaCl reduces both, the binding affinity and number of bound proteins at high ligand concentration. Interestingly, however, the authors had to use two different values for α to fit the isotherms for ionic strengths above 40 mM (11.9) and below (4.9). The authors conjectured that he low value used for the low ionic strength data reflects the formation of a double layer of proteins. For the purpose of this review, it is of interested to have a closer look on the ionic strength dependence of the apparent equilibrium constant at low protein concentrations, which varies between 8∙104 M−1 at very low (4.35 mM) and ca. 1.5∙103 M−1 at high (104 mM) ionic strength. The low ionic strength value lies well below the high affinity values that emerged from the analysis of cyto binding to CL by Kinnunen and coworkers (Rytöman et al. 1992; Rytöman and Kinnunen 1994; Domanov et al. 2005).
Binding isotherms for association of cyto with 210 μM DOPG at low and high ionic strengths. Low ionic strength: 0.21 mM (filled circle), 4.35 mM (open circle), 10 mM (filled square), 16.9 mM (star), and 29.4 mM NaCl (open square); high ionic strength: 42 mM (filled triangle), 54.4 mM (open triangle), 79.4 mM (filled rhombus), and 104.4 mM NaCl (open rhombus). Global nonlinear least-squares fits of the isotherms as described in the text yielded the full lines. Two different stoichiometries, α = 11.9 and 4.9, were used for separate fits in the high and low ionic strength regimes, respectively. Taken from ref. (Heimburg and Marsh 1995) with permission
The discovery of L-site binding
In a series of more recent studies, Nantes and coworkers reported the discovery of another binding site which operates only at pH values below 7 (Kawai et al. 2005). They termed this process L-site binding. Understanding cytochrome c–cardiolipin interactions at slightly acidic pH might be important because proton pumping can lower the pH in the intermembrane space of mitochondria below 7 (Porcellli et al. 2005). Below, we discuss the work of the Nantes group in more detail.
Experiments that first established L-site binding utilized a somewhat indirect method to probe the binding of cyto to cardiolipin containing small unilamellar micelles (SUVs) with 50 mol% 1,2-dipalmitoyl-sn-glycero-3-phosphocholine, 30 mol% 1,2-dipalmiyoyl-sn-glycerol-3-phosphoethanolamine, and 20 mol% bovine heart cardiolipin (PCPECL), namely the turbidity due to vesicle fusion caused by the cyto binding to the liposome’s surface. It had been observed before that a high occupation of liposomes by cytochrome can produce vesicle fusion (Oellerich et al. 2002). Kawai et al. found that a cyto–liposome mixture with a CL/cyt ratio of ca. 60Footnote 3 produces significant turbidity below pH 7 (Fig. 10, (Kawai et al. 2005)). At these conditions, liposomes are only partially covered with proteins (Pandiscia and Schweitzer-Stenner 2015b). They explained their finding by describing cyto as bidentate ligand with A- and L-site binding sites which are nearly opposite to each other on the protein surface (Fig. 7). The authors applied a very thorough protocol to identify the lysine residues involved in L-site binding by comparing the induced turbidity caused by the binding of acetylated and carbethoxylated cytochrome c. Acetylation blocks all lysine groups. Carbethoxylation affects deprotonated lysines and possibly histidines. By combining carbethoxylation with a MALDI-TOF analysis of the protein and of protein fragments produced by tryptic digestion, they identified K22, K27, K87, and H33 as being actively involved in L-site binding (cf. Fig. 7). The pH-dependence of the observed turbidity, which indicated an effective pK value of 7, was rationalized in terms exceptionally low pK values of the lysine residues as produced by repulsive electrostatic interaction between them. A similar interaction was ruled out for A-site lysines because of a potentially larger distance between the protonable end groups of their side chains. Zeta-potential measurements ruled out the involvement of any phosphate group protonation, in line with studies referenced above (Malyshka et al. 2014; Sathappa and Alder 2016; Kooijman et al. 2017).
Turbidity probed with 480 nm absorption of a medium containing PCPECL vesicles (open circles), PCPECL vesicles with 4 μM cyto c (solid circles), PCPEPG vesicles (open triangles), and PCPEPG vesicles with 4 μM cyto c (solid triangles). The measurement was carried out 10 min after incubation at 30 °C. Experiments were carried out with 0.25 mM lipids containing 50 mol% DPPC, 30 mol% DPPE, and 20 mol% CL or phosphatidylglycerol. Taken from ref. (Kawai et al. 2005) with permission
A more recent follow up study of Kawai et al. deserves particular attention because it sheds more light on the biological relevance of the proposed L-site binding (Kawai et al. 2009). Herein, they probed the binding of cyto to cytochrome c-depleted mitoplasts by measuring the oxygen consumption rate of the system, which is an indicator of the functionality of the respiratory chain. Mitoplasts are mitochondria devoid of the outer membrane. They found (a) that the restoration of oxygen consumption depends on the concentration of the added cyto in a hyperbolic way, so that the response curve resembles a Langmuir isotherm. As shown in Fig. 11, the pH dependence apparent equilibrium constant inferred from the above response data is in close correspondence with the pH-dependence of L-site binding obtained from the above turbidity measurements. Moreover, they also found that the detachment of cytochrome c from non-depleted mitoplasts exhibits a complementary pH-dependence. The authors interpreted their data as indicating the existence of two different binding processes, a pH-independent one with a very high affinity (108 M−1) and low efficiency with regard to the initiation of oxygen consumption and a low affinity process with a higher functional efficiency for which the apparent affinity is 2.45∙106 M−1 at pH 7.4 and 7.75∙106 M−1 at pH 6.2.
Apparent affinity of cyto c for the binding to inner mitochondrial membranes plotted as a function of pH (filled circles). As described by Kawai et al., these data were obtained from double-reciprocal plots of respiration rates versus the total concentration of cyto c added to the medium. The data were fitted by using a modified version of the Henderson-Hasselbach equation. Taken from ref. (Kawai et al. 2009) with permission
Binding studies with varying lipid concentrations
The binding studies described above have in common that varying amounts of cyto were added to a batch of liposomes, so that the lipid concentration was kept constant. An alternative strategy keeps the protein concentration constant and varies the lipid concentration (and thus the number of liposomes).
Belikova et al. undertook an attempt to explore the relationship between cyto binding to small unilamellar vesicles (SUVs) prepared with 50 mol% DOPC/50 mol% TOCL and peroxidase activity (Belikova et al. 2006). Protein binding was probed by measuring the fluorescence of the W59 residue which is an indicator of protein unfolding. The authors did not analyze their spectral response date, but the obtained curve indicates an effective binding affinity in the range of 104 M−1 which is significantly lower than the values obtained from experiments of Kinnunen group (Rytöman et al. 1992; Gorbenko et al. 2006).
Sinibaldi et al. used visible circular dichroism to probe structural changes in the heme pocket that occurred upon the binding of horse heart cyto to (unfortunately unspecified) liposomes with 100% bovine heart cardiolipin (Sinibaldi et al. 2008). In its native state, the CD signal of the Soret band is a couplet, which reflects the splitting of the excited B-state caused by a quadratic Stark effect produced by the internal electric field of the protein and associated vibronic perturbations (Schweitzer-Stenner 2008). Upon protein binding to cardiolipin, structural changes lead to a conversion of the couplet into a positive Cotton band (Fig. 12). This reflects a change of the internal electric field as well as the removal of the F82 residue from the proximity to the heme (Pielak et al. 1986), which is caused by the displacement of the native M80 ligand either by a lysine or histidine side chain. Electrostatic coupling between the heme and F82 contribute predominantly to the negative signal of the native state couplet. Figure 13 shows the ellipticity measured at 416 nm as a function of the total CL concentration. Interestingly, the authors fitted their data with a sum of two Hill-type Langmuir functions, which in terms of the reported spectroscopic data read as follows:
where χ1 and χ2 = 1-χ2 are the fractions of two different binding sites on the liposome surface. [CL] denotes the free cardiolipin concentration, Ki and ni are the equilibrium and Hill constants associated with the two binding sites. Since the authors used liposomes with 100% CL-content, the justification for assuming two different surface binding sites is not entirely clear. For two different protein binding sites, one would use a single grand partition sum that contains K[CL] terms (K: equilibrium constant) for both sites (Pandiscia and Schweitzer-Stenner 2015a). It is also unclear how the authors calculated the free protein concentrations from the respective total concentrations. Their analysis yielded binding affinities of K1 = (4.9 ± 0.5)∙104 M−1 and K2 = (2.4 ± 0.5)∙104 M−1 with respective Hill coefficients of 2 and 4. For the mole fractions, they obtained χ1 = 0.78 and χ2 = 0.22. Irrespective of the validity of this analysis, the obtained equilibrium constants clearly reflect a much lower binding affinity than the values reported by the Kinnunen and Nantes groups (Rytöman et al. 1992; Domanov et al. 2005; Kawai et al. 2009). Sinibaldi et al. found that the addition of NaCl predominantly affected the binding to site 1. This observation led them to the conclusion that sites 1 and 2 should be identified with the A- and C-site proposed by Rytömaa et al. (1994).
Soret band CD spectrum of cyt0 c recorded in the absence (solid line) and presence (dashed) of 60 μM CL at pH 7. The protein concentration was 10 μM. The temperature was 25 °C. The inset shows the corresponding absorbance spectrum of cyt0 c in (from top to bottom) the absence and presence of 25 and 60 μM CL. The wavelength range was 670–720 nm. Taken from ref. (Sinibaldi et al. 2008) with permission
Changes in the 416 nm Cotton effect induced by stepwise addition of CL to a 10 μM cyt c solution. Experimental points are averages of at least three measurements. The solid line resulted from a fit described in the text. Taken from ref. (Sinibaldi et al. 2008) with permission
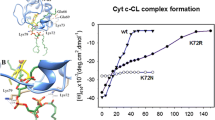
In subsequent papers, Santucci and coworkers used CD response data to investigate the influence of specific mutations on protein binding to 100% CL liposomes (Sinibaldi et al. 2013). They found that replacing K72 by R abolishes any change of the CD signal. The data seem to indicate a reduction of the binding affinity for the second step. In another study from this laboratory, the involvement of K79 in the binding of cytochrome c to CL-containing liposomes was inferred from respective mutation experiments (Sinibaldi et al. 2017).
In an attempt to correlate binding, structure, and function, Kapralov et al. combined measurements of the W59 fluorescence and the 695 nm absorption band to probe the binding of cyto to 50:50 mixture of DOPC and a variety of anionic lipids, including CL (Kapralov et al. 2007). Among their set of the latter TOCL and DOPA were found to be most effective in changing the structure of the protein. This aspect of their study will be discussed in more detail in the subsequent section of this review. If one takes their response curves as indicators of binding, the corresponding apparent equilibrium constants should lie in the 104 M range. The work of the Pletneva group on cytochrome c–cardiolipin binding generally focused on structural changes of the protein and will be discussed in detail in the next section. Here, a binding experiment reported by Hanske et al. is noteworthy (Hanske et al. 2012). They measured the change of fluorescence anisotropy of Zn-substituted cytochrome c in response to the addition of liposomes formed with 50 mol% TOCL/50 mol% DOPC. The reported size distribution of their liposomes suggest that they lie somewhere between SUVs and LUVs. Their response curve is shown in Fig. 14. The authors inferred an equilibrium constant of 1.4∙104 M−1 from their data, but they did not elaborate on the utilized algorithm used for their analysis. Apparently, this value is even lower than the K2 constant reported by Sinibaldi et al. (2008)
Fluorescence anisotropy of 2 μM zinc substituted cytochrome c (λex = 423 nm, λem = 588 nm, [cyto c] = 3 μM) with added TOCL/DOPC liposomes as a function of total lipid concentration. The addition of 150 mM NaCl to a sample with a lipid concentration of 660 μM leads to a decrease in the ZnP anisotropy value as indicated. The line resulted from a fit to the Hill binding equation with Ka 1.5∙104 M−1. Taken from ref. (Hanske et al. 2012) with permission
Several studies exploring the binding of CL-containing SUVs have been conducted in our own laboratory over the last 3 years (Pandiscia and Schweitzer-Stenner 2014, 2015a, b; Serpas et al. 2016). Figure 15 exhibits different sets of spectroscopic responses to the binding of cyto to TOCL/DOPC SUVs with different TOCL content (20, 50, and 100%) at neutral pH (7.4). Figure 15a shows the increase of W59 fluorescence which is an indication of the W59 residue moving away from the heme group during a partial unfolding process. Panel B and C show the response of the corresponding polarized fluorescence. In panel D, the data indicate changes of the circular dichroism in the Soret band region. The observable ΔΔε plotted in this figure was calculated as ΔΔε = Δε417- Δε405 to eliminate the influence of varying baselines. Panel E displays the integrated intensity of the 695 nm charge transfer band which is an indicator of the Fe3+ ligation by the sulfur atom of M80. Its disappearance at high lipid concentration reveals a conformational change involving the replacement of M80 by another ligand.
Dose response of a the integrated fluorescence intensity of sub-band F1, b the parallel and c perpendicularly polarized fluorescence intensity at 340 nm, d the ΔΔε of the couplet peak of the Soret band CD spectra, and e the oscillator strength of the 695 nm absorbance band of cyto to an increasing CL concentration for TOCL/DOPC liposomes with 20% (black), 50% (red), and 100% (blue) CL-content. The two cardiolipin concentrations omitted for the fitting procedure concentrations are symbolized by triangles. Captions and figure were taken from ref. (Pandiscia and Schweitzer-Stenner 2015b) with permission
In a first attempt to self-consistently analyze the response data for the binding to 20%TOCL/80%DOPC, Pandiscia and Schweitzer-Stenner assumed two different binding sites on the protein (Pandiscia and Schweitzer-Stenner 2015a). In this case, each of the response functions can be written as follows:
where s0 denotes the spectral parameter (fluorescence intensity, ΔΔε, etc.) of cyto in solution, Ki and ni are the equilibrium constants for the two assumed protein binding sites and the corresponding Hill coefficient. si are the spectroscopic values of the protein bound via the assumed sites 1 and 2. That model allowed a successful fit to all the response data (not shown); the obtained corresponding equilibrium constants were K1 = 1.75∙105 M−1 and K2 = 4.5∙104 M−1, the corresponding Hill coefficients were 1.45 and 3.5. The equilibrium constants are expressed with regard to cardiolipin concentration in the outer lipid layer. Apparently, these values are very close to the binding constants Sinibaldi obtained for cyto binding to 100 mol% CL (Sinibaldi et al. 2008) and thus again clearly lower than the values obtained from the Kinnunen and Nantes experiments (Rytöman et al. 1992; Domanov et al. 2005; Kawai et al. 2009, 2014).
Figure 16 depicts how the above spectroscopic response data change upon the addition of the indicated concentration of NaCl (Pandiscia and Schweitzer-Stenner 2015b). On a first view, the data seem to indicate that the addition of salt inhibits cytochrome c binding: the fluorescence intensity decreases, the circular dichroism spectra become more native like (Hagarman et al. 2008), and the 695 nm band intensity increases. However, a closer inspection of the response curves shows that the observed changes affect the amplitudes rather than the half points, suggesting limited changes of equilibrium constants, contrary to expectations from the Heimburg-Marsh model (vide supra) (Heimburg and Marsh 1995). Instead, the data seem to indicate that proteins undergo a transition from a non-native into a more native-like state. This picture did indeed emerge from the first analysis of Pandiscia and Schweitzer-Stenner (2014).
Dose response curves of the a integrated fluorescence intensity of sub-band F1, the b parallel and c perpendicular polarized fluorescence intensity at 340 nm, the d ΔΔε of the couplet peak of the Soret band CD spectra, and the e oscillator strength of the 695 nm absorption band of cyto to an increasing CL concentration for TOCL/DOPC liposomes with 20% (black), 50% (red), and 100% (blue) CL-content. These curves were obtained in the presence of 100 mM NaCl for 20% CL-content, 200 mM NaCl for 50% and 100% CL-content. The two cardiolipin concentrations omitted for the fitting procedure concentrations are symbolized by triangles. The solid lines result from a fitting procedure that is described in detail in the main manuscript. Captions and figure were taken from the Supporting Information of ref. (Pandiscia and Schweitzer-Stenner 2015b)
In a second paper, Pandiscia and Schweitzer-Stenner subjected their complete set of response data in Figs. 15 and 16 to a more thorough analysis (Pandiscia and Schweitzer-Stenner 2015b). First, they utilized the findings of Pletneva and coworkers (vide infra) who convincingly showed that cyto bound to CL-containing liposomes can exist in distinguishable sets of conformations (vide infra, (Hanske et al. 2012)). One set has been characterized as compact and is likely to contain native as well non-native proteins. The other one contains more extended conformations and is therefore entirely non-native. Therefore, Pandiscia and Schweitzer-Stenner considered the following reaction:
This model assumes an equilibrium between a native-like conformation with no W59 fluorescence (nf: non-fluorescing) and a non-native fluorescing (f) conformation. The binding itself is reduced to a single binding step. The corresponding response function reads as follows:
The equilibrium constant KC was assumed to decrease with the cyto occupancy on the liposome surface in a Hill type fashion. Thus, the value is low at low and high at high cardiolipin concentrations. This approach was justified with molecular crowding effects. At low lipid concentration, the occupancy is high and the space for the unfolding processes limited. This changes at high lipid concentrations. Even with this extension, the binding model is still Langmuir like. To arrive at a more realistic picture, Pandiscia and Schweitzer-Stenner invoked a theoretical concept of Heimburg and Marsh which considers surface bound proteins as van der Waals gas (Eq. 6).
The solid lines in Fig. 15 result from a fit of this model to the displayed data sets. In order to account for the NaCl-dependence of the response data (Fig. 16), they augmented their model by assuming that the addition of salt shifts the Kc values for high lipid concentrations to lower values. Thus, in accordance with findings of Hanske et al. (2012), the addition of NaCl stabilizes the more native states. The analysis showed that the destabilization of the f-conformation can be related to a Langmuir isotherm for NaCl binding.
Details about equilibrium constants and spectroscopic parameters can be found in the paper of Pandiscia and Schweitzer-Stenner (2015b). Here, we just focus solely on the apparent equilibrium constant Kapp = KKC. Figure 17 depicts its dependence on the cardiolipin concentration. The equilibrium constant varies between ca. 4∙104 and 4∙105 M−1. It increases substantially with increasing cardiolipin content of the liposomes, in agreement with Rytömaa et al. (1994) Concomitantly, its variation with the cardiolipin concentration increases. For liposomes with 20 and 50 mol% CL, it decreases at low lipid concentration (i.e., high liposome occupancy) owing to a decrease in Kc. For 100 mol% CL, it shows a local maximum at very low lipid concentration owing to the very high density of proteins on the liposome surface. It should be noted here that Pandiscia and Schweitzer-Stenner kept the corresponding total concentration of lipids constant so that the number of liposomes decreased with increasing CL-content.
Apparent binding constant Kapp plotted as a function of cardiolipin concentration for varying CL-content of the employed TOCL/DOPC liposomes. Captions and figure were taken from the Supporting Information of ref. (Pandiscia and Schweitzer-Stenner 2015b)
The two-state analysis of Pandiscia and Schweitzer-Stenner was recently confirmed by a resonance Raman study which explored the spin and ligation state of the heme iron of cyto on TOCL-containing liposomes. In order to distinguish between native and non-native-like conformations, Malyshka and Schweitzer-Stenner exploited that the oxidized native protein can be photo-reduced in the presence of potassium ferrocyanide, while non-native state with M80 replaced by another residue ligand remains unaffected (Malyshka and Schweitzer-Stenner 2017). Figure 18 shows the Raman difference spectra produced by subtracting the spectra of cytochrome c–liposome (20%TOCL/80%DOPC) mixtures from the spectrum of the protein in solution for different lipid to protein ratios. In solution, the use of 442 nm excitation leads to complete photoreduction of the protein. Upon adding liposomes, an increasing number of proteins remain in the oxidized state, indicating the population of a non-native state with a much lower redox potential. The authors showed that the emergence of the oxidized population can be correlated with the population of the f-state as predicted by Pandiscia and Schweitzer-Stenner (2015b).
Difference spectra in the high-frequency region of resonance Raman spectra of cyto in the presence of 20% DOPC/80% TOCL SUVs at the indicated CL concentrations, obtained as was described in the text. Positions of major bands are labeled according to band assignments of Abe et al. (1978), and arrows indicate the evolution of extrema in the difference spectra for increasing cardiolipin concentrations. Captions and figure were taken from ref. (Malyshka and Schweitzer-Stenner 2017) with permission
More recently, work in our own group made contact with the L-site studies of Nantes and coworkers. Milorey et al. probed the interaction of cyto with 20%TOCL/80%DOPC SUVs at pH 6.75 and 6.5 (Milorey et al. 2017). Figure 19 shows the spectral response of W59 fluorescence, circular dichroism (vide supra), the 695 nm band, and another charge transfer band at 620 nm, which is generally indicative of the population of high-spin states. The population of high-spin states increases with lipid concentration and pH. It does not occur at neutral pH with the indicated lipid composition, but traces of high-spin state populations have been observed before at very high concentrations of liposomes with 50 and 100% TOCL. The data of Milorey et al. clearly show that high-spin species become predominant at high lipid concentrations and mildly acidic pH (6.5). This is important because high-spin species might be much more capable to perform peroxidase reactions (Capdevilla et al. 2015b). In addition to the above binding studies, the authors carried out resonance Raman studies, which revealed that the high-spin population is a mixture of pentacoordinate and hexacoordinate species. They attributed the observed conversion of the low-spin hexacoordinate non-native state into these high-spin species to the protonation of a histidine ligand which had replaced the native M80. This assignment is supported by recent resonance Raman studies of the Smulevich group which showed that both, H33 and H26, can form bis-His complexes in non-native states of cyt0 produced by its interaction with cardiolipin (Milazzo et al. 2017). Milorey et al. combined their results with those of Pandiscia and Schweitzer-Stenner to arrive at the reaction scheme in Fig. 20, which invokes the conversion of the low-spin f-state into a non-native high-spin state owing to the protonation of the H33/H26 ligand.
Spectral response data indicating the interaction of cyto with 20% TOCL/80% DOPC at pH 6.5, 6.7, and 7.4. a Total integrated intensities of the fluorescence F-band, b difference in positive and negative maxima of the Soret band CD, c total integrated intensities of the CT1 band, and d total integrated intensities of the CT2 band. All data are plotted as a function of the cardiolipin concentration in μM units. It should be noted that the CL concentration was increased tenfold for charge transfer measurements (frames c, d) along with the protein concentration to keep the lipid to protein ratios consistent. Captions and figure were taken from ref. (Milorey et al. 2017) with permission
Reaction scheme proposed by Milorey et al. to explain the pH dependence of spectroscopic responses to cyto binding to TOLC/DOPC liposomes. Details are described in the text. The figure was taken from ref. (Milorey et al. 2017) with permission
Binding studies with reduced cytochrome c
All the above referenced studies have utilized oxidized cytochrome c. With regard to studies on cytochrome c–membrane interactions, the reduced state of the protein (cytr) has not attracted a lot of attention. That might be in part due to the much greater structural flexibility of the oxidized protein which can adopt six different states within a pH range from 1 to 12. Only the state populated between pH 4 and 8 (state III) is native (Schweitzer-Stenner 2014; Soffer and Schweitzer-Stenner 2014). The reduced native protein remains unaffected between pH 3 and 12 (Moore and Williams 1980). Its unfolding temperature lies predominantly well above 100 °C. The reason for this stability of the reduced state lies in a protein-induced strengthening of the Fe2+-M80 bond (Mara et al. 2017).
Work on cytr interactions with anionic vesicles can be summarized as follows. Iwase et al., by means of absorption spectroscopy, observed that bovine cytr becomes oxidized upon binding with micelles composed of 100% CL (Iwase et al. 1996). The authors did not provide an explanation for their findings. Nantes et al. used magnetic circular dichroism (MCD) spectra of the Soret band region to probe changes in the heme environment of reduced horse heart cytochrome c upon its interaction with PCPECL liposomes (Nantes et al. 2001). Their cardiolipin was taken from bovine heart. Though their data also seem to indicate that cytr c becomes oxidized upon binding to these liposomes, the authors dismissed this possibility on the basis of the observation that in the absence of any reductant, the addition of NaCl led to a recovery of the MCD signal assignable to the reduced state of the protein. They instead proposed the generation of a protein conformation with a ferrous high-spin state, in which the Fe–M80 linkage is broken. Their assignment seems to be consistent with the finding of Droghetti et al. that ferrous cytochrome c can switch into a pentacoordinate high-spin (pcHS) state upon interaction with sodium dodecyl sulfate micelles and phospholipid vesicles (Droghetti et al. 2006).
Kalanxhi and Wallace investigated the binding of a variety of cytr derivatives to liposomes formed with a mixture of PC, PE, and CL in a 5:4:3 ratio. For horse heart cytr, their data indicate a binding affinity of ca. 105 M−1(Kalanxhi and Wallace 2007). The observed binding process was at least partially inhibited in the presence of NaCl. The authors proposed a model that follows the Kinnunen concept by invoking a two-step process, with electrostatic binding as the primary and the subsequent insertion of a CL alkyl chain into the hydrophobic channel of the protein as the secondary process. The latter could produce the high-spin state reported by Nantes et al. (2001)
Vos and coworkers started their experiment by allowing mostly oxidized horse heart cytochrome c to bind to liposomes formed with ca. 30% CL/70% PC (Kapetanaki et al. 2009). Thereafter, they reduced the protein. In a third step, they allowed the protein to react with CO, which yielded a hexacoordinate low-spin (hcLS) protein with CO as the distal ligand of the heme iron. In this experiment, reduction is likely to produce a pentacoordinate high-spin state that easily binds CO as a ligand.
Serpas et al. carried out a somewhat more thorough study on cytr binding to SUVs with 20%TOCL/80%DOPC (Serpas et al. 2016). Figure 21 shows how the Soret band CD, absorption, and W59 fluorescence change in the presence of liposomes. The absorption band shifts to the blue and the CD signal converts into a couplet which with increasing lipid concentration converts into a positive Cotton band. These data are clearly indicative of a conversion into a native oxidized state followed by a transition into a non-native oxidized state. This observation was further corroborated by the appearance of the 695 nm charge transfer band. Figure 22 shows fluorescence and CD response data that were analyzed with an extended version of the model proposed by Pandiscia and Schweitzer-Stenner (solid lines) (Pandiscia and Schweitzer-Stenner 2015b). The effective binding constant was found to vary between 105 and 3∙105 M−1.Footnote 4
Soret band CD and absorption spectra (upper panel) and tryptophan (W59) fluorescence (lower panel) of cytr as measured for selected CL concentrations of 20% TOCL/80% DOPC mixtures (red: ferrocytochrome c in solution, orange: [CL] = 37.5 μM, yellow: [CL] = 65 μM, green: [CL] = 150 μM, cyan: [CL] = 200 μM). Each solution was prepared in an aerobic environment, with a final ferrocytochrome concentration of 5 μM. The CL concentrations are numerically identical to the lipid/protein ratios. Captions and figure were taken from ref. (Serpas et al. 2016) with permission
Upper panel, left: ΔΔε = Δε417 − Δε405 derived from Fig. 21 plotted as a function of the CL concentration of cytr c − 20% TOCL/80% DOPC liposome mixtures. Upper panel, right: integrated intensity of the main sub-band at 340 nm derived from the W59 fluorescence band decomposition plotted as a function of CL concentration. Lower panel: polarized fluorescence, IVV (right) and IVH (left), measured at 340 nm plotted as function of CL concentration. The solid lines result from a global fit described in the text. The error bars for the ΔΔε values are within the size of the used symbols. Captions and figure were taken from ref. (Serpas et al. 2016) with permission
In further experiments, Serpas et al. observed that the change of the oxidation state is inhibited at anaerobic conditions. This clearly proves the involvement of O2 → O2− reactions. Even more interesting is the observation that liposomes bound cytochrome c becomes indeed reduced, in line with the observations of Nantes et al. (2001). However, from the presented data, it is clear that this process does not involve the formation of pentacoordinate high-spin states, as proposed by Nantes et al.
Summary of binding studies
It is apparent that the above referenced binding studies do not provide a consistent picture of how cytochrome c reacts with anionic lipid membranes. Studies in which cytochrome c is added to a sample with a constant concentration of lipids (type I binding experiments) generally yields apparent binding constants that are generally two orders of magnitude higher (> 107 M−1) than those observed with experiments where the cytochrome c concentration is kept constant and the lipid (liposome) concentration is varied (104–105 M−1) (type II binding experiments). The reason for this discrepancy has yet to be determined. We could think of two possible explanations. In view of the very different experimental techniques used for the two types of binding studies, one could speculate that the high affinity binding step was not detected by the type II experiment because it does not cause any detectable conformational change of the protein. If this was the case, the experiments of Pandiscia and Schweitzer-Stenner (2015b), Sinibaldi et al. (2008), and Hanske et al. (2012) would probe binding via an alternative site. However, discrepancies remain since these studies report rather different influences of NaCl on the binding process. Alternatively, one might invoke the possibility that in type I experiments the initial phase of cytochrome binding influences the surface of liposomes, which affects the subsequent binding of proteins, subsequently added to the sample. In the type II experiments, each data point represents an independent experiment with a fresh mixture of liposomes and proteins.
Other discrepancies must be kept in mind. It is unclear how the addition of NaCl can inhibit A-site binding which indicates reversibility, whereas the addition of liposomes does not cause any redistribution of bound cytochromes (Rytömaa and Kinnunen 1995). A-site binding is clearly electrostatic, while the low affinity binding process reported by Pandiscia and Schweitzer-Stenner is only indirectly affected by the addition of salt (i.e., by stabilizing the native-like over non-native-like conformations of adsorbed proteins) (Pandiscia and Schweitzer-Stenner 2015b). We wonder whether the direct interaction of anions (Na+) stiffens the membrane, reduces the depth of protein penetration, and thus facilitates the dissociation from the liposome surface (Böckmann et al. 2003). Moreover, one has to take into account that the binding observed by Kinnunen and coworkers starts at very high lipid to protein ratios where the influence of NaCl on the overall binding affinity is higher that it is at low lipid concentrations. That could in part explain why the influence of NaCl seems to be much more pronounced in experiments where cytochrome c is added to a fixed amount of liposomes. However, even the binding affinity that Pandiscia and Schweitzer-Stenner reported for high lipid to protein ratios does not account for the high affinity binding indicated by the results of type I binding experiments (Pandiscia and Schweitzer-Stenner 2015b), so that one may doubt whether their experiments and those of the Kinnunen group involve the same binding site. The mechanism for the proposed C-site binding is somewhat unclear, because multiple lines of experimental evidence rules out a partial protonation of phosphate groups above pH 4 (Malyshka et al. 2014; Sathappa and Alder 2016; Kooijman et al. 2017). The L-site binding reported by Kawai et al. seems to be on more solid footing. Experiments with mitoplasts revealed its biological relevance (Kawai et al. 2005, 2009). The binding studies of Milorey et al. confirmed its electrostatic character and provided evidence for the concomitant population of high-spin states caused by the protonation of a second histidine ligand that replaces M80 as ligand in the non-native state population of CL-bound cytochrome c (Milorey et al. 2017).
From a biological point of view, the question arises to what extent the observed binding processes represent the interaction of cytochrome c with the inner membrane of mitochondria. Earlier work by Cortese et al. revealed a somewhat more complex picture in that it suggests an ionic strength-dependent equilibrium between populations of membrane-bound cytochromes with different electron transfer activity (Cortese et al. 1998). Their results suggest that only a small fraction of mitochondrial cytochrome c is bound to the inner membrane at physiological conditions, the rest (ca. 100 μM) diffuses freely in the intermembrane space (Gupte and Hackenbrock 1988). This clearly suggests that cytochrome c binding to the innermembrane is mostly electrostatic in nature and that it is therefore substantially reduced by electrostatic screening due to the high NaCl concentration (120 mM). In view of the slightly acidic pH at the innermembrane surface, it seems reasonable to assign a major physiological role to the only recently discovered L-site binding (Kawai et al. 2009). Generally, the relationship between the characteristics of cytochrome c binding to anionic membranes in solution and the innermembrane of mitochondria still needs to be established.
Conformations of cytochrome c on anionic membrane surfaces
Some aspects of conformational changes of cytochrome c induced by binding to anionic lipids have already been mentioned in the preceding chapter. Here, we give a more systematic account of structural studies.
Obviously, investigating structural properties of proteins bound to membrane surfaces is not a straightforward task. X-ray crystallography is not an option. Solution NMR is not either. Solid-state NMR has been recently employed (vide infra), but this method is too demanding for a systematic study of structural changes. Therefore, structural studies using a variety of spectroscopic techniques have focused on specific aspects of structural changes rather than on a complete determination of protein structures.
While focusing on structural changes of the protein, we will also briefly address proposed changes of the membrane surfaces induced by the binding of cytochrome c. Here, we will pay particular attention to recent studies on gigantic unilamelar vesicles (GUVs) which provided strong evidence for pore formation and incorporation of bound proteins into the interior of such vesicles.
Earlier structural studies
Since this review focuses on recent work on cytochrome c interactions with anionic surfaces, we mention only a few studies carried out in the 80s and 90s, which provided the framework for later research.
In the 70s and 80s, several spectroscopic studies indicated that the binding of cytochrome c to cardiolipin-containing vesicles leads to changes of the membrane structure and also alters the protein’s conformation. Vincent et al., for instance, used electron paramagnetic resonance spectroscopy at 13 K to observe that complexation of cyto with bovine cardiolipin produced a small population of high-spin states and a change of the crystal field of the heme ligands for the hexacoordinate low-spin majority of the lipid bound proteins (Vincent et al. 1987). The experiments were conducted at pH 7.0 (where the L-site binding can be expected to start at room temperature) and with rather low lipid/protein ratios. The modified hexacoordinate low-spin state was found to exhibit larger tetragonal and rhombic distortions of the crystal field. The latter is indicative of a B1g-type deformation of the heme macrocycle, which is known to particularly large in ferric hexacoordinate low-spin systems due to static Jahn-Teller deformations. The latter occur if the Fe3+ ground state with exhibits E-symmetry in an ideal D4h-symmetry (Schweitzer-Stenner et al. 2000).
In the late 80s and 90s, Hildebrandt performed a series of resonance Raman studies to explore changes of the heme’s ligation and spin state upon binding to anionic surfaces. First, Hildebrandt et al. found that the binding of horse heart cyto to negatively charged surfaces and lipid dispersions at neutral pH produces an equilibrium between two states, a low-spin state termed cyt23+ and a pentacoordinate high-spin state (Hildebrand and Stockburger 1989; Hildebrandt and Stockburger 1989; Hildebrandt et al. 1990). For the former, they reported a more open heme crevice compared with the native state III that the protein adopts in solution. Subsequently, Heimburg et al. followed up on their studies by using resonance Raman spectroscopy to examine the equilibrium between cyt23+ and high-spin species after cyto was allowed to react either with 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol (DMPG) or mixtures of DOPG with DOPC and DOG (1,2 dioleoyl-sn- glycerol) (Heimburg et al. 1991). For DMPG, the equilibrium between the modified and native low-spin species was found to shift to the former upon the membrane’s transition from a gel-like into a liquid like phase. Figure 23 shows how the contribution of different spin and ligation states to the spin marker band ν3 at 1500 cm−1 and spectral distributions in the low wavenumber region are modified when DOG lipids are added to DOPG. Apparently, DOG stabilizes the native low-spin state, which clearly indicates that the ligation state of the heme depends on the lipid composition. 31P NMR results suggest that this change of spin equilibrium might be related to increasing curvature of the membrane which at very high mol% of DOG (50 mol%) leads to an inverted hexagonal phase.
Resonance Raman spectra of a, b cyto c-DOPG and c, d cyto-DOPG-DOG excited at 407 nm. The dotted lines are the Lorentzian profiles obtained from spectral decomposition. Taken from ref. (Milorey et al. 2017) with permission
Finally, we like to briefly discuss one of the rare kinetic studies aimed at exploring the reaction of cyto with anionic lipid vesicles by Pinheiro et al.(1997). They combined the stopped-flow technique with fluorescence and absorption measurement to investigate the binding of cyto to DOPs vesicles. The lipid to protein ratio was very large (in the 103 region). Their results revealed an unfolding of the native state at a rate of 1.5 s−1, which finally lead to the formation of a lipid-inserted denatured state. The experiments were carried out at neutral pH. The denatured protein was found to still exhibit a secondary structure content very similar to that of the native state. In view of the very high lipid to protein ratio used by these authors, the result is very much in line with the model of Pandiscia and Schweitzer-Stenner (2015b), which allows for a dominance of the f-state at these conditions. It is also in agreement with structural studies of the Pletneva group, which we discuss in great detail below.
The lipid insertion model
In the preceding section of this paper, we already mentioned that Kinnunen and coworkers proposed a binding model according to which electrostatic binding is augmented by the insertion of one of the four lipid chains of cardiolipin into the hydrophobic crevice of the protein, which would have to undergo a structural change into a non-native state in order to facilitate such a step. Some direct evidence for lipid insertion was derived from fluorescence quenching experiments (vide supra). Kalanxhi and Wallace chose a more indirect approach (Kalanxhi and Wallace 2007). They compared the bonding of several yeast cytochrome c derivatives (mutants and chemical derivatives) to CL-containing liposomes, wild-type horse heart cytochrome c, and a R81Nle derivatives of the latter (Nle: norleucine). Interestingly, they used cytr for their experiments, which one would normally expect to strongly resist the required conformational changes. The utilized liposomes were 5:4:3 mixtures of PC/PE/CL. Their binding assay was unusual somewhat in that they probed changes of the difference of the absorptivity at 550 and 526.5 nm. The latter is an isosbestic point for any changes caused by a low-spin > high-spin transition. If one assumes that any non-spin-related change effects the absorptivity at both wavelengths equally, any change of the difference solely probes spectral changes induced by a spin change of the heme iron. It is not clear, however, why the authors expected such a spin state change to occur. In their paper, they report that the Q0 and Qv band of the typical cytr spectrum merged into a single band upon binding to the above liposomes. However, this was also observed by Lee et al. and clearly related to a change of the redox state without any accompanying change of the spin state. Whatever the reason for the utilized spectral change was, there is no doubt that it could be used as an indicator for binding. The authors found a substantial spread of binding constants for their cytr derivatives which they attributed to different capabilities to accommodate a lipid chain in the heme interior. Figure 24 shows the hydrophobic stretches that the authors proposed to accommodate a penetrating lipid.
Illustration of the hydrophobic channel that may allow CL-anchorage in cytochrome c. a The cleft that could accommodate one of the acyl chains in cyt c consists of non-polar residues comprising polypeptide strands 67–71 and 82–85 (ochre) with positively charged residues Arg-91 and Lys-72 (blue) at either extremity. b Space-filling model representing the two hydrophobic stretches. The protein structure was generated with the VMD molecular modeling program (Protein Data Bank accession code 1AKK). The figure and most of the caption was taken from ref. (Kalanxhi and Wallace 2007) with permission
Strong though still somewhat indirect support for a lipid penetration emerged from the work of Kagan et al., who showed that CL-bound cytochrome c catalyzes the oxidation of cardiolipin (Kagan et al. 2005). Apparently, that requires a very close proximity of the lipid to the active site of the heme. The work of Kapralov et al. (2007) indicates a close correlation between conformational change, heme nitrolysation, and peroxidase activity, but contrary to later claims (Basova et al. 2007), this work and related papers do not provide any evidence for the population of a high-spin pentacoordinate state. With regard to the lipid insertion hypothesis, their data do not provide any additional support. The difficulty to unambiguously prove the validity of the hypothesis has been emphasized by Rajaopal et al. (2012), who found that replacing several residues in the hydrophobic stretches of the yeast cytochrome c (S65, T69, N70, K72) by alanine has a very limited influence on the respective peroxidase activity. Only the replacement of R91 was found to affect the protein’s stability. Interestingly, this mutation increases the proteins peroxidase capability in solution, but this gain is nearly eliminated upon interactions with CL-containing liposomes. Altogether, it is our impression that the jury is still out about the validity of the lipid insertion model.
Binding and peroxidase activity
In 2005, Kagan et al. published a major study which firmly established the relationship between cytochrome c binding to the inner membrane of mitochondria, cardiolipin oxidation via a peroxidase reaction involving cytochrome c, the permeabilization of the outer membrane, and the release of proapoptic factors (including cytochrome c) into the cytosol, where they trigger the caspase cascades (Kagan et al. 2005). Their data clearly show that this process requires the presence of cytochrome c. EPR-data revealed the production of an amino acid radical (most likely tyrosine) during the peroxidase process and mass spectroscopy data show that unsaturated cardiolipin is the target of the protein’s peroxidase activity. In a later study from this laboratory, Kapralov et al. offered several lines of evidence for the involvement of Y67 as the likely donor in the oxygenase half-reaction of cytochrome c–cardiolipin complexes (Kapralov et al. 2011). Some results of this study seem to indicate that the gain of peroxidase activity requires a structural change of the protein and a switch from a low-spin to a high-spin state.
The Kagan group published several follow up papers of the above study. Here, we focus only on a few of them which we deem of particular relevance for illuminating the relationship between structure and function of cardiolipin-bound cytochrome c. Basova et al. conducted a comprehensive electrochemical study to address an important question, namely to what extent cytochrome c can maintain its high redox potential while bound to CL-containing membranes (Basova et al. 2007). Here, we mention only two of the numerous experiments these researchers carried out. Figure 25 shows the cyclic voltammograms of cytochrome c covalently attached to carboxylic acid terminated self-assembled monolayers covered gold electrons in and without the presence of 50 mol% TOCL/50 mol% DOPC. The reduction peak had moved from 220 to − 200 mV after 1 h. The oxidation peak shifted by ca. 50 mV to lower values. Overall, these observations are clearly indicative of a switch from a positive to a very negative redox potential, which was attributed to a change of the axial ligand.
Cyclic voltammograms of cytochrome c electrostatically attached to carboxylic acid terminated in the absence (dashed line) and the presence of 30 μM H2O2 after 3 (solid line) and 15 (dotted line) min incubation with H2O2. a Cyclic voltammograms of cytochrome c covalently attached to SAM at different concentrations of H2O2 in the absence (b) and presence of TOCL/DOPC liposomes (c). Cytochrome c was incubated with liposomes for 60 min prior to the addition of H2O2. The voltammograms were recorded after preincubation with H2O2 during 15 min under N2. The voltammograms of cytochrome c without liposomes at pH 7.0 are shown for comparison on both figures (solid line). The utilized scan rate was 20 V/s. The figure and most of the caption was taken from ref. (Basova et al. 2007) with permission
In order to identify the redox properties of TOCL-bound cytochrome c, Basova et al. carried out a titration experiment which involved the addition of sodium dithionite, the classical reducing agent for heme proteins owing to its very negative E0 value of − 564 mV (Basova et al. 2007). The experiment was performed under anaerobic conditions and in the presence of gallocyanine, a redox dye with an E0 value of 20 mV. Figure 26 shows the concentration of cytr as a function of dithionite concentration. At low TOCL/cyt ratios (25:1 and 50:1), dithionite reduced cytochrome c while the dye remained oxidized. At higher TOCL/cyt ratios, however, the reduction of the protein required rather high concentrations of dithionate while gallocyanine was reduced. These results are indicative of a relatively high redox potential (> 100 mV) at low and relatively low potential (< 100 mV) at high TOCL concentrations. The authors estimated that the fraction of proteins with a negative redox potential was about 20% at a TOCL/cyt ratio of 25 and 90% at very high ratios (200). This suggests the existence of at least two populations of liposome-bound cytochrome c. In further experiments, Basova et al. investigated the influence of ascorbate (58 mV) and the O2/O2− couple on liposome-bound cytochrome c to more precisely identify the redox potential of the protein’s non-native state. They arrived at the conclusion that the redox potential of cytochrome shifts negatively by 350–400 mV upon binding to TOCL-containing liposomes.
Concentration dependence of cytr c in the presence of TOCL and dithionite obtained by titration in the presence of gallocyanine (8 μM). The employed molar ratios of TOCL/cyt c were 25:1 (curve 1), 50:1 (curve 2), 100:1 (curve 3), and 200:1 (curve 4). The figure and most of the caption was taken from ref. (Basova et al. 2007) with permission
Structural analysis via fluorescence resonance energy transfer
This paragraph focuses on papers that the Pletneva group published nearly 6 years ago which shed some light on the conformational ensemble of liposome-bound cytochrome c. For most of their experiments, they used liposomes composed of 50 mol% TOCL and 50 mol% DOPC.
The first of the above papers by Hanske et al. utilized Zn-substituted horse heart cytochrome c (Hanske et al. 2012). Additionally, they produced proteins with different fluorescence labels. To accomplish this task, they substituted four of the protein’s amino acid residues (E4, K39, E66, and E92) with cysteine (Fig. 27). Each derivative contained only one of these mutations. Dansyl fluorophores were then attached to these cysteine residues. Spectroscopic studies revealed that all these modifications do not significantly change the structure and stability of the protein. The authors measured the dansyl emission as an indicator of fluorescence resonance energy transfer (FRET) between the heme group of cyto (acceptor) and the individual dansyl moieties (donor). In the native state, a significant fraction of the dansyl fluorescence is quenched due to FRET to the heme group. If the protein starts to unfold, distances between donor and acceptor increases, which causes the dansyl fluorescence to increase. The degree of FRET can be assessed by measuring changes of the fluorescence lifetime. If an ensemble of proteins is heterogeneous, one observes a distribution of lifetimes. This is what Hanske et al. observed for liposome-bound cyto as well as for denatured proteins. All experiments were carried out at a very high lipid/protein ratio. The CL/cyto ratio of their experiments was close to 500. This puts them into a regime where the low protein occupation of the liposomes’ surfaces should allow for some unfolding of the protein (Pandiscia and Schweitzer-Stenner 2015b).
a Structure of horse heart cyto c showing the positions of DNS labels attached to cysteine groups of the protein. b Structure of the DNS label. The figure was taken from ref. (Hanske et al. 2012) with permission
Hanske et al. used an earlier developed algorithm to translate lifetime into distance distributions. The result of this analysis is shown in Fig. 28. For the native protein, they observed rather narrow nearly Gaussian distributions that peak somewhere between 20 and 25 Å for all dansyl labels. Upon the addition of TOCL-containing liposomes, a very heterogeneous set of conformers with larger distance emerge for all four dansyl labels. This change is particularly pronounced for the dansyl attached to position 92, a residue which in the model of Englander and coworkers belong to low-stability foldon that contains M80 (Englander et al. 1998). The author observed a more drastic increase of conformational heterogeneity for the denatured protein, which indicates that the proteins on the liposome surface are not completely denatured.
Distribution of donor-acceptor distance P(r) obtained from FRET experiments four DNS-labeled variants of cyto c at pH 7.4 [native (N), gray]; with TOCL/DOPC liposomes at 660 μM total lipid [CL-bound (CL), blue]; with TOCL/DOPC liposomes at 660 μM total lipid and 150 mM NaCl (CL. salt, red); and in 5.8 0.2 M GuHCl solution at pH 7.4 [GuHCl-unfolded (UGuHCl), green]. The figure was taken from ref. (Hanske et al. 2012) and modified.
Hanske et al. invoked a two-state model to explain their results. They interpreted their results as reflecting an equilibrium between compact, more native-like states (C-conformations) and extended, more unfolded states (E-conformations). The absence of a 695-nm band in the optical absorption spectrum led them to conclude that most of the lipid-bound proteins had lost M80 ligand and were therefore at least partially unfolded. This view is corroborated by later results from our laboratory (Pandiscia and Schweitzer-Stenner 2015b). For the extended state, they invoked a rupture of non-covalent interactions between the terminal helices, which are thus allowed to interact most effectively with the lipid surface (Fig. 26). This model could be represented by a double well potential with very rugged potential surfaces. Local minima of the potential would correspond to conformational sub-states, which are all slightly different in structural terms (Hong et al. 1990).
The addition of NaCl shifts the equilibrium from E- to C-states. While the authors conclude that this involves some loss of bound proteins, their binding studies (vide supra) do not indicate a large inhibition with 150 mM NaCl. It is therefore much more likely that the observed distance changes just reflect a structural redistribution between the C and E sub-ensembles.
In a follow-up paper, Hong et al. attached another fluorophore termed bimane to the cysteine group at position 92 (Hong et al. 2012). Results from FRET studies utilizing the bimane-heme pair very much corroborated the two-state model of Hanske et al. (2012). Moreover, they found that an increase of the concentration of cytochromes and thus of the occupation density on the surface of liposomes lead to as shift toward the C-conformation ensemble. An increase of the C-content of liposomes from 10 to 50 mol% increases the E-fraction from ca. 5 to 60%. Fluorescence correlation experiments revealed a very fast (sub-ms) exchange between C- and E-conformations.
How do the results of these very comprehensive mostly structure-oriented studies of the Pletneva group fit into the picture that emerge from the thus far cited binding and structural studies? Let us start with the lipid insertion model. It is neither falsified nor verified by the FRET results. As pointed out by Hong et al. (2012), only the compact structure could really accommodate and attach the lipid to the interior of the protein. However, the results of the spectroscopic studies of Pandiscia and Schweitzer-Stenner suggest that at least a measurable fraction of compact membrane-bound protein are native-like with M80 as sixth ligand (Pandiscia and Schweitzer-Stenner 2015b). It is doubtful that such a conformation could accommodate a lipid chain. The results of Hong et al. further suggest that the addition of NaCl affects the protein’s conformational distribution by shifting it toward C. However, their data do not reveal whether the initial binding is actually affected. The observed changes are not reminiscent of the NaCl influence on A- and L-site binding. As we outlined above, the binding affinity measured by Hanske et al. is much lower than that of Kinnunen’s A-site binding.
In addition to the work referenced above, the Pletneva group conducted a very illuminating kinetic study, where they measured the fluorescence of the above introduced dansyl labels as a function of time in the presence of different concentration of 50 mol% TOCL/50 mol% DOPC liposomes. Muenzner et al. identified a burst phase (fluorescence change within the dead time of the employed stop flow instrument) and three kinetic phases with corresponding amplitudes different for the used dansyl labels (Muenzner et al. 2013). The authors found that the amplitude of the burst phase increases with increasing liposome concentration. The amplitude of the first detectable phase was found to be generally small and the related time constant did not depend on the lipid concentration. The apparent rate constant of the second phase was also found to be independent on the lipid concentration. In contrast to these observations, the authors observed a significant lipid concentration dependence of the third apparent rate constant. These and other observations led them to suggest a reaction mode that is visualized in Fig. 29. The authors considered the first binding step as electrostatically driven. Based on work of the Spiro group, they suggested that subsequent conformational changes require a rupture of the H26-P44 hydrogen bond (Balakrishnan et al. 2012). That would release H26 for ligation of the heme iron. In the last step, the linkage between the C- and N-terminal helix breaks which allows the former to unfold. Overall, this leads to the extended structure obtained from the above FRET studies.
Representation of a proposed mechanism of cyto c unfolding upon contact with CL-containing liposomes. (1) Positively charged cyt c and negatively charged liposomes interact electrostatically, leading to binding of cyt c to the membrane within milliseconds. (2) A protein surface region near K72, K73, K86, and K87 as well as N50, E66, and E92 is the likely contact site for binding to the membrane. The H26–P44 hydrogen bond breaks upon binding of cyt c to the surface. (3) The low-stability loop containing M80 rearranges, and the weak M80–heme coordination is broken. In addition, unfolding of the green and yellow foldon units loosens the protein tertiary structure. (4) The 50s helix and the beginning of the C-terminal helix insert partially into the membrane and anchor the protein, while the 60s helix interacts flat with the membrane surface. The 40s Ω loop is pulled closer to the membrane, while staying in relatively close proximity to the heme group. The end of the C-terminal helix largely unfolds and N- and C-terminal helix contacts are broken, leading to an extended protein conformer E with an easily accessible heme group. The figure and most of the caption was taken from ref. (Muenzner et al. 2013) with permission
Apparently, there are a lot of similarities between the results of the Pletneva group and those of Pandiscia and Schweitzer-Stenner, which were reported several years later. The respective apparent binding affinities are comparable. One would be tempted to relate their nf-state to the C and their f-state to E. Differences and similarities, however, are noteworthy. Both models predict an increase of the more unfolded state over the more native-like state with increasing CL-content, but the effect seems to be much more pronounced for the data derived from the FRET analysis. The E-state fraction was reported to increase from 19 for 20% to nearly 60% for 50% TOCL in the absence of NaCl. From the binding analysis of Pandiscia and Schweitzer-Stenner (2015b), a much more modest increase from 50 to 58% was obtained for the f-state at high TOCL concentrations. With regard to the influence of NaCl, the results are much more in agreement. The addition of 150 mM NaCl reduces the E-content from ca. 50% to ca. 28%.Footnote 5 For their f-state fraction on the surface of 50 mol% TOCL/50 mol% DOPC liposomes, 200 mM addition of NaCl reduces its fraction from ca. 57 to 18%. Pletneva and coworkers considered the C-state ensemble as being composed of unbound native, and non-native compact proteins. However, the 695 nm band and resonance Raman data show that the nf-ensemble of Pandiscia and Schweitzer-Stenner contains native-like (with M80) and non-native proteins. Altogether, the two ensemble pairs should be similar but might not be totally identical. Some of the C-state proteins might still fluoresce and thus count to the f-state ensemble. Generally, it can be stated that the results of the studies from the two laboratories are mutually supportive of each other.
A somewhat less biologically oriented study of cytochrome c on surfaces is noteworthy in this context. Deng et al. used electrospray ion beam deposition to identify an ensemble of folded and unfolded cytochrome c ions on Au(111) and boron-nitride nanomesh layer on Rh(111) surfaces (Deng et al. 2012). The picture that emerged from their characterization of the surface-adsorbed proteins resembles to a major extent what Hong et al. wrote about C- and E-state cytochrome on CL-containing liposomes.
In this context, it is useful to recall the results of the electrochemical studies of Basova et al. (2007). They found that their low redox potential fraction of liposome-bound cytochrome c increases from 20% at a TOCL/cyt ratio of 25 to 90% at a TOCL/cyt ratio of 200. One is tempted to assign the former to the C/nf and the latter to the E/f ensemble. For nf and f, the corresponding numbers are 32 and 65%. Apparently, the 60% E-state population that Hong et al. obtained at high lipid/protein ratios is closer to the f-fraction than to the reported fraction of proteins with a low redox potential (Hong et al. 2012). However, we should keep in mind that the 695 nm data of Pandiscia and Schweitzer-Stenner indicate that some of the nf-conformations at higher lipid/protein ratios lack M80 as axial ligand (Pandiscia and Schweitzer-Stenner 2015b). With regard to Basova et al., they would count toward the low redox potential fraction. Even if M80 is maintained as sixth heme ligand, the protein itself might adopt slightly different structures on the membrane surface due to electric field effects (Hannibal et al. 2016). Overall, we can consider the results of the functional and structural studies as consistent.
In this context, it should be emphasized that the studies by Schweitzer-Stenner and associates provided some additional structural information. Optical spectroscopy and resonance Raman data clearly suggest that the vast majority of proteins in the f-state ensemble exhibit a hexacoordinate low-spin state (Malyshka and Schweitzer-Stenner 2017), for which more recent investigations suggest a bis-histidine ligation (Milazzo et al. 2017; Milorey et al. 2017). Only for liposomes with 50% and to a slightly larger extent for those with 100% TOCL, a small fraction of high-spin species appears at very high lipid concentrations. The situation changes dramatically at acidic pH, where the low-spin f-state is replaced by a mixture of penta- and hexacoordinate high-spin species, most likely due to the protonation of the imidazole of the second histidine ligand (H33 and/or H26).(Milorey et al. 2017)
We finish this paragraph by referencing some corollary spectroscopic work aimed at investigating cytochrome c binding to anionic surface. Wackerbarth and Hildebrandt used surface-enhanced resonance Raman spectroscopy to investigate cytochrome c electrostatically adsorbed in the double layer of an Ag electrode/electrolyte interface (Wackerbarth and Hildebrandt 2003). They identified a potential dependent equilibrium between the native conformation (termed B1 by the authors) and non-native conformations (B2) where M80 is replaced by other ligands. The oxidized B2 ensemble was comprised of sub-conformations containing heme groups with hexacoordinate low-spin bis-histidine, hexacoordinate high-spin histidine-water, and pentacoordinate high-spin heme irons. The equilibrium between these sub-conformations was found to depend on the electric field provided by the electrode. In view of their results, the question arises whether the equilibrium between low- and high-spin f-conformations does also reflect changes of the electric field on the surface of CL-containing liposomes. Wackerbarth and Hildebrandt explained such an influence of the electric field with a weakening of tertiary structure stabilizing hydrogen bonds. The proposed influence of external electric fields on the tertiary structure of cytochrome c was later corroborated by MD simulations (Hannibal et al. 2016). In line with these studies, Ranieri et al. found an equilibrium between oxidized low-spin bis-histidine and high-spin pentacoordinate conformations for CL-bound cytochrome c immobilized on a SAM of decane-1-thiol (Ranieri et al. 2015).
In a more recent study, Zeng et al. produced 100% CL membrane on a gold surface to investigate the adsorption of cytochrome c with surface-enhanced infrared absorption spectroscopy (Zeng et al. 2016). For neutral pH, their results seem to indicate (a) changes of the secondary structure from α-helix to β-sheet and a somewhat combined involvement of electrostatic and hydrophobic interactions. While one is tempted to emphasize the similarity with the findings of Pandiscia and Schweitzer-Stenner (2015b), any such comparison should be cautioned since CL-membranes on a gold electrode might differ from vesicles in solution regarding to their response to protein binding.
Recent structural studies
An interesting approach to study the secondary structure of cyto on bilayer surfaces (not liposomes) formed with bovine CL and DPPG has been undertaken by Nguyen (2015). This author utilized chiral sum frequency generation vibrational spectroscopy, which is an ideal tool to examine molecules on surfaces. The measurements were performed in 20 mM phosphate buffer at pH 5.5 and 7.2. From an analysis of their amide I signal, the author concluded that the protein undergoes a structural change into a mostly β-sheet structure with different orientations at the two investigated pH values. Unfortunately, the author did not acknowledge that his results contradicted other results on the secondary structure of liposome-bound cytochrome c, which did not indicate any major formation of β-sheets (Hong et al. 2012). His results are somewhat reminiscent of findings reported by Spiro and coworkers who found that unfolded cytochrome c can form β-sheet like fibrils at pH 3 and high temperatures (Balakrishnan et al. 2012).
A more revealing study on the structure of cardiolipin-bound cytochrome c was carried out by Mandal et al. (2015). These authors employed solid-state NMR to 13C and 15N labeled cyto, which was allowed to react with LUVs containing 25 mol% TOCL and 75 mol% DOPC. The chosen lipid/protein ration of 40/1, which corresponds to an effective TOCL/cyt ration of 5, is very low. Under these conditions, the ensemble of membrane-bound proteins can be expected to be dominated by proteins with a native-like state in which M80 is still an axial ligand (Pandiscia and Schweitzer-Stenner 2015b). Interestingly, the authors observed a significant increase of peroxidase activity. The authors used two different solid-state NMR techniques involving ‘1H-13C cross polarization’ (CP) and ‘insensitive nuclei enhanced by polarization’ (INEPT) to study membrane-bound horse heart cyto and cytr. They performed measurements at 271, 257, and 236 K and found a general increase of signal intensities with decreasing temperature (Fig. 30). At 271 K, both spectra display mostly signals from lipids. With decreasing temperature, the CP spectrum becomes more populated by protein signals, while the INEPT spectrum remains mostly a lipid indicator. The authors drew the following conclusions from their results. First, the data reveal that membrane-bound cytochrome c is less mobile than the alkyl chains of the lipid which indicates a rather compact and inflexible structure. Second, the immobilization of the protein must be coupled primarily to the head groups while leaving the alkyl groups of the tails mostly unaffected. The absence of major unfolding was then confirmed by results from 2D magic angle spinning NMR spectroscopy that did not reveal any major structural disorder. FTIR spectroscopy indicated a mostly intact secondary structure. Their results further suggest that the LUVs retained their lipid bilayer structure and the penetration into the membrane is limited. In order to reconcile their findings with the observed increase in peroxidase activity, the authors hypothesized that local dynamics close to the active site could facilitate interactions with substrates.
1D MAS NMR spectra of oxidized and reduced U-13C,15N cytochrome c bound to TOCL/DOPC LUVs. a, b 1H-13C CP and direct excitation 13C spectra of oxidized and reduced membrane-bound cytochrome c measured at 271 K. The spectra in each panel are plotted with absolute intensities, and were processed with 25 Hz exponential line broadening and fourfold zero-filling. c 1H-13C CP spectrum measured at 257 K, showing mostly lipid signals. d CP spectrum at 250 K, and e 236 K, with much increased protein signals (up arrows). f 1D 1H-13C refocused INEPT spectrum at 257 K, with mostly lipid peaks (headgroups marked). g, h INEPT data at 250 and 236 K, showing loss of lipid headgroup and acyl chain signals, respectively (down arrows). i 1D 1H spectrum at 257 K, with intense liquid water peak. j 1D 1H spectrum at 250 K, after freezing of solvent (down arrow). k 1H 1D at 236 K, showing a loss of acyl chain signals (down arrow). Data obtained at 600 MHz (1H) and 8.33 kHz MAS, and processed with a cosine-bell apodization function and zero-filling to twice the original size. Selected spectra are scaled as indicated. The figure and most of the caption was taken from ref. (Mandal et al. 2015) with permission
If one relates the results of this very comprehensive study to the insight gained by spectroscopic and FRET experiments, however, they do not really come as a surprise. As indicated above, the population analysis of Pandiscia and Schweitzer-Stenner predicts a dominance of the native-like nf(C)-conformations for the experimental conditions chosen (Pandiscia and Schweitzer-Stenner 2015b). One should also emphasize that all NMR experiments were performed at temperatures significantly below room temperature, which should stabilize the native-like states further. The peroxidase measurements, however, were carried out at room temperature, where the fraction of non-native nf(C) conformations is low, but not totally negligible.
Investigating alternative structures in solution
Since structural studies on cytochrome c bound to membrane surfaces is a tedious business, some research group have investigated some non-native structures in solution instead. These investigations were aimed at finding clues about how such structures could actually resemble conformations adopted by the membrane-bound protein. Here, we focus on two most recent studies which involve X-ray crystallography of the investigated proteins.
Amacher et al. conducted a very thorough study of the oxidized state of a yeast protein derivative in which four natural amino acid residues were substituted by others (K72A/C102S/T78C/K79G) (Amacher et al. 2015). The authors focused only on the effect of the last two mutations and considered the K72A/C102S as pseudo-wild type. This is justified because none of these mutations has a significant impact on the protein’s structure which still exhibits M80 as axial ligand. The C102S replacement is routinely used to avoid dimerization of cytochrome c, while the K72A substitution prevents M80 from being replaced by K72 as heme ligand. Spectroscopic properties of the above mutant in solution all point to the C78 thiolate as the sixth ligand of the heme iron. That made cytochrome c more similar to cytochrome c P450. However, to the authors’ surprise, they found that the only near heme pocket lysine that had not been eliminated was the sixth ligand in the crystal structure of the protein. The necessary structural arrangement involved a disulfide bridge between two proteins and the formation of a β-hairpin involving P76 and G77 (Fig. 31). The observed structure was rather compact. In order to conduct some functional studies on this particular conformation, the authors carried out peroxidase assays with maleimide-modified T78C/K79G mutants, the spectroscopic properties of which suggest that it is a solution analogue to the structure explored by X-ray crystallography. They found that the peroxidase activity of these derivatives is higher than that of the corresponding pseudo-wild type and even higher than the values for M80A mutants. They also observed a high peroxidase activity for T78C/K79G solutions in crystal drops where lysine is still the axial ligands. Their results contrast finding regarding the alkaline IV states of cyto in solution which exhibit a lower peroxidase activity than even the native state. Overall, these results kept lysine as a possible ligand of liposome-bound cytochrome c in the race.
Coordination loop differences in cytochrome c structures with K73, hydroxide (PDB ID: 4MU8), or M80 (PDB ID: 2YCC) coordinated to the heme iron. Structures are colored by carbon atoms as follows: K73-ligated T78C/K79G = cyan, M80-ligated C102T = yellow, hydroxideligated WT = green. Noncarbon atoms are colored by element: N = blue, O = red, S = yellow, Fe = orange. a Alignment of the three structures by main-chain atoms reveals similar core conformations, with RMSD = 0.50 Å and 0.27/0.33 Å (A- and B-protomers), respectively. The K73 and M80 residues are highlighted (side chain stick figures) and labeled for each structure. b–d Heme coordination geometry for K73-ligated T78C/K79G (b), K80-ligated C102T (c), and hydroxide-ligated WT (d), as labeled. e, f The heme coordination loops of T78C/K79G (e) and C102T (f) are shown following superposition of the core conformations in cartoon representation (middle box) and as stick figures (left and right boxes). Stick representations of the loops are labeled by residue, and black dashed lines mark hydrogen bonds. Below, the van der Waals surfaces for T78C/K79G (cyan) and C102T (yellow) are shown to highlight the positions of the heme coordination loop. The figure and most of the caption was taken from ref. (Amacher et al. 2015) with permission
The second work to be considered is a paper by McClelland et al. (2014). They reported a 1.45-Å resolution X-ray structure of a yeast cytochrome derivative in the trimethyllysine was replaced by an alanine. As shown in Fig. 32, this led to a replacement of M80 by a water molecule as sixth ligand. As one would expect for such a conformation, this structural modification enhances the peroxidase activity of the protein. The authors discovered a water channel that facilitates the access to the active site and provide a conduit for proton translocation. From their data, they inferred that the N38 residue mediates the opening and closing of the water channel. They hypothesized that this process could serve as a gating mechanism for the peroxidase process.
Comparison of the overall structures of K72A and tmK72 cytochrome c. a Alignment of K72A cyt c (green; chain A of PDB ID code 4MU8) with tmK72 cyt c (gray; PDB ID code 2YCC; carries a C102T mutation). The heme and Met80 are shown as stick models. A close-up view of the heme and M80 is displayed (left). b Close-up view of the heme crevice showing water molecules (red spheres) in K72A cytochrome c in the space occupied by M80 in tmK72 cytochrome c. Low occupancy positions observed for two of the waters are shown in purple. c Heme crevice close-up view showing the 2jFoj − jFcj electron density map contoured at 1.2σ (blue wire) with the model used to fit the data. The figure and most of the caption was taken from ref. (McClelland et al. 2014) with permission
The studies cited above reveal that specific mutations of cytochrome c can lead to rather stable non-native structures of cytochrome c. Horse heart, human, and yeast cytochrome c differ in terms of their peroxidase activity in the native as well as in the partially unfolded alkaline state. These functional differences seem to reflect the influence of amino acid residues at the positions 72, 81, and 83 on the dynamics of the so called Ω-loop (McClelland et al. 2014; Nold et al. 2017). Whether or not they are of relevance for an understanding of cytochrome c–cardiolipin complexes remains to be seen.
Membrane changes
There is an ongoing debate about whether or not the binding of cytochrome c to CL-containing vesicles can induce major changes of the membrane. Very early NMR studies by de Kruijff and Cullis seem to indicate that cytochrome c penetrates into the interior of vesicles while the membrane adopts an inverted hexagonal phase (De Kruijff and Cullis 1980). Heimburg et al. studied cytochrome c–vesicle interactions by combining resonance Raman with 31P NMR (Heimburg et al. 1991). They found that the admixture of DOG to DOPG facilitates the increase of the bilayer curvature of vesicles upon cytochrome binding, which leads to the earlier proposed inverted hexagonal phase. Choi and Dimitriadis studied cytochrome c adsorption to DOPG with atomic force microscopy (Choi and Dimitriadis 2004). They interpreted their results as suggesting that cytochrome c does indeed penetrate into the membrane, but in their experiments the proteins were found to reside in the hydrophobic core rather than in the interior of the utilized vesicles. Gorbenko et al. analyzed FRET between lipid attached fluorophores the heme group of cyto bound to CL/PC LUVs. Based on their results, they concluded that the protein penetrates the membrane only at pH < 6 (Gorbenko et al. 2006).
The clearest direct evidence for cytochrome c penetration into the membrane of CL-containing vesicles was more recently provided by very elegant microscopic experiments performed by Bergstrom et al. (2013). These authors labeled yeast cyto with Alexa Fluor 633 maleimide attached to C102. They allowed the labeled proteins to react with gigantic unilamelar vesicles (GUVs) composed of 20 mol% CL and 80 mol% DOPC. Confocal microscopy images shown in Figs. 32 and 33 reveal that the labeled cytochrome interacted with the GUVs in a way that allowed for their penetration into the vesicles interior. In order to check the extent of pore formation, the authors took images of their GUVs mixed with fluorescent 10-kDa dextran in the presence and absence of cytochrome c. Only for the latter case did the authors observe that the probe penetrated the membrane. This result suggests that cytochrome c binding produced (metastable) membrane pores through which the labeled dextran could move into the vesicles interior. From an analysis of their data, they proposed a scenario where a negative membrane curvature is formed with respect to the membrane surface and a positive curvature with regard to the normal to the membrane surface.
Confocal microscopy images of cytochrome c interactions with CL-containing membranes of GUVs. a DOPC with NBD-PE (left vesicle) and 80:20 DOPC:CL (right vesicle), treated with Alexa Fluor 568–cyt c in solution (red). b The 80:20 DOPC:CL GUV with NBD-PE, treated with fluorescent 70-kDa dextran (green) and Alexa Fluor 633–cyt c in solution (red). c–e Time series of 80:20 DOPC:CL GUVs with NBD-PE, treated with fluorescent 10-kDa dextran (green) and Alexa Fluor 633–cyt c in solution (red). In all images, the cyan lipid dye is 0.5 mol% NBD-PE. Average normalized cytochrome c and dextran concentration for each vesicle interior vs. background is plotted. The figure and most of the caption was taken from ref. (Bergstrom et al. 2013) with permission
Another very elegant and novel approach to study cytochrome c–cardiolipin interactions has recently been presented by Kitt et al. (2017). They combined Raman spectroscopy with optical trapping to monitor simultaneously structural changes of the protein and the membrane of 40 mol% CL/60 mol% DPPC vesicles upon cytochrome c binding. In addition, they used the Raman spectra of membrane-impermeable tracer, 3-nitrobenzenesulfonate, as indicator of membrane permeabilization. The chosen excitation wavelength of 647 nm served multiple purposes. With regard to cytochrome c, it enabled the authors to monitor heme vibrations owing to pre-resonance enhancement with what is most likely the vibronic side band of the 695 nm band, which is assignable to a S(M80)-> Fe3+ transition. Moreover, protein bands from aromatic residues and amide bands could be observed for probing changes of the secondary and tertiary structure. Second, they could probe marker bands diagnostic of the lipid structure. By using fully deuterated cardiolipin chains, their contributions could be isolated. Finally, spectral changes assignable to the above tracer were used as an indicator of pore formation. They found that cytochrome binding causes solely changes of the cardiolipin alkyl chains indicating increased disordering and a decreased intensity of bands assignable to vibrational modes of cis-double bonds. Changes in the tracer spectra were diagnostic of pore formation. The reduction of contributions from heme vibrations could be attributed to the elimination of M80 ligation.
While the work of Bergstrom et al. provides clear cut evidence for cytochrome c penetration into the CL-containing membrane and pore formation (Bergstrom et al. 2013), caution has to be exercised when one compares their study with those which did not reveal any evidence for protein penetration. It must be noted that the GUV-based studies were carried out with rather low lipid/protein ratios (< 1), which allow for a significant surface pressure exerted by the bound proteins. From the description of their experiments, we could not infer the lipid/protein ratio of the Raman experiments of Kitt et al. (2017). All studies where results are inconsistent with a significant penetration of cytochrome c were carried out with much higher lipid/protein ratios (Hanske et al. 2012; Hong et al. 2012; Mandal et al. 2015; Pandiscia and Schweitzer-Stenner 2015a, b). This notion even applies to the work of Mandal et al., which was carried out in a low lipid/protein region where native-like states are favored (Mandal et al. 2015). It is likely that the propensity for penetration depends not only on the protein density on the surface but also on the lipid composition of the membrane and the size of the vesicles. Obviously, more work needs to be done to describe the conditions for membrane penetration more exactly.
Summary and outlook
This review is aimed at delineating the current status of knowledge about the relationship between cytochrome c binding to anionic lipid surface, the structures it can adopt on such surfaces, and the changes of its functional properties. We tried to emphasize some unresolved issues where further clarification is warranted.
A very important discrepancy exists regarding experimental binding constants. Experiments where cytochrome c is added to a fixed amount of lipids generally yield binding affinities that are between one and two orders of magnitude higher than those derived from experiments where a spectroscopic response was measured as a function of lipid concentration at a fixed concentration of cytochrome c. Moreover, a clear picture about the number and types of binding sites has yet to emerge. The experiments of the Kinnunen group indicate that all cytochrome c binding to CL-containing liposomes above pH 6 is electrostatic in nature (A-site binding), while hydrogen bonding dominates at more acidic pH (C-site binding) (Rytöman et al. 1992; Rytöman and Kinnunen 1994). However, several lines of experimental evidence clearly revealed the existence of at least two different mechanism of cytochrome-CL interactions at pH values below and above 7 (Kawai et al. 2005, 2009, 2014; Milorey et al. 2017). The binding below pH 7 (termed L-site binding) is clearly electrostatic in nature, but the situation is less clear for the binding at neutral pH, where electrostatics is more involved in stabilizing extended unfolded over more compact and native-like proteins. Some papers indicate that cytochrome c can penetrate into the interior of the membrane bilayer or even of the utilized vesicles themselves, while others do argue to the contrary. Here, it seems likely that penetration and pore formation requires a rather high protein density and thus very low lipid to protein ratios. Several lines of evidence point to the insertion of a lipid chain into the protein interior, but the conditions for this to occur have not been unambiguously identified. Finally, the relationship between protein conformation and peroxidase activity remains obscure. Some papers, e.g., from the Kagan group, seem to indicate the necessity of a spin conversion from a low- into a high-spin state of the heme iron (Kapralov et al. 2007), which requires the displacement of M80 as ligand. However, a more recent paper from this group and their collaborators paint a totally different picture in that it provides evidence for the enhanced peroxidase activity of CL-bound cyto, the conformation of which is still very much native like. Hanske et al., however, showed that the peroxidase activity clearly increases with increasing stabilization of extended E-state proteins (Hanske et al. 2012), in which the heme group is most likely ligated by a histidine (Milorey et al. 2017; Milazzo et al. 2017). Another paper from the same group showed that the addition of ATP to cytochrome c–50 mol% TOCL/50 mol% DOPC mixtures actually boosts peroxidase activity while simultaneously destabilizing the extended E-state in a manner reminiscent of the influence of NaCl (Snider et al. 2013). On the other hand, Ranieri et al., by investigating urea-unfolded yeast cytochrome c adsorbed on gold electrodes coated with an anionic monolayer, observed a high heme spin-mediated reduction of H2O2. The same effect was found to be much weaker for the native protein (Ranieri et al. 2015). Patriarca et al. measured the peroxidase activity of a series of horse heart cytochrome c mutants (Patriarca et al. 2012). Their results underscore the notion that conformational changes are necessary to increase the protein’s peroxidase activity from the low level of the native protein.
We believe that in order to resolve all the issues, more systematic studies which are aimed at exploring the multidimensional parameter space of cytochrome structure and function on anionic lipid membranes has to be conducted. With regard to tests of peroxidase test, a larger emphasis needs to be put on actually probing the oxidation of cardiolipin itself since this is the crucial biological process which eventually facilitates the protein’s release from the intermembrane space of mitochondria.
Notes
Contrary to the authors claim, Eq. 9 of their paper has not been reported by Jähnig (Jähnig 1976). Heimburg and Marsh use the high potential limit approximation of Jähnig for their own modeling of cytochrome c binding to anionic lipid surface (Heimburg and Marsh 1995). Their eq. 10 is clearly distinct from Eq. 9 of Grobenko et al.
It should be noted that our notation differs from that of Heimburg and Marsh.
Kawai et al. reported that they mixed 4 μm cyto with 250 μM lipids. This corresponded to a CL-concentration of 50 μM. If one assumes an equal partition of CL between the inner and outer membrane, one arrives at a CL/cyt concentration of approximately 62.
It should be noted that the K values in the Supporting Information of Lee et al. are in units of (μM)−1, not in M−1 as indicated in the axis legend.
The values for corresponding E-fractions in the papers of Hong et al. and Hanske et al. are slightly different, which might be attributable to the different donors used for the respective FRET experiments.
References
Abe M, Kitagawa T, Kyogoku Y (1978) Resonance Raman spectra in octaethylporphyrin-Ni(II) and mesodeuterated and 15N substituted derivatives. II. A normal coordinate analysis. J Chem Phys 69:4526–4534
Abe M, Niibayashi R, Koubori S et al (2011) Molecular mechanisms for the induction of peroxidase activity of the cytochrome c- cardiolipin complex. Biochemistry 50:8383–8391
Alessi M, Hagarman AM, Soffer JB, Schweitzer-Stenner R (2010) In-plane deformations of the heme group in native and nonnative oxidized cytochrome c probed by resonance Raman dispersion spectroscopy. J Raman Spectrosc 42:917–925
Alvarez-Paggi D, Hannibal L, Castro MA et al (2017) Multifunctional cytochrome c learning new tricks from an old dog. Chem Rev. https://doi.org/10.1021/acs.chemrev.7b00257
Amacher JF, Zhong F, Lisi GP, et al (2015) A compact structure of cytochrome c trapper in a lysine-ligated state: loop refolding and functional implications of a conformational switch. J Am Chem Soc 8435–8449
Assfalg M, Bertini I, Dolfi A et al (2003) Structural model for an alkaline form of Ferricytochrome c. J Am Chem Soc 125:2913–2922
Atlante A, Calissano P, Bobba A et al (2000) Cytochrome c is released from mitochondria in a reactive oxygen species (ROS)-dependent fashion and can operate as a ROS scavenger and as a repiratory substrate in cerebellar neurons undergoing excitotoxic death. J Biol Chem 275:37159–37166
Bai Y, Milne JS, Mayne L, Englander SW (1993) Primary structure effects on peptide group hydrogen exchange. Proteins 17:75–86. https://doi.org/10.1002/prot.340170110
Balakrishnan G, Hu Y, Spiro TG (2012) H26 protonation in cytochrome c triggers microsecond β-sheet formation and heme exposure: implications for apoptosis. J Am Chem Soc 134:19061–19069
Banci L, Bertini I, Gray HB et al (1997) Solution structure of oxidized horse heart cytochrome c. Biochemistry 36:9867–9877
Basova LV, Kurnikov IV, Wang L et al (2007) Cardiolipin switch in mitochondria: sutting off the reduction of cytochrome and turning on the peroxidase activity. Biochemistry 46:3423–3434
Battistuzzi G, Borsari M, Cowan JA et al (2002) Control of cytochrome c redox potential: axial ligation and protein environment effects. J Am Chem Soc 124:5315–5324
Beales PA, Bergstrom CL, Geerts N et al (2011) Single vesicle observations of the cardiolipin-cytochrome c interaction: introduction of membrane morphology changes. Langmuir 27:6107–6115
Belikova NA, Vladimirov YA, Osipov AN et al (2006) Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry 45:4998–5009
Berghuis AM, Brayer GD (1992) Oxidation state-dependent conformational changes in cytochrome c. J Mol Biol 223:959–976
Bergstrom CL, Beales PA, Lv Y et al (2013) Cytochrome c causes pore formation in cardiolipin-containing membranes. Proc Nat Acad Sci USA 110:6269–6274
Bhardwaj A, Walker-Kopp N, Wilkens S, Cingolani G (2008) Foldon-guided self-assembly of ultra-stabel protein fibers. Prot Sci 17:1475–1485
Blouin C, Guillemette JG, Wallace CJA (2001) Resolving the individual components of a pH-induced conformational change. Biophys J 81:2331–2338
Böckmann RA, Hac A, Heimburg T, Grubmüller H (2003) Effect of sodium chloride on a lipid bilayer. Biophys J 85:1674–1655
Bushnell GW, Louie G V, Brayer GD (1990) High-resolution Three-dimensional Structure of Horse Heart Cytochrome c. J Mol Biol 214
Capdevilla DA, Marmisolle WA, Tomasima F et al (2015a) Specific methionine oxidation of cytochrome c in complexes with zwitterionic lipids by hydrogen peroxide: potential implications for apoptosis. Chem Sci 6:705–713
Capdevilla DA, Rouco SA, Tomasina F et al (2015b) Active site structure and peroxidase activity of oxidatively modified cytochrome c species in complexes with cardiolipin. Biochemistry 54:7491–7504
Choi EJ, Dimitriadis EK (2004) Cytochrome c adsorption to supported, anionic lipid bilayers studied via atomic force microscopy. Biophys J 87:3234–3241
Cohen DS, Pielak GJ (1994) Stability of yeast iso-1 ferricytochrome c as a function of pH and temperature. Prot Sci 3:1253–1280
Colón W, Elöve GA, Walken LP et al (1996) Side chain packing of the N-and C-terminal helices plays a critical role in the kinetics of cytochrome c folding. Biochemistry 35:5538–5549
Cortese JD, Voglino AL, Hackenbrock CR (1998) Multiple conformations of physiological membrane-bound cytochrome c. Biochemistry 37:6402–6409
Dale R, Eisinger J, Blumberg W (1979) The orientational freedomof molecular probes. The orientation factor in intramolecular energy transfer. Biophys J 26:161–194
De Kruijff B, Cullis PR (1980) Cytochrome c specifically induces non-bilayer structures in cardiolipin containing model membranes. Biochim Biophys Acta 602:470–499
Deng Z, Thontasen N, Malinowski N et al (2012) A close look at proteins: submolecular resolution of two- and three-dimensionally folded cytochrome c at surfaces. Nano Lett 12:2452–2458
Domanov YA, Molotkovsky JG, Gorbenko GP (2005) Coverage-dependent changes of cytochrome c transverse location in phospholipid membranes revealed by FRET. Biochim Biophys Acta 1716:49–58
Döpner S, Hildebrandt P, Rosell FI, Mauk AG (1998) The alkaline conformational transitions of ferricytochrome c studied by resonance Raman spectroscopy. J Am Chem Soc 120:11246–11255
Droghetti E, Oellerich S, Hildebrandt P, Smulevich G (2006) Heme coordination states of unfolded ferrous cytochrome c. Biophys J 91:3022–3031
Edman ET (1979) A comparison of the structures of electron transfer proteins. Biochim Biophys Acta 549:107–144
Elöve GA, Bhuyan AK, Roder H (1994) Kinetic mechanism of cytochrome c folding: involvement of the heme and its ligands. Biochemistry 1994:6925–6935
Englander SW, Mayne L (2017) The case for defined protein folding pathways. Proc Nat Acad Sci USA 114:8253–8258
Englander SW, Sosnick TR, Mayne LC et al (1998) Fast and slow folding in cytochrome c. Acc Chem Res 31:737–744
Gorbenko GP, Molotkovsky JG, Kinnunen PKJ (2006) Cytochrome c interaction with cardiolipin/phosphatidylcholine model membranes: effect of cardiolipin protonation. Biophys J 90:4093
Gupte SS, Hackenbrock CR (1988) The role of cytochrome c diffusion in mitochondrial electron transport. J Biol Chem 263:5248–5253
Hagarman A, Duitch L, Schweitzer-Stenner R (2008) The conformational manifold of Ferricytochromec explored by visible and far-UV electronic circular dichroism spectroscopy. Biochemistry 47:9667–9677
Hannibal L, Tomasina F, Capdevila DA et al (2016) Alternative conformations of cytochrome c: structure, function and detection. Biochemistry 55:407–428
Hanske J, Toffey JR, Morenz AM et al (2012) Conformational properties of cardiolipin-bound cytochrome c. Proc Natl Acad Sci U S A 109:125–130. https://doi.org/10.1073/pnas.1112312108
Hartree EF (1973) The discovery of cytochrome. Biomed Ed 1:69–74
Hashimoto M, Takeda A, Hsu LJ et al (1999) Role of cytochrome c as a stimulator of α-synuclein aggregation in Lewy body disease. J Biol Chem 274:28849–28852
Heimburg T, Marsh D (1995) Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes. Biophys J 68:536–546
Heimburg T, Hildebrandt P, Marsh D (1991) Cytochrome c-lipid interactions studied by resonance Raman and 31P NMR spectroscopy. Correlation between the conformational change of the protein and the lipid bilayer. Biochemistry 30:9084–9089
Heimburg T, Angerstein A, Marsh D (1999) Binding of peripheral proteins to mixed lipid membranes: effect of lipid demixing upon binding. Biophys J 76:2575–2586
Hildebrand P, Stockburger M (1989) Cytochrome c at charged interfaces: 1. Conformational and redox equilibria at the electrode/electrolyte interface probed by surface enhanced resonance Raman spectroscopy. Biochemistry 28:6710–6721
Hildebrandt P, Stockburger M (1989) Cytochrome c at charged interfaces: 2. Complexes with negatively charged macomolecular systems studied by resonance Raman spectroscopy. Biochemistry 68:6722–6728
Hildebrandt P, Heimburg T, Marsh D (1990) Quantitative conformational analysis of cytochrome c bound to phospholipid vesicles studied by resonance Raman spectroscopy. Eur Biophys J 18:193–201
Hong MK, Braunstein D, Cowen BR et al (1990) Conformational substates and motions in myoglobin-external influences on structure and dynamics. Biophys J 58:429–436
Hong Y, Muenzner J, Grimm SK, Pletneva EV (2012) Origin of the conformational heterogeneity of cardiolipin-bound cytochrome c. J Am Chem Soc 134:18713–18723
Hunte C, Somaz S, Lange C (2002) Electron transfer between cytochrome bc1 complexand its substrate cytochrome c. Biochim Biophys Acta Bioenerg 1555:21–28
Hüttemann M, Pecina P, Rainbolt M et al (2011) The multiple functions of cytochrome c and their regulation in life an death decisions of the mammalian cell: from repiratin to apoptosis. Mitochondrion 11:369–381
Iwase H, Takatori T, Nagao M et al (1996) Monoexpoxide production from linoleic acid by cytochrome c in the presence of cardiolipin. Biochem Biophys Res Comm 222:83–89
Jähnig F (1976) Electrostatic free energy and shift of the phase transition from charged lipid membranes. Biophys Chem 4:309–318
Jiang X, Wang X (2004) Cytochrome c-mediated apoptosis. Annu Rev Biochem 73:87–106
Kagan VE, Tyurin VA, Jiang J et al (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptic factors. Nat Chem Biol 4:223–232
Kalanxhi E, Wallace CJA (2007) Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem J 407:179–187
Kapetanaki SM, Silkstone G, Husu I et al (2009) Interaction with carbon monoxide with the apoptosis inducing cytochrome c-cardiolipin complex. Biochemistry 48:16131610
Kapralov AA, Kurnikov IV, Vlasova II et al (2007) The hierachy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry 46:14232–14244
Kapralov AA, Yanamala N, Tyurina Y et al (2011) Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim Biophys Acta 1808:2147–2155
Kawai C, Prado FM, Nunes GLC et al (2005) pH-dependent interaction of cytochrome c with mitochondrial mimetic membranes: the role of an array of positively charged amino acid residues. J Biol Chem 280:34709–34717
Kawai C, Pessoto FS, Rodrigues T et al (2009) pH-sensitive binding of cytochrome c to the inner mitochondrial membrane. Implications for the participation of the protein in cell respiration and apoptosis. Biochemistry 48:8335
Kawai C, Ferreira JC, Baptista MS, Nantes IL (2014) Not only oxidation of cardiolipin affects the affinity of cytochrome c for lipid bilayers. J Phys Chem B 118:11863–11872
Kitt JP, Bryce DA, Minteer SD, Harris JM (2017) Raman spectroscopy reveals selective interactions of cytochrome c with cardiolipin that correlate with membrane permeability. J Am Chem Soc 139:3851–3860
König BW, Osherroff N, Wilms J et al (1980) Mapping an interaction domain for purified cytochrome c1 on cytochrome c. FEBS Letts 255:4732–4739
Kooijman EE, Swim LA, Garber ZT et al (2017) Magic angle spinning 31NMR spectroscopy reveals two essentially identical ionization states for the cardiolipin phosphates in phospholipid liposomes. Biochim Biophys Acta 1859:61–68
Lewis RNAH, McElhaney RN (2009) The physiochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membrane. Biochim Biophys Acta 1788:2069–2079
Louie GV, Brayer GD (1990) High-resolution refinement of yeast Iso-1-cytochrome c and comparisons with other eukaryotic cytochromes c. J Mol Biol 214:527–555
Malyshka D, Schweitzer-Stenner R (2017) Ferrocyanide-mediated photoreduction of Ferricytochrome C utilized to selectively probe non-native conformations induced by binding to cardiolipin-containing liposomes. Chem Eur J 23:1151–1156
Malyshka D, Pandiscia LA, Schweitzer-Stenner R (2014) Cardiolipin containing liposomes are fully ionized at physiological pH. An FT-IR study of phosphate group ionization. Vib Sprectrosc 75:86–92
Mandal A, Hoop CL, Kodali R et al (2015) Structural changes and proapoptopic peroxidase activity of cardiolipin-bound mitochondrial cytochrome c. Biophys J 109:1873–1884
Mara MW, Hadt RG, Reinhard ME et al (2017) Metalloprotein entatic control of ligand-metal bonds quatified by ultrafast x-ray spectroscopy. Science 356:1276–1280
McClelland LJ, Mou T-C, Jeakins-Cooley ME et al (2014) Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity. Proc Nat Acad Sci USA 111:6648–6653
Milazzo L, Tognaccini L, Howes BD et al (2017) Unravelling the non-native low-spin state of the cytochrome c−cardiolipin complex: evidence of the formation of a his-ligated species only. Biochemistry 56:1887–1898
Milorey B, Malyshka D, Schweitzer-Stenner R (2017) pH dependence of Ferricytochrome c conformational transitions during binding to cardiolipin membranes: evidence for histidine as the distal ligand at neutral pH. J Phys Chem Lett 8:1993. https://doi.org/10.1021/acs.jpclett.7b00597
Minton AP (1995) Confinement as a determinant of macromolecular structure and reactivity. II. Effects of weakly attractive interactions between confined macrosolutes and confining structures. Biophys J 68:1311–1322
Minton AP (2000) Effects of excluded surface area and adsorbate clustering on surface adsorption of proteins. I. Equilibrium models. Biophys Chem 86:239–247
Moore GR, Williams RJ (1980) Nuclear-magnetic resonance studies of ferrocytochrome c. pH and temperature dependence. Eur J Biochem 103:513–521
Muenzner J, Toffey JR, Hong Y, Pletneva EV (2013) Becoming a peroxidase: cardiolipin-induced unfolding of cytochrome c. J Phys Chem B 117:12878–12886. https://doi.org/10.1021/jp402104r
Nantes II, Zucchi MR, Nascimento OR, Faljoni-Alario A (2001) Effect of heme iron valence state on the conformation of cytochrome c and its association with membrane interfaces. J Biol Chem 276:153–158
Nguyen KT (2015) An electronically enhanced chiral sum frequency generation vibrational spectroscopy study of lipid-bound cytochrome c. Chem Comm 51:195–197
Nold SM, Lei H, Mou T-C, Bowler B (2017) Eeffect of a K72A mutation on the structure, stability, dynamics, and peroxidase activity of human cytochrome c. Biochemistry 56:3358–3368
Numata K, Sato R, Yazawa K et al (2015) Crystal structure and physical properties of Antheraea yamamai silk fibers: long poly(alanine) sequences are partially in the crystalline region. Polymers (Basel) 77:87–94
Oellerich S, Wackerbarth H, Hildebrandt P (2002) Spectroscopic characterization of nonnative conformational states of cytochrome c. J Phys Chem B 106:6566–6580
Oellerich S, Lecomte S, Paternostre M et al (2004) Peripheral and integral binding of cytochrome c to phospholipids vesicles. J Phys Chem B 108:3781–3878
Olafsson G, Sparr E (2013) Ionization constants pKa of cardiolipin. PLoS One 8:e73040
Pandiscia LA, Schweitzer-Stenner R (2014) Salt as a catalyst in the mitochondria: returning cytochrome c to its native state after it misfolds on the surface of cardiolipin containing membranes. Chem Comm 50:3674–3676
Pandiscia LA, Schweitzer-Stenner R (2015a) Coexistence of native-like and non-native partially unfolded Ferricytochrome c on the surface of cardiolipin-containing liposomes. JPhys Chem B 119:1334–1349
Pandiscia LA, Schweitzer-Stenner R (2015b) Coexistence of native-like and non-native cytochrome c on anionic lipsomes with different cardiolipin content. J Phys Chem B 119:12846–12859
Paradies G, Petrosilio G, Pistolese M, Ruggiero FM (2000) The effect of reactive oxygen species generated from the mitochondrial electron transfer chain on the cytochrome c oxidase activity and on the cardiolipin content in bovine heart submitochondrial particles. FEBS Letts 466:323–326
Patriarca A, Polticelli F, Piro MC et al (2012) Conversion of cytochrome c into a peroxidase: inhibitory mechanisms and implication for neurdegenerative diseases. Arch Biochem Biophys 522:62–69
Pielak GJ, Oikawa K, Mauk AG et al (1986) Elimination of the negative Soret Cotton effect of cytochrome c by replacement of the invariant phenylalanine using site-directed mutagenesis. J Am Chem Soc 108:2724–2727
Pinheiro TJT, Elöve G, Watts A, Roder H (1997) Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry 36:13122–13132
Porcellli AM, Ghelli A, Zanna C et al (2005) pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem Biophys Res Commun 28:799–804
Rajagopal B, Silkstone G, Nicholls A et al (2012) An investigation into a cardiolipin acyl chain insertion site in cytochrome c. Biochim Biophys Acta 1817:780–791
Ranieri A, Millo D, Di Rocco G et al (2015) Immobilized cytochrome c bound to cardiolipin exhibits peculiar oxidation state-dependent axial heme ligation and catalytically reduces dioxygen. J Biol Inorg Chem 20:531–540
Rossel FI, Ferrer JC, Mauk AG (1998) Proton-linked protein conformational switching: definition of the alkaline conformational transition of yeast iso-l-ferricytochrome c. J Am Chem Soc 120:11234–11245
Ruíz-Ramírez A, Barrios-Maya MA, López-Acosta O et al (2015) Cytochrome c release from rat liver mitochondria is compromised by increased saturated cardiolipin species induced by sucrose feeding. Am J Physiol Metab 309:E777–E786
Rytömaa M, Kinnunen PKJ (1995) Reversibility of the the binding of cytochrome c to liposomes. J Biol Chem 270:3197–3202
Rytöman M, Kinnunen PKJ (1994) Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J Biol Chem 269:1770–1774
Rytöman M, Mustonen P, Kinnunen PKJ (1992) Reversible, nonionic and pH-dependent association of cytochrome c with cardiolipin- phosphatidylcholine liposomes. J Biol Chem 267:22243–22248
Sanishvili R, Volz KW, Westbrook EM, Margoliash E (1995) The low ionic strength crystal structure of horse cytochrome c at 2.1 Å resolution and comparison with the high ionic strength counterpart. Structure 3:707–716
Sathappa M, Alder NM (2016) The ionization properties of cardiolipin and its variants in model bilayers. Biochim Biophys Acta 1858:1362–1372
Schweitzer-Stenner R (2008) The internal electric field in cytochrome C explored by visible electronic circular dichroism spectroscopy. J Phys Chem B 112:10358–10366
Schweitzer-Stenner R (2014) Cytochrome c: a multifunctional protein combining conformational rigidity with flexibility. N J Sci 484538:1–28
Schweitzer-Stenner R, Cupane A, Leone M et al (2000) Anharmonic protein motions and heme deformations in myoglobin cyanide probed by absorption and resonance Raman spectroscopy. J Phys Chem B 104:4754–4764
Serpas L, Milorey B, Pandiscia LA et al (2016) Autoxidation of reduced horse heart cytochrome c catalyzed by cardiolipin-containing membranes. J Phys Chem B 120:12219–12231
Sinibaldi F, Fiorucci L, Patriarca A et al (2008) Insights into cytochrome c-cardiolipin interaction. Role Played by Ionic Strength Biochemistry 47:6928–6935
Sinibaldi F, Howes BD, Piro MC et al (2010) Extended cardiolipin anchorage to cytochrmoe c: a model for protein-mitochondrial membrane binding. J Biol Inorg Chem 15:689–700
Sinibaldi F, Howes BD, Droghetti E et al (2013) Role of lysines in the cytochrome c - cardiolipin interactions. Biochemistry 52:4578–4588
Sinibaldi F, Milazzo L, Howes BD et al (2017) The key role played by charge in the interaction of cytochrome c with cardiolipin. J Biol Inorg Chem 22:19–29
Smith HT, Staudenmayer N, Millet F (1977) Use of specific lysine modifications to locate the reaction site of cytochrome c with cytochrome c oxidase. Biochemistry 16:4971–4974
Snider EJ, Muenzner J, Toffey JR et al (2013) Multifacet effects of ATP on cardiolipin-bound cytochrome c. Biochemistry 52:993–995
Soffer J. (2013) The folded, partially folded misfolded , and unfolded conformations of cytochrome c probed by optical spectroscopy. Drexel University
Soffer JB, Schweitzer-Stenner R (2014) Near-exact enthalpy-entropy compensation governs the thermal unfolding of protonation states of oxidized cytochrome c. J Biol Inorg Chem 19:1181–1194
Speck SH, Ferguson-Miller S, Osherroff N, Margoliash E (1979) Definition of cytochrome c binding domains by chemical modification: kinetics of reaction with beef mitochondrial reductase and functional organization of the respiratory chain. Proc Nat Acad Sci USA 76:155–159
Spooner PJR, Watts A (1992) Cytochrome c interaction with cardiolipin in bilayers: multinuclear magic-angle spinning NMR study. Biochemistry 31:10129–10138
Takahashi S, Yeh S-R, Das TK et al (1997) Folding of cytochrome c initiated by submillisecond mixing. Nat Struct Biol 44:44–50
Takano T, Dickerson RE (1981) Conformation change of cytochrome c .1. Ferrocytochrome c structure refined at 1.5 A resolution. J Mol Biol 153:79–94
Tanaka N, Yamane T, Tsukihara T et al (1975) The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 Å resolution. J Biochem 77:147–162
Theorell H, Åkessen Å (1941) Studies on cytochrome c. III. Titration curves. J Am Chem Soc 63:1818–1820
Therien MJ, Bowler BE, Selman MA et al (1991) Long-range electron transfer in heme proteins. Adv Chem 228:191–199
Trumpower B (1990) The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem 265:11409–11412
Trumpower BL, Gennis RB (1994) Energy transduction by cytochrome c complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu Rev Biochem 63:675–716
Trusova VM, Gorbenko GP, Molotkovsky JG, Kinnunen PKJ (2010) Cytochrome c-lipid interactions: new insight from resonance energy transfer. Biophys J 99:1754–1763
Tuominen EKJ, Wallace CJA, Kinnunen PKJ (2002) Phospholipid-cytochrome c interaction. J Biol Chem 277:8822
Vincent JS, Kon H, Levin IW (1987) Low-temperature electron paramagnetic resonance study of the Ferricytochrome c-cardiolipin complex. Biochemistry 26:2312–2314
Wackerbarth H, Hildebrandt P (2003) Redox and conformational equilibria and dynamics of cytochrome c at high electric fields. ChemPhysChem 4:714–724
Yeh S-R, Takahashi S, Fan B, Rousseau DL (1997) Ligand exchange during cytochrome c folding. Nat Struct Biol 4:51–56
Zeng L, Wu L et al (2016) Analyzing structural properties of heterogeneous cardiolipim-bound cytochrome c and their regulation by surface-enhanced infrared absorption spectroscopy. Anal Chem 88:11727–11733
Acknowledgements
The work from our research group reported in this paper was the product of hard work by my former graduate student, Dr. Leah Pandiscia, my current graduate student Bridget Milorey, and by two undergraduate researchers, Dmitry Malyshka and Lee Serpas.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Reinhard Schweitzer-Stenner declares that he has no conflict of interests with regards to this article.
Ethical approval
No experiments with humans or animals were performed for this study.
Rights and permissions
About this article
Cite this article
Schweitzer-Stenner, R. Relating the multi-functionality of cytochrome c to membrane binding and structural conversion. Biophys Rev 10, 1151–1185 (2018). https://doi.org/10.1007/s12551-018-0409-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-018-0409-4