Abstract
Transcription factors (TFs) are the key regulators of gene expression and play crucial roles during plant growth and development. A novel TF family, containing one or two BSD domains in a variety of organisms ranging from prokaryotes to human, was newly identified by computational analysis. In this study, one Arabidopsis gene encoding the BSD domain, designated as AtBSD1, was characterized. The AtBSD1 transcript was expressed in all Arabidopsis tissues tested. The expression level of the AtBSD1 transcripts was not controlled by exogenous application of abiotic stresses and plant hormones. When the AtBSD1::GFP construct under the control of the CaMV35S promoter was introduced into onion epidermal cells, a GFP signal was detected in the nucleus. A transcriptional activity assay of the AtBSD1 protein in yeast revealed that the AtBSD1 protein functions as a transcriptional activator, and the N-terminal region (1–204 amino acids) of the AtBSD1 protein contains a transcriptional activation domain. These results suggest that Arabidopsis genes encoding the BSD1 domain might be classified as a novel transcription factor family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Transcription factors (TFs) are proteins that are involved in the regulation of gene expression, bind to the promoter regions of genes, and control the transcription of downstream genes. TFs consist of two functional regions, a DNA-binding region and an activator region. It was recently reported that the Arabidopsis genome encodes approximately 2,000 TFs, which are classified into 68 gene families, mainly according to the structure of the DNA-binding region (Riano-Pachon et al. 2007). About 45% of Arabidopsis TFs are plant-specific, whereas the rest of them share DNA-binding domains common to other eukaryotes (Riechmann et al. 2000; Riechmann and Ratcliffe 2000; Riechmann 2002; Gong et al. 2004).
A novel TF family containing the BSD domain, which is not included among the Arabidopsis 68 TF gene families, has recently been grouped by BLAST and Hidden Markov Model (HMMer) searches (Doerks et al. 2002; Eddy 1998). The BSD domain is characterized by three α helices that are probably involved in DNA binding and by conserved tryptophan and phenylalanine residues located at the C-terminus of the domain. TFs harboring the BSD domain are known to be present in a variety of species ranging from primal protozoans to humans. The newly discovered BSD domain was named from mammalian BTF2 transcription factor, Drosophila synapse-associated proteins, and DOS2-like proteins (Doerks et al. 2002; Fischer et al. 1992). Arabidopsis harbors 11 genes encoding the BSD domain, but their functional roles and domains have not been investigated.
In this study, we found that the AtBSD1 containing the BSD domain functions as a transcriptional factor. The AtBSD1 protein was predominantly localized in the nucleus. By using a β-galactosidase assay in yeast, we demonstrated that the N-terminal region (1–203 AA) of the AtBSD1 protein contains a transcriptional activation domain. Potential roles of AtBSD1 in transcriptional regulation in Arabidopsis are discussed.
Materials and Methods
Plant Materials
Arabidopsis thaliana (ecotype Columbia-0) plants were grown in the culture room under a photocycle of 16 h of light (24°C) and 8 h of darkness (22°C). The atbsd1 mutant seeds were germinated on the 1/2 MS media with a supplement of 50 μg/ml kanamycin. For exogenous application of abiotic stresses and plant hormones, Arabidopsis seeds were germinated on 1/2 MS media for 3 days and then transferred to 1/2 MS liquid media supplemented with 1 μM indole-3-acetic acid, 1 μM gibberellic acid, 1 μM jasmonic acid, 1 μM zeatin, 1 μM abscisic acid, 200 mM NaCl, 200 mM mannitol, or 20% PEG, and incubated for 6 h.
RNA Extraction and RT-PCR
Total RNAs were isolated using TRIzol reagent (Invitrogen, USA) according to the instructions of the manufacturer. Reverse transcription was performed as described in the protocol of the manufacturer (Invitrogen, USA). Primers used in RT-PCR are shown in Table 1. The PCR reaction was conducted in a final volume of 20 μl containing 100 ng of cDNA, 1× i Taq buffer (iNtRON, Republic of Korea), 2.5 mM of each dNTP, 1 unit of i-Taq polymerase (iNtRON, Republic of Korea), and 10 pmol of each primer.
Isolation of the AtBSD1 cDNA and Subcellular Localization Assay
The coding region of the AtBSD1 cDNA was amplified using the AtBSD1-gene specific primers, At1g10720FF and At1g10720FR, and cloned into pGEMT-easy vector (Promega, USA). The entire nucleotide sequence was confirmed by sequencing. The cloned AtBSD1 cDNA was digested with BamHI and subcloned into the BamHI-digested pBIN35S-mGFP4 vector (Davis and Vierstra 1998). The resultant plasmid was linearized with EcoRI. Particle bombardment was performed as described (Kim et al. 2007) using a biolistic helium gun device (Bio-Rad PDS-1000/He). After bombardment, samples were incubated for 24 h at 25°C in the dark and were observed under a confocal laser-scanning microscope (Olympus BX51, Japan).
β-Galactosidase Assay in Yeast
To construct recombinant plasmids (pAtBSD1-F, pAtBSD1-N, and pAtBSD1-C), the AtBSD1 cDNA in pGEMT-easy vector (Promega, USA) was amplified using the following primer sets: the At1g10720FF and At1g10720FR2 primers for pAtBSD1-F, the At1g10720FF and At1g10720YR primers for pAtBSD1-N, and the At1g10720YF and At1g10720FR2 primers for pAtBSD1-C. Each PCR fragment was cloned into pGEMT-easy vector (Promega, USA) and was subsequently sequenced. The resultant vectors were digested with BamHI, and approximately 1.3-, 0.6-, and 0.7-kb DNA fragments were eluted and cloned into the BamHI-digested pGBKT7 vector (BD Biosciences Clontech, USA). The resultant constructs, pAtBSD1-F, pAtBSD1-N, and pAtBSD1-C, were transformed into yeast strain Y190 (MATa, HIS3, lacZ, trp1, leu2, cyhr2) according to the instructions of the manufacturer (BD Biosciences Clontech, USA). Transformants were selected on selective medium (SD-Trp) supplemented with 25 mM 3-amino-1, 2, 4-aminotriazole (3-AT). For β-galactosidase assays, transformants were cultured on selective medium (SD-Trp-His) for 1 day at 30°C, and filter-lift assays for blue color development were performed for 4 h at 37°C, as described by Breeden and Nasmyth (1985).
Isolation of the atbsd1 Mutant and Development of Transgenic Lines Overexpressing AtBSD1
T-DNA tagged atbsd1 Arabidopsis mutant seeds (SALK_069095) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). Kanamycin-resistant seedlings were screened in order to isolate homozygote lines by PCR screening using the left border primer (LBa1) and the atbsd1 gene-specific primers, At1g10720F and At1g10720R.
For the development of transgenic lines overexpressing AtBSD1, pAtBSD1-F was digested with BamHI. Approximately 1.3-kb DNA fragments were eluted and ligated into BamHI-digested pCAMBIA1300.1 vector. The constructed binary vectors were transformed into Agrobacterium strain GV3101 using the freeze–thaw method (An 1987). Arabidopsis wild type was then transformed according to the vacuum infiltration method, as described by Clough and Bent (1998). Seeds that had been bulk-harvested from each pot were sterilized and were then germinated on MS agar medium supplemented with 50 μg/ml kanamycin or 30 μg/ml hygromycin. Surviving T1 seedlings were transferred to soil, and T1 plants were used for genetic and phenotypic analyses. The identification of the introduced gene was carried out by PCR using CaMV35SF1 and At1g10720R primers.
Results and Discussion
Isolation and Characterization of the AtBSD1 cDNA
BSD domain genes have been reported to exist in the Arabidopsis genome (Doerks et al. 2002). However, no reports have systematically analyzed the BSD domain genes in Arabidopsis. We performed BLAST searches to find the BSD domain genes and found that there were 11 BSD domain genes present.
We selected an Arabidopsis gene (At1g10720), designated as AtBSD1, containing a BSD domain in order to characterize the function of the BSD domain genes. The AtBSD1 cDNA was isolated from Arabidopsis seedlings, where the AtBSD1 gene was expressed, according to the information in the Arabidopsis microarray analysis database (http://www.arabidopsis.org). As shown in Fig. 1a, the AtBSD1 protein (429 amino acids) contains acidic amino-acid-rich sequences, a characteristic feature of a transcriptional activation domain and one BSD domain in its center (Fig. 1a). No putative nuclear localization signal sequence was identified.
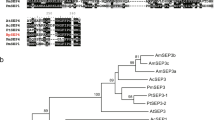
AtBSD1 gene. a Nucleotide and deduced amino acid (AA) sequences of the AtBSD1 gene. The numbering on the right refers to the AA sequences. The putative BSD domain is boxed. The location of the acidic AA sequences is underlined. b Phylogenetic tree of AtBSD1 homologues from various plant species, moss, amoeba, and green alga and alignment of the deduced amino acid sequences of their BSD domains. The conserved amino acid residues in the BSD motif are shown in the shaded region. The conserved phenylalanine (F) and tryptophan (W) residues are indicated by inverted triangles
Homology searches revealed that the AtBSD1 was matched to proteins with a BSD domain from various plant species (Vitis vinifera, Oryza sativa, Picea sitchensis, Medicago truncatula, and Populus trichocarpa), moss (Physcomitrella patens), amoeba (Dictyostelium discoideum), and green alga (Chlamydomonas reinhardt), suggesting that AtBSD1 is evolutionarily conserved (Fig. 1b).
Expression of the AtBSD1 Gene
To investigate the expression of AtBSD1 gene, RT-PCR was performed. The AtBSD1 transcripts were constitutively expressed in all Arabidopsis tissues tested (Fig. 2a). Then, we examined the effects of stresses and hormones on the expression of AtBSD1 gene. The results showed that the AtBSD1 expression was not regulated by salt and drought stresses in the rd29A gene-induced condition as well as various plant hormones (Fig. 2b, c). This finding suggests that it may be a housekeeping gene. This observation is consistent with the Arabidopsis microarray analysis from various tissues and 10-day-old seedlings treated with an exogenous application of abiotic and biotic stresses (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
Expression of the AtBSD1 gene. a Total RNAs were isolated from various Arabidopsis tissues, converted into cDNA, and amplified by RT-PCR using gene-specific primers. The Arabidopsis actin2 (At3g18780) gene was used as an internal control. b, c RT-PCR analysis of AtBSD1 gene expression by exogenous application of various plant hormones and salt and drought stresses (see “Materials and Methods”). The Arabidopsis actin7 (At3g18780) gene was used as an internal control. The rd29A gene (At5g09810), which is known as a drought stress-inducible gene, was used as a control in the condition of abiotic stresses
Subcellular Localization of the AtBSD1 Protein
To investigate the subcellular localization of the AtBSD1 protein, AtBSD1::mGFP was transiently expressed in onion epidermal cells. After incubation in the dark for 24 h, the onion cells were visualized under a confocal laser scanning microscope. As shown in Fig. 3, the AtBSD1::mGFP fusion protein was predominantly localized to the nuclei, whereas mGFP was found throughout the cell. In plants, the typical nuclear localization signal (NLS) was known to be a Pro-Lys-Lys-Lys-Arg-Lys (PKKKRK) (Jans 1995). However, the NLS was not present in the AtBSD1, suggesting that unknown sequences might be involved in the targeting of the AtBSD1 to the nucleus.
AtBSD1 Functions as a Transcriptional Activator, and the N-terminal Region of the AtBSD1 Protein Contains the Transcriptional Activation Domain
Since some BSD domain proteins function as a basal transcription factor and AtBSD1 was localized in the nucleus, we speculated that AtBSD1 may be a transcription factor that has a transcriptional activity. To determine whether AtBSD1 protein acts as a transcriptional activator and to investigate where its activation domain is if it acts as a transcriptional activator, the full-length (1–429 amino acids) gene and the N-terminal (1–204 amino acids) and C-terminal (205–429 amino acids) regions of the AtBSD1 were subcloned into pGBKT7 plasmid (Fig. 4a). Three resultant plasmids were transformed into yeast. Growth and filter-lift assays showed that full-length and N-terminal region of AtBSD1 had a transcriptional activity (Fig. 4b). In the N-terminal half of AtBSD1, there are acidic amino-acid-rich sequences (amino acids 151–169). It is known that these sequences are among the transcriptional activation domains, which include glutamine- and proline-rich domains depending on the type of amino acid enrichment in the domain (Triezenberg 1995). This result revealed that AtBSD1 functions as a transcriptional activator and that the N-terminal region of AtBSD1 protein contains the transcriptional activation domain.
Transcriptional activation assay of AtBSD1 protein in yeast. a Schematic diagram of full-length and deletion constructs of the AtBSD1 in the pGBKT7 vector. b Growth of yeast cells in medium SD/-Trp (left) and SD/-Trp-His (middle) containing 25 mM 3-AT. The X-gal lift assay was carried out using SD/-Trp-His plate (right). V Yeast transformed with the pGBKT7 vector; F yeast transformed with the full-length of the AtBSD1 in the pGBKT7 vector; C yeast transformed with the C-terminal region of the AtBSD1 in the pGBKT7 vector; N yeast transformed with the N-terminal region of the AtBSD1 in the pGBKT7 vector
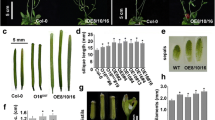
The BSD domain is present in diverse organisms, including primal protozoans, plants, yeasts, and humans. This suggests a conserved function for the domain (Doerks et al. 2002). BTF2 is a transcription factor that is essential for the initiation of transcription by RNA polymerase B (Fischer et al. 1992). Synapse-associated proteins function as important molecular components of the nervous system (Reichmuth et al. 1995). DOS2 is known to be required for silencing of all heterochromain regions in yeast. It has been found that deletion of DOS2 results in defects in chromosomal segregation and telomere clustering (Li et al. 2005). Therefore, the BSD domain is found in proteins with diverse functions. This prompted us to question the function of AtBSD1. To determine function of AtBSD1, we examined the atbsd1 knockout mutant and generated transgenic plants overexpressing AtBSD1 gene. In atbsd1, T-DNA is inserted into the third exon of the AtBSD1 gene (Fig. 5a). RT-PCR analysis showed that AtBSD1 is not expressed in the atbsd1 mutant (Fig. 5b). The mutant was indistinguishable from wild-type plants under normal growth conditions (Fig. 5c). In addition, AtBSD1 was overexpressed under the control of 35S promoter. RT-PCR confirmed the overexpression of AtBSD1 (Fig. 5d). However, the transgenic plants were essentially normal, which suggests that AtBSD1 may not be the limiting factor for the growth and development of Arabidopsis or require other proteins for its function.
Genotype and phenotype of the atbsd1 mutant and transgenic plants overexpressing AtBSD1. a The position of the T-DNA insertion site in the atbsd1 mutant. b RT-PCR analysis of the AtBSD1 gene in the atbsd1 mutant. c Phenotype of the atbsd1 mutant. The photograph was taken 25 days after sowing. d The overexpression of AtBSD1 in transgenic plants was analyzed by RT-PCR. The Arabidopsis actin2 (At3g18780) gene was used as an internal control. WT Wild-type plant
Because AtBSD1 is constantly expressed and has a transcriptional activity, we propose that AtBSD1 may be a basal transcription factor that is involved in the initiation of transcription, as suggested in BTF2 (Fischer et al. 1992). It is also possible that other BSD domain proteins may have redundant functions with AtBSD1. Generation of multiple knockout mutants of BSD domain genes will help to reveal their exact functions.
References
An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153:292–305
Breeden L, Nasmyth K (1985) Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol 50:643–650
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36:521–528
Doerks T, Huber S, Buchner E, Bork P (2002) BSD: a novel domain in transcription factors and synapse-associated proteins. Trends Biochem Sci 27:168–169
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Fischer L, Gerard M, Chalut C, Lutz Y, Humbert S, Kanna M, Chambon P, Eqly JM (1992) Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science 257:1392–1395
Gong W, Sen YP, Ma LG, Pan Y, Du YL, Wang DH, Yang JY, Hu LD, Liu XF, Dong CX, Ma L, Chen Y-H, Yang X-Y, Gao Y, Zhu D, Tan X, Mu J-Y, Zhang D-B, Liu Y-L, Dinesh-Kumar SP, Li Y, Wang X-P, Gu H-Y, Qu L-J, Bai S-N, Lu Y-T, Li J-Y, Zhao J-D, Zuo J, Huang H, Deng XW, Zhu YX (2004) Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol 135:773–782
Jans DA (1995) The regulation of protein transport to the nucleus by phosphorylation. Biochem J 311:705–716
Kim MJ, Kim JK, Shin JS, Suh MC (2007) The SebHLH transcription factor mediates trans-activation of the SeFAD2 gene promoter throught binding to E- and G-box elements. Plant Mol Biol 64:453–466
Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ (2005) Two novel proteins, Dos1 and Dos2, interact with Rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol 15:1448–1457
Riano-Pachon DM, Ruzicic S, Dreyer I, Mueller-Roeber B (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics 8:42–51
Riechmann JL (2002) Transcriptional regulation: a genomic overview. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville, MD, pp 1–46
Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3:423–434
Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G-L (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–110
Reichmuth C, Becker S, Benz M, Debel K, Reisch D, Heimbeck G, Hofbauer A, Klagges B, Pfluqfelder GO, Buchner E (1995) The sap47 gene of Drosophila melanogaster codes for a novel conserved neuronal protein associated with synaptic terminals. Brain Res Mol Brain Res 32:45–54
Triezenberg SJ (1995) Structure and function of transcriptional activation domains. Curr Opin Genet Dev 5:190–196
Acknowledgments
We thank the Salk Institute for Genomic Analysis Laboratory for providing sequence-indexed Arabidopsis T-DNA insertion mutants of atbsd1 (SALK_069095). This work was supported by grants from the Agricultural Plant Stress Research Center (R11-2001-09205001-0) of the Korea Science and Engineering Foundation and the Rural Development Administration, Republic of Korea. Acknowledgement is also made to the Bioenergy Research Center, Chonnam National University for its support of this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s12374-009-9055-5
Rights and permissions
About this article
Cite this article
Park, J., Kim, M.J., Jung, S.J. et al. Identification of a Novel Transcription Factor, AtBSD1, Containing a BSD Domain in Arabidopsis thaliana . J. Plant Biol. 52, 141–146 (2009). https://doi.org/10.1007/s12374-009-9015-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-009-9015-0