Abstract
Cytochrome c undergoes structural variations upon binding of cardiolipin, one of the phospholipids constituting the mitochondrial membrane. Although several mechanisms governing cytochrome c/cardiolipin (cyt c/CL) recognition have been proposed, the interpretation of the process remains, at least in part, unknown. To better define the steps characterizing the cyt c–CL interaction, the role of Lys72 and Lys73, two residues thought to be important in the protein/lipid binding interaction, were recently investigated by mutagenesis. The substitution of the two (positively charged) Lys residues with Asn revealed that such mutations cancel the CL-dependent peroxidase activity of cyt c; furthermore, CL does not interact with the Lys72Asn mutant. In the present paper, we extend our study to the Lys → Arg mutants to investigate the influence exerted by the charge possessed by the residues located at positions 72 and 73 on the cyt c/CL interaction. On the basis of the present work a number of overall conclusions can be drawn: (i) position 72 must be occupied by a positively charged residue to assure cyt c/CL recognition; (ii) the Arg residues located at positions 72 and 73 permit cyt c to react with CL; (iii) the replacement of Lys72 with Arg weakens the second (low-affinity) binding transition; (iv) the Lys73Arg mutation strongly increases the peroxidase activity of the CL-bound protein.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome c (cyt c) is a mitochondrial peripheral membrane protein which acts in between the inner and the outer membrane, mediating electron transfer from cyt c reductase to cyt c oxidase in the respiratory chain. Cyt c is a single chain polypeptide of 104 amino acids which contains three major and two minor α-helices in the structure, and the heme as prosthetic group. The heme lies within a crevice lined with hydrophobic residues and is covalently attached to the polypeptide chain by two thioether bridges with residues Cys14 and Cys17. His18 and Met80 are the axial ligands of the six-coordinated low spin heme iron in the native state.
In recent years, the role played by cyt c in cell apoptosis after its release from the mitochondrion, has renewed interest in this protein [1–3]. About 15% of the protein is tightly bound to cardiolipin (CL), one of the phospholipids constituting the mitochondrial membrane [4–10]. In contrast with free cyt c, which participates in electron transfer and prevents oxidative stress, the tightly bound protein shows peroxidase activity, an event crucial for initiating cell apoptosis [4]. During apoptosis cyt c is released from the mitochondrial interface space into the cytosol, which implies protein dissociation from the mitochondrial membrane [11, 12]. In the cytoplasm, cyt c binds to the apoptosis protease activation factor (APAf-1) and forms a complex which, activating pro-caspase 9, gives rise to an enzymatic reaction cascade leading to apoptosis in cell [13]. Therefore, the cyt c–CL recognition plays a critical role in switching the protein function from electron transfer to a pro-apoptotic agent.
The interaction with CL induces a heterogeneous ensemble of non-native protein species [14, 15]. CL-bound cyt c shows altered tertiary structure, a perturbed heme crevice and the displacement of Met80 from the sixth coordination position of the heme-Fe atom [5–8, 16–19]. Depending on the experimental conditions, the sixth coordination position of the heme iron remains unbound or is occupied by another side chain (likely a Lys or a His residue) [7, 20, 21]. The partially unfolded conformation of membrane-bound cyt c induces a drastic change of the redox potential and favors its peroxidase activity [22], facilitating the access of small molecules (such as hydrogen peroxide) into the heme pocket region [23].
As a powerful CL-specific peroxidase, the cyt c/CL complex generates CL hydroperoxides, which actively participate in the release of cyt c and other pro-apoptotic factors from the mitochondrial membrane [3]. Thus, if the native fold endows cyt c with an electron carrier activity, the non-native conformation favors peroxidase activity of the CL-bound protein, representing a crucial event for the execution of the apoptotic program.
The mechanism of cyt c/CL complex formation has been widely investigated. Kinnunen and collaborators hypothesized that cyt c binds to CL through two binding sites [24]. At the site indicated as the A-site the interaction is of electrostatic nature and involves positively charged residues of cyt c (possibly Lys72 and Lys73) and the deprotonated negatively charged phosphate group of CL. Conversely, at the C-site, the cyt c/CL interaction is hydrophobic and is stabilized by the H-bond formed by Asn52 with the protonated phosphate group of CL. Ionic strength modulates the cyt c/CL interaction around neutrality; the cyt c/CL complex spontaneously forms at low ionic strength but tends to dissociate as the ionic strength increases [5, 6]. The “extended lipid conformation” model [24, 25] hypothesizes that at the C-site one acyl chain of CL accommodates into the protein interior by extending from the surface to the heme pocket region through a hydrophobic channel close to the small helix comprising the invariant residue Asn52. Conversely, the other chain of CL points in the opposite direction from the head group.
Despite the large body of studies performed to identify the protein site(s) involved in the cytc/CL interaction, the location of these region(s) still remains largely unknown. One such binding region has been proposed to be located in the segment containing the positively charged residues Lys72, Lys73, Lys86 in the neighborhood of the heme-binding region. The cleft formed by these side chains is thought to facilitate penetration of the acyl chain into the protein [6]. Based on the observation that the cyt c/CL binding reaction is characterized by two distinct transitions [7, 24], and that CL is the only phospholipid of the mitochondrial membrane able to bind cyt c tightly [8, 10, 27], the hypothesis that two acyl chains of CL, instead of one, may penetrate inside the protein during complex formation has been recently formulated [26]. According to this model, the electrostatic interaction between the Lys79 and Lys72 residues and the phosphate groups of CL favors protein binding to the phospholipid.
As invariant (Lys72) or largely invariant (Lys73) residues, these two lysines located in the Met80-containing loop adjacent to the heme pocket region are expected to play a critical role in determining the stabilization and the correct functionality of the native protein. The replacement of one (or both) Lys residues is expected to bring about the partial unfolding of the Ω loop and alteration of the heme pocket region, weakening the Met80-Fe axial bond strength [6]. We have recently shown that the Lys72 → Asn and Lys79 → Asn mutations prevent CL binding to the protein and that, unlike the CL-bound wt protein, the CL-bound Lys73Asn mutant does not show peroxidase activity [28]. These results were, at least in part, unexpected; therefore, to better define the role played by residues located at positions 72 and 73 in the cyt c/CL recognition process, we have extended our study to the reaction of Lys72Arg, Lys72Ala, Lys73Arg, and Lys73Ala mutants of horse heart cyt c with CL. We also attempted to obtain the Lys79Arg mutant, but this variant did not fold. In contrast to the previous investigation, where a positively charged residue was replaced by a polar side chain carrying no net charge (Lys → Asn mutation), in the present study the Lys residues have been replaced by Arg carrying a positive charge at neutral pH as Lys, but with a different structure, and the non-polar, aliphatic amino acid, Ala. The present study has been performed on ferric horse heart cyt c, which behaves similar to the human counterpart [29] and actively participates in cell apoptosis [4, 30–33].
Materials and methods
Materials
Horse heart cytochrome c (type VI, oxidized form) and cardiolipin, as sodium salt from bovine heart (approx 98% purity, lyophilized powder), were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used without further purification. All reagents were of analytical grade.
Liposomes preparation
Aqueous dispersions of CL liposomes were prepared as follows: a film of lipid was prepared on the inside wall of a round bottom flask, by evaporation of a chloroform solution containing the proper amounts of lipid. The films obtained were stored in a desiccator overnight under reduced pressure, then 1 ml of a 20 mM Hepes buffer solution was added to obtain a 2.5 mM lipid dispersion. Solutions were vortex-mixed and then freeze-thawed six times from liquid nitrogen to 30 °C. Dispersions were then extruded (10 times) through a 100 nm polycarbonate membrane. Extrusions were carried out at 30 °C. The vesicle size was determined by light scattering measurements. In the buffer, the vesicle diameter is approx 130 nm.
Construction of horse heart cyt c expression system
A version of the cyt c synthetic gene was designed on the basis of the sequence of a previously reported cyt c synthetic gene [34] and its synthesis performed by Primm srl (Milano, Italy). The synthetic gene was flanked by the NcoI and BamHI restriction sites, at the 5′ and 3′ ends, respectively. The pBTRI plasmid was converted to the cyt c expression plasmid by removing the yeast iso-1-cyt c gene and replacing it with the new synthetic horse cyt c gene, using the unique NcoI and BamHI sites. The sequence of the expression construct (pHCyc) was confirmed by DNA sequence analysis (M-Medical, Milano, Italy). The plasmid pHCyc was then subjected to one round of mutagenesis, which introduced either Lys72Arg or Lys72Ala and either Lys73Arg or Lys73Ala substitutions into the horse cyt c gene.
Cell growth and purification of recombinant proteins
The expression plasmids of horse cyt c were introduced into E. coli JM 109 as wild-type (wt) or its variants. Protein expression and purification of the recombinant protein were then conducted as previously described [35]. Briefly, bacteria containing the pBTRI (or the mutated) plasmid were grown at 37 °C, in 2 l of SB medium containing 100 µg/ml ampicillin to an absorbance of 0.3 OD at 600 nm. Induction was accomplished by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.75 mM. Cells were then incubated at 37 °C overnight, harvested by centrifugation and frozen at –80 °C. After thawing, the reddish pellets were resuspended in 50 mM Tris–HCl buffer, pH 8.0 (3–4 ml/g of wet cells). Lysozyme (1 mg/ml) and DNase (5 µg/ml) were then added to the homogenized cells. The suspension was left in ice for 1 h and then sonicated for 1 min, at medium intensity. After centrifugation, the supernatant was dialyzed overnight against 10 mM phosphate buffer pH 6.2, and loaded on a CM 52 column (40 ml bed volume) equilibrated with the same buffer. Purification was performed by eluting the protein with one volume of 45 mM phosphate pH 6.8, 250 mM NaCl, following a previously published procedure [36]. After purification, the recombinant proteins (~500 µM) had a purity >98% (determined by SDS-PAGE analysis and reverse phase HPLC, not shown) and stored at –80 °C in 200 µl aliquots.
Electronic absorption measurements
Electronic absorption measurements were carried out at 25 °C using a Jasco V-530 (Tokio, Japan) or a Cary 60 (Agilent Technologies, CA) spectrophotometer. The cyt c concentration was determined on the basis of the extinction coefficient ε = 106 mM−1 cm−1 at 408 nm.
Circular dichroism (CD) measurements
CD measurements were carried out at 25 °C using a Jasco J-710 spectropolarimeter (Tokyo, Japan). Binding of CL liposomes to wild-type and mutant cyt c was investigated by following the changes induced in the far-UV (215-250 nm) and Soret CD spectrum (400-450 nm) of the protein by stepwise addition of few μl of a 2.5 mM CL buffered solution to a 10 μM cyt c buffered solution. The buffer was 25 mM Hepes + 0.1 mM EDTA, pH 7.0. Dichroic spectra were recorded 10 min after mixing.
Resonance Raman measurements
The resonance Raman (RR) spectra were obtained using a 5-mm NMR tube and by excitation with the 406.7 nm line of a Kr+ laser (Coherent, Innova 300 C, Santa Clara, CA). Backscattered light from a slowly rotating NMR tube was collected and focused into a triple spectrometer (consisting of two Acton Research SpectraPro 2300i and a SpectraPro 2500i in the final stage with a 3600 grooves/mm grating) working in the subtractive mode, equipped with a liquid nitrogen-cooled CCD detector. Spectral resolution was 1 cm−1 calculated theoretically on the basis of the optical properties of the spectrometer. However, for the moderately broad experimental RR bands observed in the present study (ca. 10 cm−1), the effective spectral resolution will in general be lower. To improve the signal/noise ratio, a number of spectra were accumulated and summed only if no spectral differences were noted. The RR spectra were calibrated with indene and CCl4 as standards to an accuracy of 1 cm−1 for intense isolated bands. Due to their sensitivity to laser irradiation, all the mutants were cooled by a gentle flow of N2 gas passed through liquid N2, to minimize local heating of the protein by the laser beam, and the sample was changed every 15 min. A relatively low laser power of 2 mW was used for the Arg and Ala mutants, compared to 6 mW used previously for the Asn mutants [28].
Kinetic measurements
Kinetics of CL binding to cyt c mutants have been investigated by stopped-flow (Applied Photophysics, Salisbury, UK), following the absorption change at 408 nm upon mixing cyt c mutants with CL liposomes. Kinetic traces have been analyzed according to the following equation:
where ODobs is the observed optical density at a selected wavelength and at a given time-interval, OD0 is the optical density at t = 0, r is the number of exponentials, DOD i is the optical density change associated to the exponential i, i k is the rate constant of the exponential i and t is the time.
Peroxidase activity assay
The peroxidase activity of CL-free and CL-bound wild-type and mutant cyt c was determined by measuring the H2O2-dependent oxidation of guaiacol as a function of guaiacol concentration. The steady-state kinetics of guaiacol oxidation to give its tetramer was measured spectrophotometrically at 470 nm and 25 °C (ε470 = 26.6 mM−1 cm−1).
Results
Structural properties and stability of the Lys72Arg and Lys73Arg mutants
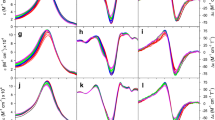
Interestingly, the substitution of Lys72 or 73 affects the protein in the same manner, but the spectroscopic properties depend on the substituting residue. Figure 1 compares the UV–Vis spectra of the Lys72 mutants under investigation, while the corresponding figure for the Lys73 mutants is reported in Supplemental Information (Fig. S1). The Arg mutation does not affect the protein, as the spectrum is identical to that of wt cyt c, while a slight decrease of the CT band at 695 is observed in the Ala mutant. Although significant changes have been observed previously for the Asn mutants [28], the CD properties of the Lys72Arg and Lys73Arg mutants are similar to those of the wt protein. The far-UV dichroic spectra (not shown) are typical of proteins possessing α-helix secondary structure, and the ellipticity is comparable to that of the native form. Figure 2 compares the Soret (400–450 nm) CD spectra of the Lys → Arg, Lys → Asn [28], and Lys → Ala mutants. The Soret CD spectrum of the Lys72Arg mutant is very similar to that of the wt protein; in fact, the typical ferric cyt c dichroic band at 416-nm shows the same intensity with approximately a 2 nm blue-shift. Conversely, the 414-nm dichroic band of the Ala mutants decreases in intensity, and in the Lys72Asn and Lys73Asn mutants this band is significantly weaker. As illustrated in the figure, the dichroic Soret band of ferric cyt c is characterized by a pronounced couplet, the two components resulting from the coupling of the heme group with different residues in the heme pocket [37, 38]. In particular, the negative component (centered at 416 nm in the wt protein) is considered a probe of the native state, since it decreases, or disappears, as the protein rearranges into a less folded state [39, 40]. Thus, the reduction of the negative component of the Soret CD band is indicative of an altered heme pocket, as a consequence of rearrangements regarding the 70-to-85 loop [28]. In the present study, CD Soret spectra show that the Lys72Arg and Lys73Arg mutants have structure similar to that of the wt protein, while the Lys72Asn and Lys73Asn mutants are the variants that most differ from it. The decreased rotational strength of the 414-nm band observed for the Asn mutants, is likely attributable to the presence of a minor heterogeneous subpopulation with non-native X-Fe-His18 axial coordination (where X is a misligated endogenous ligand) in equilibrium with the major Met80-Fe-His18 form [14, 15, 17]. Upon mutation, also the near-UV (270–300 nm) dichroic band decreases on passing from Arg to Ala to Asn (Fig. S2). In the near-UV wavelength range the signal originates from aromatic amino acids which, for cyt c, are four Tyr, four Phe, and one Trp. The Trp being the one that generates the strongest signal. Thus, the decreased intensity of the CD spectrum shown by the mutants can likely be ascribed to a weaker Trp59-heme propionate H-bonding [41] and an increased distance between the two interacting groups (a slight increase for the Arg mutant and a further increase upon passing from Ala to Asn). Contrary to the behavior previously observed for the Lys → Asn mutants [28], the spectroscopic properties of the Lys → Arg variants do not highlight significant differences with respect to those of native cyt c. Moreover, the CD spectra of the Lys → Ala mutants are intermediate between those of the Lys → Arg and Lys → Asn mutants, consistent with the presence of slight rearrangements in the heme region of cyt c.
UV-Vis electronic absorption spectra of Lys72Arg (red), Lys72Ala (blue), and Lys72Asn (green) mutants compared with wt cyt c (black). The 470–650 nm region and the 650–800 nm are expanded 5-and 20-fold, respectively. The spectra have been shifted along the ordinate axis to allow better visualization. The experimental conditions were 20 mM Hepes + 0.1 mM EDTA, pH 7.0, and 25 °C
Soret CD spectra of the Lys72Arg and Lys73Arg (long dashed lines), Lys72Ala and Lys73Ala (short dashed lines), and Lys72Asn and Lys73Asn (dash with dots) mutants. For each type of mutation, the Lys72 and Lys73 variants show almost identical spectra. The concentration for all mutants was 10 μM. The CD spectrum of wt cyt c (straight line) is reported for comparative purposes. Other experimental conditions as followed for Fig. 1
In agreement with the UV–Vis and CD spectra, the RR spectra in the high frequency region of the Lys72Arg (Fig. 3a) and Lys73Arg (Fig. S3A) mutants are relatively insensitive to the mutations, showing essentially no significant variation with respect to the spectrum of wt cyt c; in contrast, the Lys72Asn and Lys73Asn mutants show an upshift of the core size marker bands. Accordingly, in contrast with the Lys72Asn and Lys73Asn mutants where a downshift of ν8 at 349 cm−1 and an increase in the intensity of the 413 cm−1 band (Fig. 3b, S3B), the latter being a distinctive feature of Lys bound cyt c derivatives, led us to conclude the presence of a small subpopulation with an alternative low spin heme ligation (Lys-Fe-His) [28], the Lys72Arg and Lys73Arg mutants give rise to RR spectra very similar to wt cyt c. Conversely, the Lys72Ala and Lys73Ala mutants display spectral variations similar to those of the Asn mutants, although less marked. In particular, the upshift of the ν2 core size marker band, the slight decrease in intensity of the bands at 568 and 701 cm−1, downshift of ν8 at 349 cm−1, an increase in the intensity of the 413 cm−1 band and decrease of the CT band intensity in the UV–Vis spectra (Figs. 1, S1) may suggest the presence of a small amount of a subpopulation with an alternative low spin Lys-Fe-His ligation [42].
RR spectra in the high (a) and low (b) frequency regions of Lys72Arg (red), Lys72Ala (blue), and Lys72Asn (green) mutants compared with wt cyt c (black). Experimental conditions: 20 mM Hepes + 0.1 mM EDTA, pH 7.0, 25 °C, excitation wavelength 406.7 nm; WT laser power at the sample 5 mW, average of 5 spectra with 25 min integration time (a) and average of 4 spectra with 20 min integration time (b); K72R average of 6 spectra with 30 min integration time (a) and average of 8 spectra with 40 min integration time (b); K72A average of 6 spectra with 30 min integration time (a) and average of 8 spectra with 40 min integration time (b); K72N average of 5 spectra with 25 min integration time (a, b). The intensities are normalized to that of the ν4 band. The spectra have been shifted along the ordinate axis to allow better visualization
Since the spectroscopic characterization indicates that only the Arg mutants show a structure similar to that of the wt protein with the Met80 residue coordinating the heme-Fe atom, the stability of the Arg mutants was investigated by CD following the (Gdm-HCl)-induced protein unfolding process. Unlike the wt protein, but similar to the Lys → Asn variants, the mutants display very similar biphasic unfolding profiles (Fig S4). The first transition is characterized by a c 1/2 = 1.3 M, the second transition by a c 1/2 = 2.5 M. Analysis of data indicates that approximately 12% of the residues constituting the protein α-helical segments undergo unfolding during the first denaturation transition; this corresponds to about five residues, as the total number of residues constituting the helical segments in equine cyt c is 41. Thus, the number of residues unfolding in the (Lys → Arg) mutants is lower than that observed for the (Lys → Asn) variants (16 residues, corresponding to approximately 40% [28]).
Mutant binding to liposomes
As shown above, only the Arg mutants are able to maintain a structure similar to the wt protein with the Met80 bound to the heme-Fe atom; hence, the interaction with CL of only these two mutants was studied. Furthermore, the Ala mutants showed a very low stability in the presence of CL; at the CL:cyt c molar ratio of 4:1, more than the 40% of α-helical content was already lost (data not shown). Figure 4 shows the titration curve of the Lys72Arg and Lys73Arg mutants with large (100 μm) unilamellar CL liposomes. A marked decrease of the 414-nm Cotton effect is observed upon addition of CL to the Lys73Arg mutant, indicating that CL binding alters the heme pocket region. Formation of the mutant cyt c/CL complex occurs via two distinct transitions, as for the wt protein; the progressive growth of a species with non-native heme coordination upon increasing CL concentration was observed. The affinity of CL for the Lys73Arg mutant is comparable to that for the wt protein; at a 6:1 CL/cyt c molar ratio, the 416-nm Cotton effect fully disappears, indicative of non-native heme coordination [7, 40]. The Lys72Arg mutant also reacts with CL, but a higher CL concentration is required to achieve full complexation. This finding is intriguing, since the Lys72Asn mutant does not bind CL [28]. This suggests that the residue located at position 72 reacts with CL liposomes only when positively charged. In particular, while the first binding transition (see Fig. 4) proceeds similar to wt cyt c and the Lys73Arg mutant, the second transition is characterized by a lower affinity of the Lys72Arg mutant for CL. This is confirmed by the kinetic data shown in Fig. 5. At a 6:1 CL/cyt c molar ratio, CL liposomes do not affect the absorption spectrum of Lys72Arg (curve a), while they induce a remarkable change of optical signal in Lys73Arg (curve c). To observe a comparable change in the absorption spectrum of Lys72Arg, a higher CL liposome concentration is necessary (curve b). It should be pointed out that the observed increase of optical density at 408 nm (Fig. 5) should be related to the interaction of CL liposomes with cyt c mutants and indicative of a structurally linked effect. In fact, no time dependent effect is observed in the absence of protein (data not shown). The apparent lack of a signal change when Lys72Arg reacts with 60 μM CL liposomes (curve a of Fig. 5), in spite of the significant extent of interaction (see Fig. 4), indeed suggests that the observed optical signal is to be attributed to the second binding transition (as observed by CD).
Titration curves of CL binding to the Lys72Arg (filled circle) and Lys73Arg (unfilled circle) mutants. Experimental points refer to the change of the 414 nm (416 nm for the wt protein) Cotton effect induced by the stepwise addition of CL to a 10 μM protein solution. Experimental points are averages of at least three measurements. The titration curve of wt cyt c (filled inverted triangle) is reported for comparative purposes. Other experimental conditions as followed for Fig. 1
Kinetics of CL binding to the Lys72Arg and Lys73Arg mutants, followed by the absorption at 408 nm. Kinetic progress curves: 10 μM Lys72Arg mixed with 60 μM CL liposomes (a), 10 μM Lys72Arg mixed with 200 μM CL liposomes (b), 10 μM Lys73Arg mixed with 60 μM CL liposomes (c). Other experimental conditions as followed for Fig. 1
The variation of the α-helical content of the two mutants upon addition of CL liposomes is shown in Fig. S5. The far-UV spectrum of the two mutants (215–250 nm range) is similar to that of the wt protein; upon addition of CL, at a 4:1 CL/protein molar ratio, a decrease of the 222 nm-centered dichroic band is observed corresponding to approximately 20% of the helical content (i.e., 8 residues out of the 41 constituting the α-helical segments). According to the “foldons” theory [43], the 50–54 and the 72–74 α-helical residues are the segments assumed to unfold under the conditions investigated. The mutants maintain this helical conformation up to the 11:1 CL/protein molar ratio. A further decrease of helix content is observed only at the 13:1 CL/protein molar ratio, with loss of approximately 40% of α-helical content, which involves unfolding of the 61-to-68 residues.
Peroxidase activity of the mutants
To estimate the effects of mutation(s) on the cyt c/CL interaction, the steady-state kinetics of the free and CL-bound mutants was investigated. As shown in Table 1, in the absence of CL the two mutants exhibit k cat /K m values for guaiacol higher than the wt protein. This is consistent with a higher exposure of the heme pocket region, a view confirmed by the high accessibility of small molecules (such as hydrogen peroxide) to the metal center. The addition of CL at a 6:1 CL/cyt c molar ratio brings about a strong enhancement of the k cat /K m ratio for both wt cyt c and the Lys73Arg mutant (Fig. S6), which suggests that significant structural changes occur during complex formation; no significant change was instead observed for the CL/Lys72Arg.
Discussion
UV–Vis, CD and RR measurements show that the substitution of the invariant Lys72 and the largely invariant Lys73 with the positively charged residue Arg does not alter significantly the stabilization of the native Met80-Fe(III) axial bond, while their substitution with Asn gives rise to more dramatic changes, inducing misligation in approximately 25–28% of the protein population [7]. An intermediate situation is observed upon replacing the Lys residues with Ala.
However, as previously observed for the Lys → Asn mutants [28], the Lys → Arg mutations have a “long range” effect. The stability measurements reveal that the Lys72Arg and Lys73Arg mutations affect the protein structure; in particular, the rearrangement of the 66–92 region slightly decreases the protein stability. The plot of the (Gdm-HCl)-induced mutant denaturation is biphasic; it shows that approximately 12% of the helical content is lost at the end of the first transition (Fig. S4). Of the 104 residues constituting the cyt c polypeptide chain, 41 residues form the two major (N- and C-terminal) and the three minor α-helical segments. According to the “foldons” theory [43], the 50–54 helix is the segment identified as the one unfolding during the first denaturation transition. The C-terminal helix (residues 88–102) is close to the mutated region; however, the region constituted by the C-terminal and the N-terminal helices is very stable, being the first region to fold and the last to unfold in the folding-unfolding process of cyt c [43, 44].
The discovery that cyt c bound to the mitochondrial membrane plays an important role in determining the cell fate, has stimulated studies on the cyt c-mitochondrial membrane interaction. However, the mechanisms governing the process still remain unclear and the several hypotheses proposed to date have been widely debated [45]. In particular, the rupture of the heme-Fe-M80 coordination and the tertiary rearrangement of cyt c, involving the 40’s Ω loop and the M80-containing Ω loop, have been reported to occur in CL-bound cyt c [7, 20, 21]. The difficulties encountered in the study of the cyt c/CL binding process are correlated with several factors, such as the ionic strength of the solution, the membrane curvature, the CL/cyt c molar ratio at which the binding reaction is followed, which significantly affect the results obtained [7, 17, 19, 45, 46]. Moreover, the finding that CL-bound cyt c is present in solution as a heterogeneous ensemble of forms characterized by different heme coordination represents a further complication [15, 16, 28].
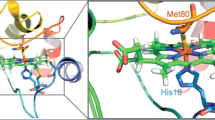
The models proposed to describe the CL/cyt c interaction identified some regions involved in the protein-liposome interaction [5, 6, 17, 18, 26, 47]; among others, the Met80-containing 66–92 region, which comprises the cleft formed by the 67-71 and 82-85 residues and a network of positively charged residues (i.e., Lys72, Lys73, Lys86) that may facilitate the insertion of the acyl chain, is particularly noteworthy [5, 6, 26]. This prompted us to focus our studies on this region, which is considered crucial for the cyt c/CL binding process. Recently, we have shown that replacement of either Lys72 or Lys79 by a Asn residue hinders formation of the cyt c/CL complex [28]. In the wt protein these residues are directed towards the solvent, as shown in Fig. 6 (panel A); therefore, they are considered important in the initial steps of cyt c/CL recognition. Since both residues are positively charged at neutral pH, the critical role of positive charges at positions 72 and 79 in the protein/CL binding reaction has been investigated. Unfortunately, the Lys79Arg mutant could not be characterized as the protein does not fold; therefore, only the CL-dependent properties of Lys72Arg and Lys73Arg variants have been studied. The overall reaction of the Lys73Arg mutant with CL is very similar to that observed for the wt protein and the Lys73Asn mutant [28]; this is in line with the hypothesis that Lys73 is not involved in the initial cyt c/CL recognition process. Of note, Lys73 is characterized by an opposite orientation with respect to Lys72 and Lys79 and is anchored to Glu66 and Glu69 by salt bridges (Fig. 6, panel a). The behavior of the Lys72Arg mutant is instead more interesting; unlike the Lys72Asn mutant which did not interact with CL [28], the binding reaction of Lys72Arg with the lipid is a two-step process, as observed for the Lys73Arg mutant and the wt protein. This suggests that the protein-lipid interaction requires positively charged residues at position 72. Indeed, the model of the cyt c/CL complex proposed by Sinibaldi et al. [26] is compatible with a role of Lys72 and Lys79 in the interaction with the negatively charged CL phosphate groups (Fig. 6, panel b). Nevertheless, the nature of the side chain is equally important as it affects the affinity of the protein for CL (Fig. 4). By comparing the present data with those of the Lys → Asn mutants, it appears clear that Lys72 (or a non-Lys positively charged residue at this position) and Lys79 [28] are critical to achieve the protein/CL interaction; CL binding does not occur if residues at positions 72 and 79 are uncharged [28]. On the other hand, the affinity of CL for cyt c decreases if the positively charged residue at position 72 is not a lysine; the protein/lipid reaction is similar for wt cyt c, Lys72Arg and Lys73Arg mutants during the first transition, whereas the Lys72Arg mutant behaves differently during the second transition (Fig. 4). This suggests that replacement of Lys72 and Lys73 by another positively charged residue does not influence the initial protein/lipid recognition process, but strongly affects the process associated with the acyl chain insertion into the protein. The decreased affinity of CL for the Lys72Arg mutant suggests that the structural alterations induced in the 71–85 Ω-loop region by complex formation, hinder insertion of the CL acyl chain.
Schematic representation of the three-dimensional structure of horse heart cyt c in complex with CL (as proposed in [26]). a Overall view of the complex showing the location of Lys72, Lys73 (and interacting acidic residues Glu66 and Glu69), and Lys79. b Detailed view of the interactions of Lys72 and Lys79 with the CL phosphate and carbonyl groups. For clarity CL and heme carbon atoms are colored in green and yellow, respectively
On the basis of the above considerations, we propose the following model for the cyt c/CL binding process: in the first step, the positively charged Lys72 and Lys79 facilitate protein-lipid recognition by electrostatic interaction with the deprotonated, negatively charged phosphate groups located on the liposome surface. This view is supported by the recent finding that both the phosphate groups of CL are negatively charged at neutral pH [48]. During this step the protein properly orients on the liposome surface to facilitate the successive step, consisting of a tight protein-lipid binding characterized by the insertion of one (or two [26]) CL acyl chain into the protein interior.
Rearrangements in the heme pocket region begin to occur even at low CL/cyt c molar ratios (Fig. 4). This is in line with the “foldons” theory, which asserts that the heme crevice is one of the first regions to unfold [39]. Conversely, the α-helix structure remains unchanged during the first transition (which is complete at approximately 3:1 CL/protein molar ratio), and slightly decreases, by approximately 20%, during the second transition (which proceeds up to 11:1 CL/protein molar ratio). Only at CL/protein molar ratio higher than 13:1 (conditions not considered in the present study) the decrease of helix content becomes approximately 40%. Consequently, in this study CL-bound cyt c possesses a compact conformation [14, 15]. The present data agree well with previously reported models for the yeast protein [6, 9]; such models associated insertion of the CL acyl chain into cyt c with rearrangements occurring in the 66–92 region of the protein, characterized by the rupture of hydrogen bonds and/or perturbation of electrostatic interactions. Analysis of the horse cyt c structure (PDB codes 1AKK and 1OCD) shows that the Arg91 residue interacts with the carboxyl group of Glu69 and with the sulfur atom/carbonyl group of Met65. Furthermore, (i) Asn70 forms hydrogen bonds with the carbonyl group of Glu66 and the side chain amine group of Lys86, and (ii) Leu68 backbone amide group is hydrogen bonded to the backbone carbonyl groups of Met65 and Leu64. All these interactions are expected to be perturbed upon the insertion of the CL acyl chain into the hydrophobic cleft surrounding Met80 (whose walls are formed by residues Leu64, Tyr67, Leu68, Pro71, Phe82, Leu94) with consequent structural rearrangements in this region, leading to the rupture of the Met80-Fe coordination and to the (above mentioned) partial unfolding of the 66–92 segment.
Unlike wt cyt c, the Lys72Arg and Lys73Arg mutants show peroxidase activity in the absence of CL. After addition of the lipid (CL/protein molar ratio of 6:1), the peroxidase activity of Lys73Arg significantly increases (Table 1), whereas that of Lys72Arg remains essentially unchanged. It is evident from the titration curves (Fig. 4) that at the CL/Lys72Arg molar ratio of 6:1 the second binding transition has not yet started; this suggests that the increased peroxidase activity of CL-bound cyt c derives from the insertion of the lipid acyl chain into the protein.
On the basis of the present and previous work a number of overall conclusions can be drawn: (i) Lys72 and Lys79 are crucial for the cyt c/CL complex formation [28]; (ii) position 72 must be occupied by a positively charged residue to assure cyt c/CL recognition; (iii) non-lysine positively charged residues located at position 72 allow cyt c to react with CL; (iv) the replacement of Lys72 by Arg weakens the second (i.e., low-affinity) transition, i.e., the insertion of the CL acyl chain into the protein; (v) the Lys73Arg mutation strongly increases the peroxidase activity of the CL-bound protein; (vi) insertion of the CL acyl chain into cyt c enhances the peroxidase activity of the cyt c/CL complex.
In conclusion, present results indicate that the Lys72 and Lys73 residues play a crucial role in the stability of cyt c and the apoptotic process catalysing CL peroxidation.
Abbreviations
- cyt c :
-
Cytochrome c
- CD:
-
Circular dichroism
- CL:
-
Cardiolipin
- RR:
-
Resonance Raman
References
Garrido G, Galluzi L, Brunet M, Puig PE, Didelot C, Kroemer G (2006) Cell Death Diff 13:1423–1433
Santucci R, Sinibaldi F, Patriarca A, Santucci D, Fiorucci L (2010) Exp Rev Proteom 7:507–517
Hüttermann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, Doan JW, Lee I (2011) Mitochondrion 11:369–381
Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG (2005) Nat Chem Biol 1:223–232
Rytömaa M, Kinnunen PK (1995) J Biol Chem 270:3197–31202
Kalanxhi E, Wallace CJA (2007) Biochem J 407:179–187
Sinibaldi F, Fiorucci L, Patriarca A, Lauceri M, Ferri T, Coletta M, Santucci R (2008) Biochemistry 47:6928–6935
Schug ZT, Gottlieb E (2009) Biochim Biophys Acta 1788:2022–2031
Rajagopal BS, Silkstone GG, Nicholls P, Wilson MT, Worrall GA (2012) Biochim Biophys Acta 1817:780–791
Santucci R, Sinibaldi F, Polticelli F, Fiorucci L (2014) Curr Med Chem 21:2702–2714
Ott M, Zhivotovsky B, Orrenius S (2007) Cell Death Diff 14:1243–1247
Caroppi P, Sinibaldi F, Fiorucci L, Santucci R (2009) Curr Med Chem 16:4058–4065
Zou H, Li Y, Liu X, Wang X (1999) J Biol Chem 274:11549–11556
Hong Y, Muenzner J, Grimm SK, Pletneva EV (2012) J Am Chem Soc 134:18713–18723
Hanske J, Toffey JR, Morenz AM, Bonill AJ, Schiavoni KH, Pletneva EV (2012) Proc Natl Acad Sci 109:125–130
Muenzner J, Toffey JR, Hong Y, Pletneva EV (2013) J Phys Chem B 117:12878–12886
Muenzner J, Pletneva EV (2014) Chem Phys Lipids 179:57–63
Pandiscia LA, Schweitzer-Stenner R (2015) J Phys Chem B 119:1334–1349
Pandiscia LA, Schweitzer-Stenner R (2015) J Phys Chem B 119:12846–12859
Nantes IL, Kawai C, Pessoto FS, Mugnol KC (2010) Methods Mol Biol 606:147–165
Godoy LC, Muñoz-Pinedo C, Castro L, Cardaci S, Schonhoff CM, King M, Tortora V, Marin M, Miao Q, Jiang JF, Kapralov A, Jemmerson R, Silkstone GG, Patel JN, Evans JE, Wilson MT, Green DR, Kagan VE, Radi R, Mannick JB (2009) Proc Natl Acad Sci 106:2653–2658
Basova LV, Kurnikov IV, Wang L, Ritov VB, Belikova NA, Vlasova II, Pacheco AA, Winnica DE, Peterson J, Bayir H, Waldeck DH, Kagan VE (2007) Biochemistry 46:3423–3434
Vladimirov YA, Proskurnina EV, Izmailov DY, Novikov AA, Brusnichkin AV, Osipo AN, Kagan VE (2006) Biochemistry (Moscow) 71:998–1005
Rytömaa M, Kinnunen PKJ (1994) J Biol Chem 269:1770–1774
Tuominem EK, Wallace CJ, Clark-Lewis I, Craig DB, Rytömaa M, Kinnunen PK (2002) J Biol Chem 277:8822–8826
Sinibaldi F, Howes BD, Piro MC, Polticelli F, Bombelli C, Ferri T, Coletta M, Smulevich G, Santucci R (2010) J Biol Inorg Chem 15:689–700
Schlame M, Ren M (2009) Biochim Biophys Acta 1788:2080–2083
Sinibaldi F, Howes BD, Droghetti E, Polticelli F, Piro MC, Di Pierro D, Fiorucci L, Coletta M, Smulevich G, Santucci R (2013) Biochemistry 52:4578–4588
Patel CN, Lind MC, Pielak GJ (2001) Protein Expr Purif 22:220–224
Kluck LM, Ellerby HM, Najem S, Yaffe MP, Margoliash E, Bredesen D, Mauk AG, Sherman F, Newmeyer DD (2000) J Biol Chem 275:16127–16133
Sinibaldi F, Mei G, Polticelli F, Piro MC, Howes BD, Smulevich G, Santucci R, Ascoli F, Fiorucci L (2005) Protein Sci 14:1049–1058
Sinibaldi F, Droghetti E, Polticelli F, Piro MC, Di Pierro D, Ferri T, Smulevich G, Santucci R (2011) J Inorg Biochem 105:1365–1372
Bergstrom CL, Beales PA, Lv Y, Vanderlick TK, Groves JV (2013) Proc Natl Acad Sci 110:6269–6274
Sinibaldi F, Howes BD, Piro MC, Caroppi P, Mei G, Ascoli F, Smulevich G, Santucci R (2006) J Biol Inorg Chem 11:52–62
Sinibaldi F, Piro MC, Howes BD, Smulevich G, Ascoli F, Santucci R (2003) Biochemistry 42:7604–7610
Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG (1998) Biochemistry 37:6124–6131
Schweitzer-Stenner R (2008) Phys Chem B 112:10358–10366
Schweitzer-Stenner R (2014) New J Sci 2014:484538
Pielak GJ, Oikawa K, Mauk AG, Smith M, Kay CM (1986) J Am Chem Soc 108:2724–2727
Santucci R, Ascoli F (1997) J Inorg Biochem 68:211–214
Moore GR, Pettigrew GW (1990) Cytochromes c. In: Evolutionary, structural and physicochemical aspects, Chapter 4. Springer-Verlag Eds, Berlin, Heidelberg
Döpner S, Hildebrandt P, Rosell FI, Mauk AG (1998) J Am Chem Soc 120:11246–11255
Maity H, Maity M, Krishna MM, Mayne L, Englander SW (2005) Proc Natl Acad Sci 102:4741–4746
Krishna MM, Englander SW (2005) Proc Natl Acad Sci 102:1053–1058
Hannibal L, Tomasina F, Capdevila DA, Demicheli V, Tortora V, Alvarez-Paggi D, Jemmerson R, Murgida DH, Radi R (2015) Biochemistry 55:407–428
O’Brien ES, Nucci NV, Fuglestad B, Tommos C, Wand AJ (2015) J Biol Chem 290:30879–30887
Kawai C, Prado FM, Nunes GLC, Di Mascio P, Carmona-Ribeiro AM, Nantes IL (2005) J Biol Chem 280:34709–34717
Malyshka D, Pandiscia LA, Schweitzer-Stenner R (2014) Vib Spectro 75:86–92
Acknowledgements
This work was supported by Ente Cassa Risparmio di Firenze (Grant Nr. 2014-0100 to G.S.).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sinibaldi, F., Milazzo, L., Howes, B.D. et al. The key role played by charge in the interaction of cytochrome c with cardiolipin. J Biol Inorg Chem 22, 19–29 (2017). https://doi.org/10.1007/s00775-016-1404-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-016-1404-5