Abstract
Nonparametric estimation of cumulative distribution function and probability density function of continuous random variables is a basic and central problem in probability theory and statistics. Although many methods such as kernel density estimation have been presented, it is still quite a challenging problem to be addressed to researchers. In this paper, we proposed a new method of spline regression, in which the spline function could consist of totally different types of functions for each segment with the result of Monte Carlo simulation. Based on the new spline regression, a new method to estimate the distribution and density function was provided, which showed significant advantages over the existing methods in the numerical experiments. Finally, the density function estimation of high dimensional random variables was discussed. It has shown the potential to apply the method in classification and regression models.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Estimation of cumulative distribution function (CDF) and probability density function (PDF) to random variables is a classical and basic problem in statistics, which is essential to describe some random phenomena and has significant application in signal processing [1], pattern recognition [2], machine learning [3] and so on. With the known distribution of the continuous random variable, such as Gaussian, Rayleigh, log-normal or exponential distribution, CDF and PDF can be estimated with the maximum likelihood estimation and Bayes estimation [4]. But nonparametric approach will be employed here if the distribution is not well assumed.
To estimate PDF more exactly in nonparametric approach is still a challenging problem to researchers. As the most widely used method, kernel density estimation is proposed by Rosenblatt [5] in 1956 and Parzen [6] in 1962. Many discussions have been performed to further implement such method via optimize the kernel function and bandwidth, e.g., based on the normal distribution, normal scale rules is proposed by Silverman [7] to determine the best bandwidth; Over smoothed bandwidth selection rules from Terrell [8] is more flexibility and larger application; Alexandre [9] provided iterative algorithm used when solve the equation and Plug-In estimator to give the best bandwidth corresponding to the least mean integrated square error. For the large samples with high complexity, fast Parzen density estimation by Jeon and Landgrebe [10], weighted Parzen window by Babich and Camps [11], optimally condensed data samples by Girolami and He [12], etc. are based on the subset of the large sample to reduce the running time without reduce the accuracy. What’s more, some other approaches have also been proposed to estimate PDF. Such as the sum of gamma densities [13] or a sum of exponential random variables [14] was used to substitute the kernel function to express the PDF in different fields, and orthogonal series [15], Haar’s series [16], wavelets [17]. Recently, some methods based on characteristic function [18, 19] were presented. However, all these methods are based on the series to express the PDF, in which the complexity of function would be increased with the sample size. And the accuracy of estimation was determined by the form of series, which meant one method was just suitable for some certain distributions. However, prior the process of estimation, there are little information available for us to the distribution, it will be very hard to have the proper series introduced in the estimation.
Spine functions have been widely applied in interpolation [20], smoothing of observed data [21], regression [22] and PDF estimation [23,24,25]. Inspired with the characteristics of spline function, which is a continuous function piecewise-defined by polynomial functions and possesses a high degree of smoothness where the pieces connect, and to overcome such shortness, a new method to estimate CDF or PDF is introduced here in this paper. Spline Not as previous methods, in our proposed method of spline regression, the spline function was not always defined by polynomial functions or B-splines, but could be set freely and consisted of totally different types of functions in each segment. With the method here, a new method to estimate CDF and PDF was introduced, which showed advantages in these aspects:
- 1.
The PDF is expressed by piecewise functions instead of series. The estimated accuracy increases with the sample size, but the complexity of function does not increase.
- 2.
The method is suitable for most types of continuous distributions, and the form of spline function and other parameters does not need to be updated unless the distribution is quite special.
- 3.
The estimation is accurate for most types of distributions and is superior to kernel density estimation.
- 4.
The PDF is always smooth and is not influenced by parameters.
- 5.
The values of estimated CDF are less than 1, positive and monotone increasing. The values of estimated PDF are positive and the integration of PDF is about 1.
- 6.
It is easy to find a subset from the large sample to reduce the running time and get similar accuracy simultaneously.
The paper is organized as below in the following sections, the new spline regression is introduced first, and then the application of proposed approach is described in the estimation of CDF and PDF. After that, comprehensive numerical experiments with Monte-Carlo simulation were made to illustrate the characteristic and advantage. At last, the PDF estimation of high dimensional random variables is discussed, and its potential application in classification and regression models is presented.
2 Method
Let \(F\left( x \right)\) and \(f\left( x \right)\) denote the CDF and PDF of random variable X, respectively, \(y\left( x \right)\) and \(y^{'\left( x \right)}\) denote the estimated CDF and PDF, respectively.
With ascending sorted samples from random variable X,
the CDF of \(x_{i}\) is \(F\left( {x_{i} } \right) = P\left( {X \le x_{i} } \right) \approx i/\left( {n + 1} \right)\), which means the probability of event \(\left\{ {X \le x_{i} } \right\}\) is almost to be \(i/\left( {n + 1} \right)\). If we let \(y_{i} = i/\left( {n + 1} \right)\), the data points \((x_{i} ,y_{i} ) i = 1 \cdots n\) can be fitted with spline regression. Then the PDF of \(x_{i}\) can be estimated with the one order deviation of \(y\left( x \right)\), noted as \(y^{'\left( x \right)}\). Instead of spline interpolation, spline regression is used in the paper, which means that \(F\left( {x_{i} } \right)\) is not always equal to \(y_{i}\).
To avoid the large error, which is resulted from the process of derivation, transformation of random variables is employed here in the paper.
2.1 Spline Regression
Inspired with the characteristics of spline function, A new method of spline regression is introduced here, in which the spline function can be set freely and the basis functions may be totally different for each segment.
The spline function is defined as
There are u segments in this function. For each segment to the interval \(x \in \left[ {s_{i} ,s_{i + 1} } \right] , i = 1,2, \ldots ,u\), v basis functions \(\varphi_{i1} \left( x \right), \ldots ,\varphi_{iv} \left( x \right)\) are set here, which are smooth for each segment and with nonzero derivate for each knot \(s_{2} ,s_{3} , \ldots ,s_{u}\) for their any order derivative. With the request of smoothness to the spline function, the following constrained conditions are introduced here:
where \(\varphi_{ij}^{\left( k \right)} \left( x \right)\) is the kth order derivative of \(\varphi_{ij} \left( x \right)\).
For these \(u \cdot v\) parameters \(a_{11} , \ldots ,a_{uv}\) in the spline function, \(\left( {u - 1} \right) \cdot \left( {v - 1} \right)\) linear constrained equations should be met, which means that there are \(u + v - 1\) free variables in total, noted as \(I_{1} ,I_{2} , \ldots ,I_{u + v - 1}\). As equation set here is homogeneous linear equations, based on the form of solutions, all \(a_{ij}\) can be rewrote as
The spline function will be available if we can get the values of all \(b_{ijk}\) and \(I_{k}\), which will be basis to the value of \(a_{ij}\).
- (1)
Values of \(b_{ijk}\)
After transposition of the constrained equation set, we can get
in which \(i = 1,2, \ldots ,u - 1\).
Take note that if we know the values of \(a_{11} ,a_{12} , \ldots ,a_{1v} ,a_{21} ,a_{31} , \ldots ,a_{u1}\), all \(a_{ij}\) will be derived accordingly with Eq. (4), all these \(u + v - 1\) variables \(a_{11} ,a_{12} , \ldots ,a_{1v} ,a_{21} ,a_{31} , \ldots ,a_{u1}\) will be set as free variables.
If some \(I_{k} = 1\) and all others are 0 for the equation \(a_{ij} = \sum\nolimits_{k = 1}^{u + v - 1} {b_{ijk} I_{k} }\), \(a_{ij} = b_{ijk}\). \(b_{ijk}\) is derived as below: let one of \(a_{11} ,a_{12} , \ldots ,a_{1v} ,a_{21} ,a_{31} , \ldots ,a_{u1}\) be 1 and the others are 0, substitute them into Eq. (4), we can get all other \(a_{ij}\) and all \(b_{ijk}\).
- (2)
Values of \(I_{k}\)
For each \(x_{i} \in \left[ {s_{k} ,s_{k + 1} } \right]\),
Let
Substitute it into the above equation, and then \(y\left( {x_{i} } \right)\) can be represented as
According to the constrained equations, the v − 2 order derivative of spline function is continuous, but the v − 1 order derivative is not continuous. Nevertheless, it is hoped there is stronger smoothness for the spline function, with De Boor’s smoothing spline [26] and define as
Let
Then
We should minimize both the value of G and the sum of squared residues, so we define
where σ is a parameter called smooth factor that we should set.
Based on the least square method,
After transposition
Update that into matrix form, it can be rewrote to
where \(B = \left( {B_{im} } \right)_{{n \times \left( {u + v - 1} \right)}}\), \(A = \left( {A_{im} } \right)_{{n \times \left( {u + v - 1} \right)}}\), n is the sample size, σ is smooth factor, \(I = \left( {\begin{array}{*{20}c} {I_{1} } & {I_{2} } & \cdots & {I_{u + v - 1} } \\ \end{array} } \right)^{\text{T}}\) and \(Y = \left( {\begin{array}{*{20}c} {y_{1} } & {y_{2} } & \cdots & {y_{n} } \\ \end{array} } \right)^{\text{T}}\).
The value of \(I_{k}\) can be derived with the following steps:
- 1.
For each \(x_{i},\) calculate the values of \(B_{im} = \mathop \sum \nolimits_{j = 1}^{v} b_{kjm} \varphi_{kj} \left( {x_{i} } \right)\) and \(A_{im} = \mathop \sum \nolimits_{j = 1}^{v} b_{kjm} \varphi_{kj}^{{\left( {v - 1} \right)}} \left( {x_{i} } \right),\)\(i = 1, \ldots ,n\quad m = 1, \ldots ,u + v - 1.\)
- 2.
Set appropriate smooth factor σ and solute the equation \(\left( {B^{\text{T}} B + \sigma /n \cdot A^{\text{T}} A} \right)I = B^{\text{T}} Y\), values of \(I_{1} ,I_{2} , \ldots ,I_{u + v - 1}\) will be derived accordingly.
- 3.
All parameters of the spline function can be derived with \(a_{ij} = \mathop \sum \nolimits_{k = 1}^{u + v - 1} b_{ijk} I_{k}\).
Take note that we suppose all matrices above are full rank. If some matrix is not full rank, another group of basis functions or knots will be used.
2.2 Transformation of Random Variable X
In the case of that some CDFs are not easily estimated with general spline function, such as, \(F\left( x \right) = \sqrt x ,x \in \left[ {0,1} \right]\), in which \(\mathop {\lim }\limits_{x \to 0} f\left( x \right) = + \infty\). CDF cannot be estimated with polynomial spline function to fit the data. However, if we set \(\hat{x} = { \ln }x\), then \(F\left( x \right) = e^{{\hat{x}/2}} ,\hat{x} \in \left( {0, + \infty } \right)\), which is much easier to be estimated.
For random variable X, set \(\hat{X} = \psi \left( X \right)\) in which ψ is a monotone increasing and analytic function, and let \(\hat{F}\left( {\hat{x}} \right)\) and \(\hat{f}\left( {\hat{x}} \right)\) are the distribution function and density function of \(\hat{X}\), respectively.
Then
Using spline function to fit the data points \(\left( {\hat{x}_{i} ,y_{i} } \right)i = 1 \ldots n\) in which \(\hat{x}_{i} = \psi \left( {x_{i} } \right)\), we can get \(\hat{F}\left( {\hat{x}} \right)\) and \(\hat{f}\left( {\hat{x}} \right)\), and then we can get \(F\left( x \right)\) and \(f\left( x \right)\) based on the above equations.
In this paper, we transformed the random variables based on the following steps:
For ordered samples: \(x_{1} ,x_{2} , \ldots ,x_{n} \left( {x_{1} \le x_{2} \le \cdots \le x_{n} } \right)\), noted the the a quantile as \(x_{an}\).
Step 1:
If \(\frac{{x_{0.02n} - x_{1} }}{{x_{0.2n} - x_{1} }} < 0.02\) and \(\frac{{x_{n} - x_{0.98n} }}{{x_{n} - x_{0.8n} }} \ge 0.02\), \(\psi_{1} = { \ln }\left( {x - x_{1} } \right)\).
If \(\frac{{x_{0.02n} - x_{1} }}{{x_{0.2n} - x_{1} }} \ge 0.02\) and \(\frac{{x_{n} - x_{0.98n} }}{{x_{n} - x_{0.8n} }} < 0.02\), \(\psi_{1} = - { \ln }\left( {x_{n} - x} \right)\).
If \(\frac{{x_{0.02n} - x_{1} }}{{x_{0.2n} - x_{1} }} \le 0.02\) and \(\frac{{x_{n} - x_{0.98n} }}{{x_{n} - x_{0.8n} }} \le 0.02\), \(\psi_{1} = { \ln }\left( {x - x_{1} } \right) - { \ln }\left( {x_{n} - x} \right)\).
And in else situation, we do not transform the random variable.
Step 2:
If \(\frac{{x_{0.05n} - x_{1} }}{{x_{0.5n} - x_{0.05n} }} > 1\) and \(\frac{{x_{n} - x_{0.95n} }}{{x_{0.95n} - x_{0.5n} }} > 1,\)\(\psi_{2} = \ln \left( {cx - cx_{0.5n} + \sqrt {1 + \left( {cx - cx_{0.5n} } \right)^{2} } } \right).\)
If \(\frac{{x_{0.05n} - x_{1} }}{{x_{0.5n} - x_{0.05n} }} > 1\) and \(\frac{{x_{n} - x_{0.95n} }}{{x_{0.95n} - x_{0.5n} }} \le 1,\)\(\psi_{2} = - { \ln }\left( {1 + cx_{n} - cx} \right).\)
If \(\frac{{x_{0.05n} - x_{1} }}{{x_{0.5n} - x_{0.05n} }} \le 1\) and \(\frac{{x_{n} - x_{0.95n} }}{{x_{0.95n} - x_{0.5n} }} > 1,\)\(\psi_{2} = { \ln }\left( {1 + cx - cx_{1} } \right).\)
Do the transformations again and again until \(\frac{{x_{0.05n} - x_{1} }}{{x_{0.5n} - x_{0.05n} }} \le 1\) and \(\frac{{x_{n} - x_{0.95n} }}{{x_{0.95n} - x_{0.5n} }} \le 1\).
where c is the value that makes \(Q = \mathop \sum \limits_{i = 1}^{n} \left[ {y_{i} - y\left( {x_{i} } \right)} \right]^{2}\) get the minimum.
Step 3:
In all situations, do the transformation \(\psi_{3} = \frac{{5\left( {x - x_{0.5n} } \right)}}{{x_{0.95n} - x_{0.05n} }}\).
After the three steps, most distributions can be estimated by the spline function. Take note that these transformations focus on the discontinuity of the two ends, but if the discontinuity is in the middle, we should separate the samples to several parts and take the spline regression for each part.
2.3 Spline Function
To define the spline function, basis functions can be set as below:
With such predefined basis functions, segments from the middle are quartic spline function, but in the first and last segments, the special basis functions are employed to describe the asymptotic approximation of the distribution function.
The knots \(s_{2} ,s_{3} , \ldots ,s_{u}\) are set as the 0.05, 0.23, 0.41, 0.59, 0.77, 0.95 quantile of \(x_{1} ,x_{2} , \ldots ,x_{n} ,\) respectively. If knot s is the a quantile of \(x_{1} ,x_{2} , \ldots ,x_{n}\), then \(s = x_{k}\) with k is as the approximate number of \(a \cdot n\). with such assumptions above, the first and last segment cover 5% of all sample and the other segments cover 18%, respectively.
Here, we assumed that u = 7 segments in all, and v = 5 basis functions in each segment, it can easily be obtained that the third order derivative of these functions are still continuous. The value of u may be greater than 7 for the cases of that the distribution function is more complex or the sample size is very large, However, the complexity of function will not increase with the sample size when we take any other parameters, which is quite different from most methods.
As an important parameter, the smooth factor σ will influence the performance of estimation greatly. The proper value of σ to different distribution and different sample size will be discussed in the following sections.
2.4 Adjustment of the Spline Function
With the definition or requirement to CDF, which should not be more than 1, be positive or monotone increasing, and PDF may take negative value, some constrained requirements/conditions should be introduced when we try to estimate CDF or PDF with spline regression, which may lead to much more complex in the calculation. In this paper, a simple method is introduced to resolve such problem by adjusting the spline function after regression.
In most cases, only the first and last segments of the spline function are required be adjusted. In the first segment, the constrained conditions are
with v unknowns in these v − 1 equations, and only one free variable. For simple, a11 is set as the free variable, and Eqs. (12) can be updated as
Values of a1j, \(j = 2, \ldots ,v\) can be derived based on the initial set \(a_{11}\), different preset value of \(a_{11}\) will be repeated until we got the reasonable estimated CDF and PDF based on all samples calculated by the spline function. The last segment is adjusted in the same approach. In some cases of that the reasonable result is not available via one free value, the constrained conditions should be reduced to have two free values introduced.
The algorithm to estimate of probability distribution with spline regression model is summarized as Algorithm 1.

2.5 Method Evaluation and Comparison
For these 40 distributions (Table 1) with different types or parameters, the characteristics of these most widely used statistics has been considered to evaluate the performance of estimated CDF or PDF. However, with the increase of sample size, integrated square error (ISE), mean absolute error (MAE) and mean square error (MSE) are not valid statistics to evaluate the estimate PDF. For the example of distribution \(F\left( x \right) = \sqrt[3]{x}\,{\text{and}}\,f\left( x \right) = \frac{1}{3}x^{{ - \frac{2}{3}}} ,x \in \left( {0,1} \right)\), sample data \(x_{i} = \left( {i/n} \right)^{3} i = 1, \ldots ,n - 1\), and estimated PDF \(y^{\prime}\left( x \right) = \left\{ {\begin{array}{*{20}c} {\frac{1.01}{3}x^{{ - \frac{2}{3}}} ,x \in \left( {0,\frac{1}{8}} \right]} \\ {\frac{0.99}{3}x^{{ - \frac{2}{3}}} ,x \in \left( {\frac{1}{8},1} \right)} \\ \end{array} } \right.\) when error is 1%,
then \({\text{IAE}} \to 0.01, {\text{ISE}} \to + \infty , {\text{MAE}} \to + \infty , {\text{MSE}} \to + \infty\) when \(n \to \infty\). Integrated absolute error (IAE) is used as the statistics to evaluate the estimated PDF. Similarly, IAE and ISE are not convergent with the increase of sample data, root mean square error \({\text{RMSE}} = \sqrt {\frac{1}{n}\mathop \sum \nolimits_{i = 1}^{n} \left( {y\left( {x_{i} } \right) - F\left( {x_{i} } \right)} \right)^{2} }\) is to evaluate the estimated CDF.
And
For example, if random variable X follows the distribution:
is a set of samples taken from random variable X.
In the case of estimated error is in 1%, and the estimated PDF is \(y^{\prime}\left( x \right) = \left\{ {\begin{array}{*{20}c} {\frac{1.01}{3}x^{{ - \frac{2}{3}}} ,x \in \left( {0,\frac{1}{8}} \right]} \\ {\frac{0.99}{3}x^{{ - \frac{2}{3}}} ,x \in \left( {\frac{1}{8},1} \right)} \\ \end{array} } \right.\), for each different kind of statistics to evaluate the performance of PDF estimation as below.
When \({\text{IAE}} \to 0.01, {\text{ISE}} \to + \infty , {\text{MAE}} \to + \infty , {\text{MSE}} \to + \infty\), only IAE is a valid statistics to evaluate the estimation. Similarly, IAE and ISE are not suitable to evaluate the performance of estimated CDF due to that they are not always convergent.
3 Result
3.1 Basis Functions
For distributions normal (0,1), exponential (1) and Rayleigh (1), 1000 random samples were generated with Monte-Carlo simulation. Six different sets of basis functions as below were employed in the estimation of CDF and PDF.
E1:
E2:
E3:
E4:
E5:
E6:
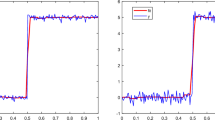
With results in Table 2, we can see the RMSECDF is not sensitive to the basis functions but IAEPDF is influenced by the basis functions significantly. Based on Fig. 1, we can also find that the two ends of the distribution are usually hard to be estimated but basis function of E1 shows the best estimation and successfully describes the asymptotic approximation, which has significant advantage over pure polynomial spline function (E4 and E6). Then we use E1 as the basis functions, which has been mentioned in Sect. 2.3.
3.2 Smooth Factor
With Monte Carlo simulation, IAEs for each batch of generated random samples were repeated for 50 times and averaged to evaluate the estimated PDF. In Fig. 2a, with the increase of smooth factor \(\sigma\), the averaged IAEs for most distributions decreases at first and increases after getting the minimum, but the averaged IAE to uniform distribution is in decreases with the increase of smooth factor \(\sigma\). The minimum averaged IAE is for different values to smooth factor \(\sigma\). and when σ = 2, the averaged IAE to most distributions is in the nearby area of minimum averaged IAE. In Fig. 2b, σ corresponding to the minimum averaged IAE is identical with different sample sizes, and with the increase of sample size, the averaged IAE is in smaller with the increase number of simulated sample. The minimum value of averaged IAEs are in the nearby area of σ = 2. Then σ = 2 is chosen as the optimal smooth factor for each different distribution and different sample size.
3.3 Evaluation for Well-Proportioned Samples
In the extreme case of well-proportioned samples \(x_{1} ,x_{2} , \ldots ,x_{n} \left( {x_{1} \le x_{2} \le \cdots \le x_{n} } \right)\), which means \(F\left( {x_{i} } \right) = y_{i} = i/\left( {n + 1} \right)\) can be well estimated. With Monte Carlo simulation, all these 40 distributions from Sect. 2.5 were evaluated using this well-proportioned sample. As Fig. 3, RMSEs to these estimated CDFs are all smaller than 0.00016 and IAEs to these estimated PDFs are all smaller than 0.008, which means that the proposed method estimated most distributions well.
3.4 Evaluation for Random Samples
With Monte Carlo simulation, for each distribution from Sect. 2.5, RMSE and IAE for each batch of generated random samples were repeated for 50 times and averaged to evaluate the estimated CDF and PDF. Each batch random samples were constructed in the steps as below:
- (1)
Sort the sampled n random samples from standard uniform distribution \(U\left( {0,1} \right)\).
- (2)
Calculate F−1(x) (inverse function of the distribution function) with these n samples, as the random samples for each distribution.
As the CDF is follow the standard unit distribution \(U\left( {0,1} \right)\), for each set of random samples,
is to evaluate the deviation from distribution of these samples, which can also be seen as the error of empirical distribution function with \(i/\left( {n + 1} \right)\) as the estimation of \(F\left( {x_{i} } \right)\). In Fig. 4, RMSErand for our proposed method is almost always smaller than RMSErand for any distribution, which indicates that our method is superior to empirical distribution function.
In each set of random samples, red dots are the RMSErand for each set of random numbers generated by step (1). Corresponding series of random samples to each set of random sample from step (1) is generated for each different distribution by step (2). RMSE to each estimated CDF was represented by black dots. a The sample size is 300. b The sample size is 1000
In PDF estimation, compared to kernel density estimation in Fig. 5 and Table 3, our proposed method is superior in the estimation of PDF for any distribution. Most of the IAE is in the range of 0.05–0.06 on average, while kernel density estimation is generated larger error than the proposed method. In Fig. 6, both normal distribution and Rayleigh distribution can be well estimated with both kernel density estimation with normal kernel function and spline regression, but spline regression is much more accuracy than kernel density estimation with normal kernel function in the estimation of PDF for all of these distributions.
3.5 Large Sample Size
With the much large sample size, CDF and PDF are estimated by a subset of the sample. With Monte Carlo simulation, the similar accuracy is for both our method for the subset sample and the full sample. In the simulated samples, the subset sample is obtained via the every 100 data for this ascending sorted sample when the sample size is n = 100,000, then the subset sample of these 1000 observations are used to estimate CDF and PDF. With Table 4, the RMSE to estimated CDF and IAE to estimated PDF are quite similar between the full sample and the subset samples.
4 Discussion
In this section, the PDF estimation to high dimensional random variables and the application in classification and regression models will be discussed.
4.1 Probability Distribution of n Dimensional Random Variables
It is almost impossible to estimate the joint probability distribution of n dimensional random variables by limited number of samples because of the curse of dimensionality, but the problem can be simplified as the linear correlations of every variables which will be a rough but quite practical approach in the estimation.
To simplify the problem, the following assumption is to be hold:
If random variables \(Y_{1} ,Y_{2} , \ldots ,Y_{n}\) follow normal distribution, n dimensional random variable \(\left( {Y_{1} ,Y_{2} , \ldots ,Y_{n} } \right)\) follows n dimensional joint normal distribution approximately.
This approximation uses normal distribution as a bridge to construct high dimensional probability distribution. Then, for any n dimensional random variable \(\left( {X_{1} ,X_{2} , \ldots ,X_{n} } \right)\), set the marginal distribution functions as \(F_{1} \left( {x_{1} } \right),F_{2} \left( {x_{2} } \right), \ldots ,F_{n} \left( {x_{n} } \right)\) and define
Then \(P\left( {\hat{X}_{i} \le \hat{x}_{i} } \right) = P\left( {\varPhi^{ - 1} \left( {F_{i} \left( {X_{i} } \right)} \right) \le \hat{x}_{i} } \right) = P\left( {X_{i} \le F_{i}^{ - 1} \left( {\varPhi \left( {\hat{x}_{i} } \right)} \right)} \right) = F_{i} \left( {F_{i}^{ - 1} \left( {\varPhi \left( {\hat{x}_{i} } \right)} \right)} \right) = \varPhi \left( {\hat{x}_{i} } \right)\), where \(\varPhi \left( x \right)\) is the distribution function of standard normal distribution. And we can see \(\hat{X}_{1} ,\hat{X}_{2} , \ldots ,\hat{X}_{n}\) follow normal distribution.
Based on the previous assumption, the joint distribution function of n dimensional random variable \(\left( {X_{1} ,X_{2} , \ldots ,X_{n} } \right)\) can be estimated as
And the joint density function is
where \(\hat{x} = \left( {\hat{x}_{1} ,\hat{x}_{2} , \ldots ,\hat{x}_{n} } \right)\), \(R = \left( {\rho_{ij} } \right)\) is the correlation coefficient matrix of \(\left( {\hat{X}_{1} ,\hat{X}_{2} , \ldots ,\hat{X}_{n} } \right)\), \(\lambda_{i}\) and \(\alpha_{i}\) is the eigenvalues and eigenvectors of R, respectively, and \(\left( {\beta_{1} ,\beta_{2} , \ldots ,\beta_{n} } \right) = \hat{x}\left( {\alpha_{1} ,\alpha_{2} , \ldots ,\alpha_{n} } \right)\).
\(\varphi_{n} \left( x \right) = \left( {2\pi } \right)^{{ - \frac{1}{2}}} \left| R \right|^{{ - \frac{1}{2}}} { \exp }\left( { - \frac{1}{2}xR^{ - 1} x^{\text{T}} } \right),x = \left( {x_{1} ,x_{2} , \ldots ,x_{n} } \right)\) is the density function of n dimensional standard normal distribution, and \(\varPhi_{n} \left( x \right) = \mathop \smallint \limits_{ - \infty }^{{x_{1} }} \cdots \mathop \smallint \limits_{ - \infty }^{{x_{n} }} \varphi_{n} \left( {t_{1} , \ldots ,t_{n} } \right){\text{d}}t_{1} \cdots {\text{d}}t_{n}\) is the distribution function.
4.2 Application in Bayesian Classification
For the sample with n features \(\left( {x_{1} ,x_{2} , \ldots ,x_{n} } \right)\), based on Bayes’ theorem, the probability that it belongs to class Ck is
where \(P\left( {C_{k} } \right)\) is the probability that any sample belongs to class Ck, \(P\left( {x_{1} ,x_{2} , \ldots ,x_{n} |C_{k} } \right)\) is prior and \(P\left( {C_{k} |x_{1} ,x_{2} , \ldots ,x_{n} } \right)\) is posterior.
With the assumption of that, every features are independent of each other for a given class label, which is independence of conditional probability. Then
Take note that \(P\left( {x_{1} ,x_{2} , \ldots ,x_{n} } \right)\) is independent of Ck, so we can get the predictive classification of \(\left( {x_{1} ,x_{2} , \ldots ,x_{n} } \right)\)
This method is naïve Bayes classifier, a basic algorithm in machine learning and shows implausible efficacy in many complex real-world situations [27,28,29].
But release such strong assumption and based on the estimation of the density function of n dimensional random variables, the predictive classification can be calculated straight forwardly:
With such update, we not only include the correlation of features into the model prediction, but also greatly extend the application of Bayesian classification.
4.3 Application in Maximum Likelihood Regression
With the proposed approach in the CDF and PDF estimation, maximum likelihood estimation can be extended from parameter estimation [30] to regression models.
The maximum likelihood function to sample \(\left( {x_{1} ,x_{2} , \ldots ,x_{n} } \right)\) is \(L\left( y \right) = P\left( {Y = y |x_{1} ,x_{2} , \ldots ,x_{n} } \right)\) with L(y) get the maximum value at the point y.
Then
where \(F_{Y} \left( y \right)\) and \(f_{Y} \left( y \right)\) is the distribution and density function of Y, respectively, \(\hat{x} = \left( {\hat{x}_{1} ,\hat{x}_{2} , \ldots ,\hat{x}_{n} } \right)\), \(\hat{y} = \varPhi^{ - 1} \left( {F_{Y} \left( y \right)} \right)\), R is the correlation coefficient matrix of \(\left( {\hat{X}_{1} ,\hat{X}_{2} , \ldots ,\hat{X}_{n} } \right)\) and r is the correlation coefficient vector between \(\hat{Y}\) and \(\hat{X}_{1} ,\hat{X}_{2} , \ldots ,\hat{X}_{n}\).
With \(\left( {\begin{array}{*{20}c} R & {r^{\text{T}} } \\ r & 1 \\ \end{array} } \right)^{ - 1} = \left( {\begin{array}{*{20}c} {\left( {R - r^{\text{T}} r} \right)^{ - 1} } & { - \left( {R - r^{\text{T}} r} \right)^{ - 1} r^{\text{T}} } \\ { - r\left( {R - r^{\text{T}} r} \right)^{ - 1} } & {1 + r\left( {R - r^{\text{T}} r} \right)^{ - 1} r^{\text{T}} } \\ \end{array} } \right)\), the log-likelihood function can be rewritten as
MLE y will be derived with equation as below.
If Y follows normal distribution N (μ, σ), the above equation can be simplified as
Then
which is the value \(\hat{y}\) as we got for linear regression.
5 Conclusion
In this study, we proposed a new method to estimate CDF and PDF based on a new spline regression, in which the spline function is not always defined by polynomial functions or B-splines, but can be set freely and consists of totally different types of functions in each segment. In this method, the PDF is expressed by piecewise functions instead of series, and with the increase of sample size, the estimated accuracy increases but the complexity of function does not increase. This method is suitable for most types of continuous distributions, and the form of spline function and other parameters does not need to be changed unless the distribution is quite special. The estimation is accurate for various types of distributions and is superior to kernel density estimation. The PDF is always smooth and is not influenced by parameters. The values of estimated CDF are less than 1, positive and monotone increasing. The values of estimated PDF are positive and the integration of PDF is about 1. And it is easy to find a subset from the large sample to reduce the running time and get similar accuracy simultaneously. PDF estimation of high dimensional random variables was also discussed and its potential application in Bayesian classification models and maximum likelihood regression models was presented.
References
Candy JV (2009) Bayesian signal processing: classical, modern and particle filtering methods. Wiley-Interscience, New York
Bishop CM (1996) Neural networks for pattern recognition. Oxford University Press, New York
Mitchell TM, Carbonell JG, Michalski RS (1986) Machine learning: a guide to current research. Kluwer Academic Publishers, Norwell
Mood AM, Graybill FA, Boes DC (1974) Introduction to the theory of statistics, 3rd edn. McGraw-Hill Education, New York
Rosenblatt M (1956) Remarks on some nonparametric estimates of a density function. Ann Math Stat 27(3):832–837
Parzen E (1962) On estimation of probability density function and mode. Ann Math Stat 33(3):1065–1076
Silverman BW (1986) Density estimation for statistics and data analysis. Chapman & Hall, London
Terrell GR (1990) The maximal smoothing principle in density estimation. J Am Stat Assoc 85(410):470–477
Alexandre LA (2008) A solve-the-equation approach for unidimensional data kernel bandwidth selection. University of Beira Interior, Covilhã
Jeon B, Landgrebe DA (1994) Fast Parzen density estimation using clustering-based branch andbound. IEEE Trans Pattern Anal Mach Intell 16(9):950–954
Babich GA, Camps OI (1996) Weighted Parzen windows for pattern classification. IEEE Trans Pattern Anal Mach Intell 18(5):567–570
Girolami M, He C (2003) Probability density estimation from optimally condensed data samples. IEEE Trans Pattern Anal Mach Intell 25(10):1253–1264
Bowers NL (1966) Expansion of probability density functions as a sum of gamma densities with applications in risk theory. Trans Soc Actuar 18 PT.1(52):125–147
Van Khuong H, Kong HY (2006) General expression for pdf of a sum of independent exponential random variables. IEEE Commun Lett 10(3):159–161
Schwartz SC (1967) Estimation of probability density by an orthogonal series. Ann Math Stat 38(4):1261–1265
Engel J (1990) Density estimation with Haar series. Stat Probab Lett 9(2):111–117
Vannucci M (1998) Nonparametric density estimation using wavelets; Discussion Paper 95–26, ISDS. Duke University, Durham
Howard RM (2010) PDF estimation via characteristic function and an orthonormal basis set. In: Wseas international conference on systems
Xie J, Wang Z (2009) Probability density function estimation based on windowed fourier transform of characteristic function. In: International congress on image and signal processing
Wold S (1974) Spline functions in data analysis. Technometrics 16(1):1–11
Reinsch CH (1967) Smoothing by spline functions. Numer Math 10(3):177–183
Marsh L, Cormier DR (2002) Spline regression models. J R Stat Soc 52(3):49–58
Zong Z, Lam KY (1998) Estimation of complicated distributions using B-spline functions. Struct Saf 20(4):341–355
Mansour A, Mesleh R, Aggoune EHM (2015) Blind estimation of statistical properties of non-stationary random variables. J Adv Signal Process 51(1):309–314
Kitahara D, Yamada I (2015) Probability density function estimation by positive quartic C 2 -spline functions. In: IEEE international conference on acoustics, speech and signal processing
De Boor C (1978) A practical guide to splines. Springer, New York
Zhang H (2005) The optimality of Naive Bayes. In: Seventeenth international florida artificial intelligence research society conference, Miami Beach, Florida, USA
Rennie JDM, Shih L, Teevan J, Karger D (2003) Tackling the poor assumptions of Naive Bayes text classifiers. In: Proceedings of the twentieth international conference on machine learning, Washington, DC, USA
Caruana R, Niculescu-Mizil A (2006) An empirical comparison of supervised learning algorithms. In: Proceedings of the 23rd international conference on machine learning, Pittsburgh, PA, USA
Pfanzagl J, Hamböker R (1996) Parametric statistical theory. J Am Stat Assoc 91(433):269–287
Acknowledgements
This work are supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National High-Tech R&D Program of China (863 Program Contract no. 2012AA020307), the National Basic Research Program of China (973 Program) (Contract no. 2012CB721000), and Ph.D. Programs Foundation of Ministry of Education of China (Contract no. 20120073110057), the Young Scholars (Grant no. 31400704) of Natural Science Foundation of China, also computing resources provided by Center for High Performance Computing, Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dai, H., Wang, W., Xu, Q. et al. Estimation of Probability Distribution and Its Application in Bayesian Classification and Maximum Likelihood Regression. Interdiscip Sci Comput Life Sci 11, 559–574 (2019). https://doi.org/10.1007/s12539-019-00343-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-019-00343-w