Abstract
Microsomal oleic acid desaturase (FAD2) catalyzes the first extra-plastidial desaturation in plants, converting oleic acid to linoleic acid, which is a major constituent in all cellular membranes as well as in seed storage oils. Seed-specific FAD2 (SeFAD2) produced 40% of linoleic acids in the total fatty acids of sesame (Sesamum indicum) seeds. The expression of SeFAD2 transcripts was spatially and temporally controlled during seed development. To investigate the regulatory mechanism controlling seed-specific SeFAD2 expression, we isolated a well-matched sequence homologous to the basic region/helix-loop-helix proteins by yeast one-hybrid screening and named it SebHLH. SebHLH transcripts were expressed in developing seeds and roots of sesame. SebHLH:GFP fusion protein localized in the nucleus. Recombinant SebHLH protein bound E-box (CANNTG) and G-box (CACGTG) elements in the region from −179 to −53 of the SeFAD2 gene promoter, and the external C and G nucleotides in the E- and G-box motifs were essential for SebHLH protein binding. The SebHLH gene, under the CaMV35S promoter, and the GUS reporter gene driven by E- and G-box motifs were co-expressed in developing sesame seeds and Arabidopsis transgenic leaves. This co-expression demonstrated that SebHLH protein mediates transactivation of the SeFAD2 gene promoter through binding to E- and G-box elements. E- or G-box elements frequently occur in the 5′-flanking region of genes that are involved in triacylglycerol biosynthesis and that exhibit seed-specific expression in Arabidopsis and other plants, suggesting that bHLH transcription factors play a key role in the transcriptional regulation of genes related to storage lipid biosynthesis and accumulation during seed development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants, de novo fatty acid synthesis from acetyl-CoA to palmitic acid (16:0), stearic acid (18:0) or oleic acid (18:1Δ9) occurs in plastids. The fatty acids are further reduced to linoleic acid (18:2Δ9,12) and linolenic acid (18:1Δ9,12,15) via a prokaryotic pathway in plastids or a eukaryotic pathway in endoplasmic reticulum (Somerville et al. 2000). The synthesized polyunsaturated fatty acids serve as an essential component in all cellular membranes and in storage oils. Oleic acid desaturase (FAD2) on the endoplasmic reticulum catalyzes the formation of a double bond between carbons 12 and 13 of oleic acid at both the sn-1 and sn-2 positions of phosphatidylcholine (Shanklin and Cahoon 1998). Arabidopsis harbors a single copy of the FAD2 gene, but crops including sesame (Sesamum indicum), soybean (Glycine max), cotton (Gossypium hirsutum), corn (Zea mays), and canola (Brassica napus) contain at least two FAD2 isoforms; one is constitutively expressed and the other is tightly regulated during seed development (Okuley et al. 1994; Heppard et al. 1996; Jin et al. 2001; Pirtle et al. 2001; Kinney et al. 2002; Tang et al. 2005). Seed-specific FAD2 is the key enzyme responsible for the production of linoleic acid in seeds (Liu et al. 2000; Jin et al. 2001). Furthermore, the ratio of the various saturated and unsaturated fatty acids is reported to be one of the critical factors that influence the quality and commercial application of seed oils (Cahoon et al. 2002; Kinney et al. 2002; Tang et al. 2005).
During seed development, storage lipids accumulate in abundance in the middle and late phases of embryogenesis. Coincident with storage lipid accumulation, some genes involved in biosynthesis and accumulation of storage lipids, including seed-specific FAD2 gene, are spatially and temporally regulated in developing seeds (Cahoon et al. 1992; Slocombe et al. 1992; Kinney 1994, 1996; Suh et al. 1999; Liu et al. 2000; Moon et al. 2001; Rossak et al. 2001; Bao et al. 2002; Tang et al. 2005; Kim et al. 2006). The transcriptional control of these genes through a complex set of promoter DNA or promoter DNA-specific regulatory proteins has been studied. In microspore-derived cell suspension culture of Brassica napus, the seed-specific activator ABI3 promotes oleosin gene expression via interaction with the ABA response element at −160 bp of the oleosin gene promoter (Crowe et al. 2000). Promoter regions at −117 and −312 of acyl carrier protein (Cs-ACP1) and Δ4-palmitoyl-acyl carrier protein desaturase (Cs-4PAD) genes, respectively, both of which are involved in biosynthesis of petroselinic acid in coriander (Coriandrum sativum) endosperms, include cis-regulatory elements such as the GCN4-like motif (GTCA), E-box, G-box, AACA motif, and Prolamin-box. These elements are required for endosperm-specific expression of Cs-ACP1 and Cs-4PAD genes (Kim et al. 2005). In sesame, the region from −179 to −53 in the SeFAD2 promoter, including E-box (CANNTG), G-box (CACGTG), ABRE motif (ACGTGKC), G-box-like element (ACGT), RY repeat element (CATGCA), and Prolamin-box (AAAG), is essential for expression of the SeFAD2 gene in developing seeds (Kim et al. 2006).
The basic region/helix-loop-helix (bHLH) proteins are a superfamily of transcription factors that have been found in all eukaryotes. The proteins bind as homodimers or heterodimers to specific target sequences, E-box (CANNTG) or G-box (CACGTG) motifs, and function in cell proliferation and cellular differentiation pathways (Littlewood and Evan 1998; Atchley et al. 1999; Ledent and Vervoort 2001). The bHLH domain contains two functionally distinct regions. The basic region consists of approximately 15 amino acids with a high number of basic residues and is involved in DNA binding. The HLH region is comprised of hydrophobic residues that form two α-helices separated by a loop region of variable sequence and length and functions as the dimerization domain (Murre et al. 1989; Ferre-D’Amare et al. 1994; Nair and Burley 2000). Outside of the conserved bHLH domain, these proteins exhibit considerable sequence divergence (Atchley et al. 1999). Since the R gene product, Lc, which is involved in the control of anthocyanin synthesis, was reported to possess the bHLH motif in plants (Ludwig et al. 1989), several bHLH transcription factors have been identified including PIF1, PIF3, PIF4, HFR1, SPT, ALC, TT8, GL3, AMS, AtMYC2, ATR2, BEE1, BEE2, BEE3, and ICE1. These proteins regulate a broad range of growth and developmental processes at all phases of the plant life cycle (see review in Toledo-Ortiz et al. 2003). Arabidopsis and rice genome sequencing analyses provided genetic information for 147 and 167 bHLH transcription factors, respectively (Toledo-Ortiz et al. 2003; Li et al. 2006). However, approximately 10% of these proteins in Arabidopsis have been characterized through mutational or functional analysis (Toledo-Ortiz et al. 2003).
To understand the transcriptional control of the SeFAD2 gene, which is spatially and temporally regulated during seed development, a trans-acting factor that interacts with E- and G-box elements in the region from −179 to −53 in the SeFAD2 gene promoter was isolated from developing sesame seeds. The isolated gene, named SebHLH, had good sequence homology with the basic region/helix-loop-helix proteins. SebHLH protein was targeted to the nucleus, and the N-terminal region of the protein functioned as a transcriptional activator. Finally, SebHLH protein induced transcriptional activity from E- and G-boxes of the SeFAD2 promoter in developing sesame seeds and transgenic Arabidopsis leaves.
Materials and methods
Plant materials
Sesame (Sesamum indicum cv. Yangbaeck) plant was grown in soil in the green house at 26 ± 3°C under a 16-h light/8-h dark regimen. Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) was grown in soil in the Arabidopsis room under a photocycle of 16 h of light (24°C) and 8 h of darkness (22°C). To facilitate harvesting of seeds at predefined developmental stages, flowers were tagged on the day of flower opening. The developing seeds were then collected via dissection from sesame fruits and Arabidopsis siliques.
Yeast one-hybrid screening
Yeast one-hybrid screening was performed according to the manufacturer’s instructions (Clontech). The region from −179 to −53 of the SeFAD2 gene promoter was amplified by PCR using SiW6-YF4/SiW6-YR4 and inserted into the EcoRI/SacI site of the pHIS2 vector (SeYF4:HIS2). Plasmid SeYF4:HIS2 containing the SeFAD2 promoter region was linearized with KpnI and cotransformed into yeast strain Y187 (Clontech) with developing sesame seed cDNAs (10–30 DAF) in pGADT7-Rec2 vector. Recombinants with positive one-hybrid interaction were selected on SD/-His-Leu-Trp medium supplemented with 45 mM 3-AT (3-amino-1,2,4-triazole). Recombinants containing inserts larger than 500 bp were further selected by colony PCR screening using 5′LD and 3′LD amplimers. The selected plasmids isolated from recombinant yeast cells were re-transformed into E. coli XL1-Blue, and nucleotide sequencing was performed. The genetic information of the sequenced clones was obtained from BLASTX and TRANSFAC analyses (http://www.ncbi.nlm.nih.gov; http://www.gene-regulation.com).
The full-length cDNA encoding the SebHLH protein was isolated by 5′-cRACE (circular first-strand cDNA-mediated rapid amplification of cDNA ends) as described by Maruyama et al. (1995). The sequence information of all primers used in 5′-cRACE is listed on Table 1. Total RNA isolated from sesame endosperm (10–30 DAF) was reverse-transcribed, self-ligated, and then subjected to PCR. After subcloning the PCR products, the nucleotide sequence was analysed.
Northern blot analysis and RT-PCR
Total RNAs were isolated from various sesame tissues using TRIzol reagent (Sigma, USA) according to the manufacturer’s instructions. Approximately 20 μg of total RNA was fractionated via electrophoresis on 1.2% agarose gels with formaldehyde, blotted onto nitrocellulose membranes (Amersham Biosciences), and hybridized at 63°C. Radioactive 32P-dCTP-labeled hybridization probes were generated via random priming (Rediprime II Random Prime Labelling System, Amersham Biosciences) using SebHLH gene-specific DNA fragments (SF4-FF2 and SebHLH-CR primer set, 761 bp of DNA fragments), including the unique 3′-UTR sequence. After hybridization, the membranes were washed twice with 2× SSPE and 0.1% SDS for 20 min at room temperature, twice with 0.2× SSPE and 0.1% SDS for 15–30 min at 63°C, and finally exposed to X-ray film or to a PhosphorImager (Bio-Rad) for 15–32 h.
Reverse transcription was carried out as described by the manufacturer (Invitrogen). PCR was performed using gene-specific SF4-FF2 and SebHLH-ER primers. The sesame elongation factor gene (MC01037C03, Suh et al. 2003) was amplified using the Se-Elon-F and the Se-Elon-R primers to determine the quantity and quality of the cDNAs. PCR was conducted in a final volume of 30 μl containing 500 ng of cDNA, 1× i-Taq buffer (iNtRON, Korea), 10 mM of each dNTP, 1 unit of i-Taq polymerase (iNtRON, Korea), and 10 pmol of each primer. The following amplification program was used: 1 cycle of 94°C for 5 min (denaturation step), 35 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 1 min, and then 1 cycle of 72°C for 7 min (elongation step).
Subcellular localization assay
The coding region of the SebHLH cDNA was amplified using the SebHLH-GFPS and SebHLH-GFPA primers and subcloned into the pBIN35S-smGFP vector under the control of the CaMV35S promoter (Davis and Vierstra 1998). The resultant plasmid was linearized with EcoRI restriction enzyme. As described in Klein et al. (1988), gold particles (1.0 μm, Bio-Rad) were coated with the plasmid DNA mixture (2 μg of the linearized plasmid DNA, 50 μl of 2.5 M CaCl2, and 20 μl of 0.1 M spermidine) and introduced into onion epidermis via a gene gun (Bio-Rad, USA). The tissues were bombarded twice at 1,100 psi with a biolistic helium gun device (Bio-Rad PDS-1000/He). The stopping screen was positioned 3 cm below the rupture disk, and the target tissue was positioned 6 cm below the stopping screen. After bombardment, samples were incubated for 24 h at 25°C in the dark and observed under a confocal laser scanning microscope (OLYMPUS BX51).
Electrophoretic mobility shift assays
The coding region of the SebHLH cDNA was amplified using the SebHLH-EF and SebHLH-ER primers, and subcloned between EcoRI and SalI sites in pET32b expression vector (Novagen, Germany). After transformation of the resultant pSebHLH:His plasmid into E. coli strain BL21(DE3), expression of His-tagged SebHLH protein was induced with 0.2 mM IPTG for 4 h. Cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM Imidazole, pH 8.0) containing 1 mM PMSF and 1 mg/ml lysozyme, and then sonicated. Supernatants obtained by centrifugation were loaded on Ni-NTA resin columns. The column was washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl and 30 mM Imidazole, pH 8.0) and eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl and 250 mM Imidazole, pH 8.0) as described in the manufacturer’s instructions of the His-Bind Buffer Kit (Novagen).
The region from −179 to −53 in the SeFAD2 gene promoter was amplified by PCR using the SeMF4 and SeMR4 primers, and eluted. To generate double-strand oligonucleotides, single-strand oligonucleotides (SeEbox-F, SeGbox-F, mSeEbox-F, mSeGbox-FA, and mSeGbox-FB) and their complementary oligonucleotides (SeEbox-R, SeGbox-R, mSeEbox-R, mSeGbox-RA, and mSeGbox-RB) were synthesized and annealed at room temperature (25 ± 2°C). DNA probes were end-labeled with γ-32P [dATP] using T4 polynucleotide kinase and purified on a Sephadex G-50 column. Labeled probes were incubated for 30 min at 25°C with 5 μg of purified SebHLH:His protein in binding buffer (10 mM Tris–HCl, pH 7.6, 50 mM NaCl, 1 mM Na2EDTA, 5 mM DTT, and 5% glycerol) supplemented with 100 ng poly(dI–dC) in the presence or absence of non-radiolabeled competitors. The resulting DNA–protein complexes were electrophoresed on 5% polyacrylamide gels in 0.5× TBE buffer (44.5 mM Tris–HCl, 44.5 mM boric acid, 1 mM EDTA). Gels were dried on Whatman 3 MM paper and visualized by Fuji BAS-2500 phosphorimager.
β-galactosidase assay in yeast
To construct recombinant plasmids, pGBKT7-SebHLH, pGBKT7-SebHLHn, and pGBKT7-SebHLHc, the SebHLH cDNA was amplified using the following primer sets: the SebHLH-NF1 and SebHLH-CR1 primers for pGBKT7-SebHLH, the SebHLH-NF1 and SebHLH-NR1 primers for pGBKT7-SebHLHn, and the SebHLH-CF1 and SebHLH-CR1 primers for pGBKT7-SebHLHc. Each PCR fragment was digested with EcoRI and SalI restriction enzymes and cloned into the same restriction sites of the pGBKT7 vector (BD Biosciences Clontech, USA). The resultant constructs were transformed into yeast strain Y190 (MATα, HIS3, lacZ, trp1, leu2, cyh r 2). Transformants were selected on selective medium (SD-Trp) supplemented with 30 mM 3-amino-1,2,4-aminotriazole [3-AT]. For β-galactosidase assays, transformants were cultured on selective medium (SD-Trp-His) for 1 day at 30°C, and filter-lift assays for blue color development were performed for 4 h at 37°C as described in Breeden and Nasmyth (1985).
Transactivation assay in developing sesame seeds and Arabidopsis transgenic plants
To develop the pPF4 construct, the region from −179 to −53 of the SeFAD2 gene promoter was PCR-amplified using the SiPF4-TF1 and SiPF4-TR1 primers. The resulting 143-bp DNA fragments were eluted. For the pEGbox and the pmEGbox constructs, single-strand oligonucleotides (EGbox-TF1 and EGbox-TR1) and mutated single-strand oligonucleotides (mEGbox-TF1 and mEGbox-TR1) were synthesized and annealed to the complementary strand. These three DNA fragments were double-digested with EcoRI and HindIII and transcriptionally fused to 35S minimal promoter of pCAMBIA 1305.1 plasmid containing β-glucuronidase (GUS) reporter gene, followed by the 3′non-coding region of nopaline synthase gene (NOS). To construct the effector plasmid, the coding region of SebHLH cDNA was PCR-amplified using the SebHLH-CF-Xb and SebHLH-CR-Bg primers and inserted between the CaMV35S promoter and the NOS terminator.
For transient assays, 2 μg of reporter plasmid, alone or in combination with the effector plasmid, was introduced into developing sesame seeds (25–35 DAF) by particle bombardment. The tissues were bombarded twice at 1,350 psi with a biolistic helium gun device (Bio-Rad PDS-1000/He). The stopping screen was positioned 3 cm below the rupture disk, and the target tissue was positioned 3 cm below the stopping screen. After bombardment, samples were incubated for 20 h at 26°C in the dark. For β-glucuronidase (GUS) and luciferase (LUC) assays, 100 μg and 50 μg of total proteins were used to measure GUS and LUC activity, respectively. GUS activity was measured by fluorometric assays as described in Jefferson (1987). Luciferase assays were performed using the Luciferase Assay System Kit (Promega) according to the manufacturer’s instructions. Protein concentrations were determined by the Bradford method (1976) using bovine serum albumin (BSA) as a standard.
For transactivation assays in transgenic Arabidopsis plants, each reporter plasmid contained the hygromycin resistant marker gene, and the effector plasmid contained the kanamycin resistant marker gene. These plasmids were transformed into Agrobacterium strain GV3101 via the freezing-thaw method (An 1987). Using the floral dip method, Arabidopsis plants were then transformed with Agrobacterium transformed with each reporter plasmid alone or in combination with Agrobacterium transformed with the effector plasmid (Clough and Bent 1998). Seeds were harvested, sterilized in a mixture of 50% bleach and 0.02% Triton X-100 for 7 min with agitation, and then germinated on selective MS medium containing 50 mg/l kanamycin and/or 30 mg/l hygromycin. The antibiotic-resistant seedlings were transferred to soil, and the T1 transgenic leaves were used to measure GUS activity.
Results
SebHLH, which interacted with the region from −179 to −53 of the SeFAD2 promoter, was isolated from developing sesame seeds
To isolate novel proteins that bind to cis-elements in the fragment from −179 to −53 of the SeFAD2 promoter, yeast one-hybrid analysis was performed. The -179 to -53 fragment of the SeFAD2 promoter was PCR-amplified using SiW6-YF4/SiW6-YR4 primers, inserted into the pHIS2 vector (SeYF4:HIS2), and used as bait. Amplified cDNAs from developing sesame seeds (10–30 DAF) were inserted into the pGADT7-Rec2 plasmid that contained the GAL4 activation domain and used as prey. We screened 3 × 106 transformants, and selected 400 colonies on medium lacking Leu, Trp, and His. Plasmids containing inserts larger than 500 bp were further analyzed by nucleotide sequencing. From the BLASTX and TRANSFAC analyses, one clone was identified that encoded a novel protein with the basic helix-loop-helix domain (bHLH) motif. A full-length cDNA clone was obtained by 5′-cRACE and named SebHLH. Nucleotide and deduced amino acid sequences of SebHLH cDNA are shown in Fig. 1A.
Nucleotide and deduced amino acid sequences of the SebHLH cDNA (A) and alignment of deduced amino acid sequences of bHLH domains from sesame and other plants (B). A putative NLS and calcium binding sequences are single and double-underlined, respectively. The conserved 60 amino acid residues corresponding to the bHLH motif are shown in the shaded region. The conserved H5, E9 and R12 residues are shown in inverted red-triangles
The SebHLH cDNA consists of an open reading frame encoding 400 amino acids. Amino acid sequence homology searches against the NCBI public database revealed that SebHLH cDNA had 60% sequence homology with Arabidopsis bHLH transcription factor (At1g10120), whose biological function was not identified. The amino acid sequence of the N-terminal region (residues 1–227) of SebHLH protein contains a transcription activation domain featuring specific patterns of hydrophobic and aromatic amino acids and acidic amino acid-rich sequences. The C-terminal region (residues 228–400) of the SebHLH protein contains the bHLH motif (residues 241–261) that is highly conserved in the eukaryote bHLH transcription factor family. The histidine (H5), glutamate (E9), and arginine (R12) amino acid residues are also conserved in the bHLH motif of the SebHLH protein (Fig. 1B). A putative nuclear localization signal sequence (KRKRK) identified by the PSORT program (http://www.psort.org) and the putative calcium-binding domain (residues 314–334) are shown in Fig. 1A.
SebHLH transcripts were expressed in sesame developing seeds and roots
To investigate the expression of SebHLH transcripts, total RNAs were isolated from sesame roots, stems, leaves, flowers, seed coats, seeds, and seedlings. Northern blot analysis was then performed using the C-terminal domain of the SebHLH cDNA as a probe. However, no signal was detected (data not shown), suggesting that the SebHLH transcript expression level is very low.
To determine the expression level of SebHLH transcripts, RT-PCR was carried out. The sesame elongation factor gene was used to determine the quantity and quality of cDNAs. Following 35 cycles of amplification, SebHLH transcripts were detected in developing seeds and roots, and SebHLH expression in stems, leaves, and flowers was much lower than in developing seeds and roots. No SebHLH expression was detected in seed coats and seedlings (Fig. 2).
The SebHLH:smGFP fusion protein was localized in the nucleus
To investigate the subcellular localization of SebHLH protein, full-length SebHLH cDNA without a translation termination codon was translationally fused with the C-terminus of the smGFP gene and transformed into onion epidermal cells by particle bombardment. After incubating in the dark for 24 h, the bombarded tissues were visualized under a confocal laser scanning microscope. As shown in Fig. 3, the SebHLH:smGFP fusion protein was localized to the nucleus of onion epidermal cells whereas smGFP was found in the cytoplasm. Therefore, this result suggests that SebHLH protein might act as a transcription factor in the nucleus.
Recombinant SebHLH protein binds to E- and G-box elements in the region from -179 to -53 of the SeFAD2 promoter
To investigate which cis-elements interacted with SebHLH protein, the coding region of SeFAD2 cDNA was amplified by PCR and ligated into the pET-32b expression vector. His-tagged SebHLH proteins were purified from soluble E. coli extracts and incubated with the −179 to −53 fragment of the SeFAD2 promoter (Fig. 4A), which had been end-labeled with γ-32P[dATP]. Binding of recombinant SebHLH protein to the probe was determined by electrophoretic mobility shift assay (EMSA).
EMSA of SebHLH protein. (A) Nucleotide sequences of the region from −179 to −53 in the SeFAD2 promoter and DNA fragments containing E- or G-box motifs and mutated E- or G-box motifs. (B) Interaction between SebHLH protein and 32P-labeled SeFAD2 promoter (−179 to −53). (C) In vitro interaction between SebHLH protein and 32P-labeled SeFAD2P-E and SeFAD2P-G DNA fragments. The SeFAD2P-mE, SeFAD2P-mGa, and SeFAD2P-mGb were generated by site-directed mutagenesis and used as competitors. 100× and 500× indicate 10 pmol and 50 pmol molar excess of double-stranded oligonucleotides, respectively. An arrow indicates protein-DNA complexes. FP: Free probe
As shown in Fig. 4B, one specific DNA–protein complex (indicated by arrow) was observed. A 250-fold or 500-fold molar excess of unlabeled probe eliminated the strongly shifted complex. Based on previous reports that bHLH transcription factors bind to E- or G-box elements, we determined whether E-box and/or G-box elements in this region of the SeFAD2 promoter were involved in specific binding between recombinant SebHLH protein and the region from -179 to -53 of the SeFAD2 promoter. Double-stranded DNA fragments containing the E-box motif (5′-GTGCACACTCCATGTGGGCCAATG-3′; 24mer) or G-box motif (5′-GCGGATGACACGTGGCGGGCAACT-3′; 24mer) in this region of the SeFAD2 promoter were generated, named SeFAD2P-E and SeFAD2P-G, respectively, and used as competitors. A 500-fold molar excess of each unlabeled competitor was not enough to eliminate the strongly shifted complex. However, the strong DNA/protein complex was completely eliminated when 500-fold molar excess of both SeFAD2P-E and SeFAD2P-G were used as competitors.
To further determine whether the E-box and G-box sequences are essential for specific binding between recombinant SebHLH protein and the region from −179 to −53 of the SeFAD2 promoter, one mutated E-box (SeFAD2P-mE, 5′-GTGCACACTCaATGTcGGCCAATG-3′) and two mutated G-boxes (SeFAD2P-mGa, 5′-GCGGATGACtctaGGCGGGCAACT-3′ and SeFAD2P-mGb, 5′-GCGGATGAaACGTtGCGGGCAACT-3′) were generated. In EMSA of recombinant SebHLH protein and end-labeled SeFAD2P-E fragments, a 500-fold molar excess of unlabeled SeFAD2P-E fragments completely eliminated the protein/DNA complex, but a 500-fold molar excess of unlabeled SeFAD2P-mE fragments did not eliminate the protein/DNA complex. Similar results were observed in EMSA of recombinant SebHLH protein and end-labeled SeFAD2P-G fragments when SeFAD2P-mGb fragments were used as a competitor. When a 500-fold molar excess of SeFAD2P-mGa fragments were used as a competitor, the DNA/protein complex was partially eliminated (Fig. 4C).
These observations revealed that SebHLH recombinant proteins bind to E-box (CANNTG) and G-box (CACGTG) elements in the −179 to −53 region of the SeFAD2 gene promoter and that the external C and G sequences in these motifs are essential for SebHLH protein binding.
The N-terminal region of SebHLH protein contains transcriptional activation domain
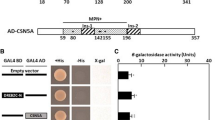
To determine whether SebHLH protein acts as a transcriptional activator, and where its activation domain is if it acts as a transcriptional activator, the full-length (1–400 amino acids) protein and the N-terminal (1–229 amino acids) and C-terminal (230–400 amino acids) regions of the SebHLH protein were PCR-amplified and translationally fused with the GAL4 DNA-binding domain in the pGBKT7 plasmid.
Three resultant plasmids, pGBKT7-SebHLH, pGBKT7-SebHLHn, and pGBKT7-SebHLHc, were introduced into yeast Y190 strain harboring the GAL1 promoter and HIS3 and lacZ reporter genes. Transformants were then selected on SD (-His/-Trp) medium supplemented with 30 mM 3-AT. Transformants with pGBKT7-SebHLH or pGBKT7-SebHLHn grew faster than those with pGBKT7-SebHLHc on selection medium. Filter-lift assays revealed β-galactosidase activity in transformants with pGBKT7-SebHLH and pGBKT7-SebHLHn, but not in transformants with pGBKT7-SebHLHc and pGBKT7 (Fig. 5). This result revealed that SebHLH protein functions as a transcriptional activator and that the N-terminal region of SebHLH protein contains the transcriptional activation domain.
Transcriptional activity assay of SebHLH protein in yeast. (A) Schematic diagram of full-length and deletion constructs of SebHLH protein in pGBKT7 vector. (B) Growth of yeast cells in medium SD/-W (left) and SD/-W-H (middle) containing 30 mM 3-AT. The X-gal lift was carried out using SD/-W plate (right)
SebHLH protein activates transcription from E- and G-box elements of the SeFAD2 gene promoter
The functional relevance of the interaction observed in vitro between SebHLH protein and E- and G-box elements of the SeFAD2 gene promoter were determined by transient expression assays in co-bombarded developing sesame seeds and by transgenic approaches with effector and reporter constructs. Figure 6A depicts the scheme of reporter and effector constructs used in transactivation studies of the SeFAD2 promoter. The region from -179 to -53 of the SeFAD2 promoter was PCR-amplified, and 41-bp DNA fragments (EG-box) containing E- and G-box elements, as well as 41-bp DNA fragments (mEG-box) containing mutated E- and G-box elements, were generated by annealing the synthesized single-strand oligonucleotides. The DNA fragments were transcriptionally fused to the 35S minimal promoter of pCAMBIA 1305.1 plasmid containing the β-glucuronidase (GUS) reporter gene, followed by the 3′ non-coding region of nopaline synthase gene (NOS). The resultant plasmids were named pPF4, pEGbox, and pmEGbox. An effector plasmid containing the SebHLH cDNA between the CaMV35S promoter and the NOS terminator was also constructed. Developing sesame seeds (25–35 DAF) were transiently transformed by particle bombardment with the pPF4, pEGbox, or pmEGbox reporter, alone or in combination with the effector construct. A construct with the luciferase gene between the CaMV35S promoter and the NOS terminator was transiently cotransformed to monitor transformation efficiency.
Transactivation assay in developing sesame seeds and transgenic Arabidopsis plants. (A) Scheme of reporter and effector constructs. The pMin35S contains the GUS reporter gene between the minimal CaMV35S promoter and the NOS terminator in the pCAMBIA 1305.1 vector. The pPF4, pEGbox, and pmEGbox were constructed by insertion of the region from −179 to −53, DNA fragments containing the E- and G-box motifs, and DNA fragments containing mutated E- and G-box motifs upstream of the minimal CaMV35S promoter in the pMin35S. Effector construct contains the SebHLH cDNA between the CaMV35S promoter and the NOS terminator in pMBP1 vector. (B) Transactivation assay in developing sesame seeds. Developing sesame seeds (25–35 DAF) were bombarded with each reporter construct alone or in combination with the effector construct and incubated. GUS activity was measured by fluorometric assays as described by Jefferson et al. (1987). Luciferase expression was used to normalize GUS activity. (C) Transactivation assay in transgenic Arabidopsis plants. Arabidopsis was transformed with each reporter construct alone or in combination with the effector construct, and T1 transgenic leaves were used to measure GUS activity
As shown in Fig. 6B, co-transfection of pPF4 and effector or pEGbox and effector resulted in 11- to 12-fold increase in GUS activity over that directed by pPF4 or pEGbox alone. In addition, mutation of the E- and G-boxes decreased trans-activation by the effector approximately 2-fold. We further confirmed the interaction between SebHLH protein and E- and G-box elements of the SeFAD2 promoter in planta. Arabidopsis plants were transformed with each reporter construct alone or in combination with the effector construct. T1 transgenic leaves of each transformant were used to measure GUS activity. When pPF4 reporter and effector were co-expressed, a 5-fold increase in GUS activity level was observed compared to the activity measured when the pPF4 reporter was expressed without an effector. Similarly, co-expression of pEGbox and effector resulted in more than 2-fold increase in GUS activity over that directed by pEGbox alone. The effector did not transactivate the reporter construct pmEGbox with mutations in the E- and G-box-binding site. Taken together, these results indicate that SebHLH protein mediates trans-activation of the SeFAD2 gene promoter through binding to E- and G-box elements.
Discussion
Seed-specific FAD2 functions in the committed step of polyunsaturated fatty acid synthesis during triacylglycerol accumulation. SeFAD2 mRNA levels are quite low in early embryogenesis, achieve maximum levels in seeds during mid- and late embryogenesis, and fall to low levels as seeds mature (Kim et al. 2006). In this study, we investigated the transcriptional control mechanism for expression of the SeFAD2 gene during seed development. We determined that the SebHLH trans-acting factor activates transcriptional activity from E- and G- boxes in the SeFAD2 promoter. This result strongly implicates the bHLH protein as an important trans-acting factor in combinatorial regulation of genes that are involved in the biosynthesis and accumulation of storage lipids in developing seeds.
X-ray crystallography analyses of Max, E47, MyoD, Pho4, and USF bHLH transcription factors have shown that the interaction between two identical or similar monomers, each with two helical segments separated by a loop, leads to the formation of a four helix bundle of homo- or heterodimers. The invariant glutamate (Glu) in the basic region of each partner binds to conserved CA bases in each palindromic half of the CAN/NTG (E-box) motif. The arginine (Arg) confers specificity for CACGTG versus CAGCTG E-boxes by directly contacting the central G of the G-box. Histidine (His) has asymmetrical contact and also interacts with the G residue complementary to the first C in the G-box (Ellenberger et al. 1994; Ferre-D Amare et al. 1994; Ma et al. 1994; Shimizu et al. 1997; Fuji et al. 2000). SebHLH protein has the conserved His (H5), Glu (E9), and Arg (R12) residues that constitute the G-box (CACGTG) recognition motif in its basic region (Fig. 1). As shown in Fig. 4, SebHLH protein bound E-box (CATGTG) and G-box (CACGTG) elements in the region from −179 to −53 in the SeFAD2 gene promoter. The external C and G sequences in the E- and G-box motifs were essential for the interaction between SebHLH protein and E- and G-box motifs, but mutation of the internal ACGT core sequence of the G-box motif did not influence their interaction. This result suggests that the external C and G nucleotides in the palindromic halves (CAN/NTG) of the E- and G-box motifs are the most critical sequences for SebHLH protein binding to E- and G-box motifs. Moreover, the flanking nucleotides outside of the hexanucleotide core have been shown to play a role in binding specificity (Littlewood and Evan 1998; Atchley et al. 1999; Massari and Murre 2000), and there is evidence that a loop residue in the protein functions in DNA binding through elements that lie outside of the core recognition sequence (Nair and Burley 2000). The flanking nucleotide sequences of the E- and G-box motifs in the region from −179 to −53 in the SeFAD2 gene promoter that are involved in the interaction with SebHLH protein remain to be investigated.
The N-terminal region (amino acids 1 to 229) of the SebHLH protein, which shares moderate sequence conservation with other plant bHLH proteins, contains a putative nuclear localization signal (NLS), the KRKRK sequence, at amino acids 163–167 (Fig. 1). Subcellular localization assays revealed that SebHLH protein was targeted to the nucleus, suggesting that the KRKRK sequence probably functions in nuclear targeting (Fig. 3). Typically, the NLS is the sequence Pro-Lys-Lys-Lys-Arg-Lys (PKKKRK) (Jans 1995). The N-terminal domain of the R gene product Lc, which is involved in the control of anthocyanin synthesis in maize, was identified as the sequence DRRAAPARPA from residues 100 to 109 (Ludwig et al. 1989). However, this NLS is not conserved among other maize B, Antirrhinum DEL, and bean PG1 bHLH transcription factors (Radicella et al. 1991; Goodrich et al. 1992; Kawagoe and Murai 1996).
Transcriptional activators are composed of distinct domains including separable DNA-binding and activation domains. The activation domains have been classified into acidic-type activation domains, glutamine-rich domains, and proline-rich domains, according to the type of amino acid enrichment in this domain (Triezenberg 1995). In acidic-type activation domains, specific patterns of hydrophobic and aromatic amino acids were reported to be equally, or even more, critical than the acidic amino acids (Regier et al. 1993; Drysdale et al. 1995). Similar hydrophobic and aromatic amino acid patterns and acidic amino acid-rich sequences were scattered in the N-terminal region of SebHLH protein (Fig. 1). The N-terminal region (amino acids 1–229) of SebHLH protein functioned as a transcription activation domain in yeast (Fig. 5). When SebHLH protein under the CaMV35S promoter was co-expressed with GUS reporter gene driven by E- and G-box motifs in developing sesame seeds and transgenic Arabidopsis leaves, GUS activity increased approximately 11-fold and 2-fold, respectively (Fig. 6B, C). The acidic-type activation domain was observed in the N-termini of several bHLH transcription factors, including Arabidopsis MYC (AtMYC2), MYB (AtMYB2), and ICE1 proteins (Urao et al. 1993; Abe et al. 1997; Chinnusamy et al. 2003). The AtMYC2 and AtMYB2 transcription factors specifically bound the MYC and MYB recognition sites, respectively, in the 67-bp regions of the rd22 and AtADH1 genes and these sites function as cis-acting elements in drought and ABA-induced expression of these genes. Both AtMYC2 and AtMYB2 activated the transcription of the GUS gene fused to the 67-bp region of the rd22 promoter in Arabidopsis leaf protoplasts (Abe et al. 1997, 2003). ICE1 bound specifically to MYC recognition sequences (CANNTG) in the CBF3 promoter and activated the transcription of CBF genes in the cold (Chinnusamy et al. 2003).
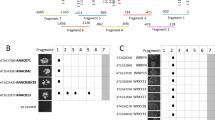
In Arabidopsis, the AtbHLH protein family consists of at least 147 members. Based on the amino acid sequence of the bHLH domain in the AtbHLH proteins, 109 and 11 proteins are predicted to be E-box binders and non-E-box binders, respectively. Eighty-nine of 109 E-box binders are categorized as G-box binders (Toledo-Oritz et al. 2003). Based on the conservation of amino acid sequence in the bHLH domain and of DNA-binding capacity, SebHLH protein is considered a typical G-box binder. The SebHLH gene was preferentially expressed in non-photosynthetic tissues, seeds and roots (Fig. 2). According to previous reports, the E- and G-box elements in the SeFAD2, Cs-ACP1, and Cs-4PAD promoters were essential cis-elements for seed-specific expression (Kim et al. 2005, 2006). Arabidopsis fatty acid elongase gene (FAE1), which catalyzes very-long-chain fatty acid biosynthesis, was specifically expressed in embryonic tissues. Two E-box elements are found at −105 to −100 (CACATG) and at −358 to −353 (CACATG) in the FAE1 promoter (Rossak et al. 2001). Interestingly, the E- and G-box elements are concentrated in the region from −250 to −50 in the promoters of acyl-CoA-diacylglycerol acyltransferase (At2g19450), phosphatidylcholine:diacylglycerol acyltransferase (At3g44830), several oil-body oleosins (At3g01570, At3g18570, At3g27660, At5g40420, and At5g51210) and two caleosins (At4g26740 and At5g55240) genes, which are involved in triacylglycerol synthesis (http://www.plantbiology.msu.edu/lipid/genesurvey/EST_data.htm; Beisson et al. 2003; http://www.arabidopsis.org) (Fig. 7). Based on the Arabidopsis microarray analysis database, GENEVESTIGATOR (https://www.genevestigator.ethz.ch/), these genes were specifically expressed in developing Arabidopsis seeds. In conclusion, SebHLH transcription factor binds to E- or G-box elements in the SeFAD2 gene promoter and contains domains for DNA binding and transcriptional activation. Through their interaction with E- and G-box cis-elements, the bHLH transcription factors, including SebHLH protein, probably play a key role in transcriptional regulation of genes involved in storage lipid biosynthesis and accumulation during seed development.
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153:292–305
Atchley WR, Terhalle W, Dress A (1999) Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol 48:501–516
Bao X, Katz S, Pollard M, Ohlrogge JB (2002) Carboxylic fatty acids in plants: biochemical and molecular genetic characterization of cyclopropane fatty acid synthesis of Sterculia foetida. Proc Natl Acad Sci USA 99:7172–7177
Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, Mhaske VB, Cho Y, Ohlrogge JB (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132:681–697
Bradford MM (1976) A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Breeden L, Nasmyth K (1985) Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol 50:643–650
Cahoon EB, Shanklin J, Ohlrogge JB (1992) Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA 89:11184–11188
Cahoon EB, Ripp KG, Hall SE, McGonigle B (2002) Transgenic production of epoxy fatty acids by expression of a cytochrome P450 enzyme from Euphorbia lagascae seed. Plant Physiol 128:615–624
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Crowe AJ, Abenes M, Plant A, Moloney MM (2000) The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci 151:171–181
Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36:521–528
Drysdale CM, Dueñas E, Jackson BM, Reusser U, Braus GH, Hinnebusch AG (1995) The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol 15:1220–1233
Ellenberger T, Fass D, Arnaud M, Harrison SC (1994) Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop helix dimer. Genes Dev 8:970–980
Ferre-D’Amare AR, Pognonec P, Roeder RG, Burley SK (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J 13:180–189
Fuji Y, Shimizu T, Toda T, Yaganida M, Hakoshima T (2000) Structural basis for the diversity of DNA recognition by bZip transcription factors. Nat Struct Biol 7:889–893
Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68:955–964
Heppard EP, Kinney AJ, Stecca KL, Miao GH (1996) Developmental and growth temperature regulation of two different microsomal ω-6 desaturase genes in soybeans. Plant Physiol 110:311–319
Jans DA (1995) The regulation of protein transport to the nucleus by phosphorylation. Biochem J 311:705–716
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jin UH, Lee JW, Chung YS, Lee JH, Yi YB, Kim YK, Hyung NI, Pyee JH, Chung CH (2001) Characterization and temporal expression of a ω-6 fatty acid desaturase cDNA from sesame (Sesamum indicum L.) seeds. Plant Sci 161:935–941
Kawagoe Y, Murai N (1996) A novel basic region/helix-loop-helix protein binds to a G-box motif CACGTG of the bean storage protein b-phaseolin gene. Plant Sci 116:47–57
Kim MJ, Shin JS, Kim JK, Suh MC (2005) Genomic structures and characterization of the 5′-flanking regions of acyl carrier protein and Δ4-palmitoyl-ACP desaturase genes from Coriandrum sativum. Biochim Biophys Acta 1730:235–244
Kim MJ, Kim HJ, Shin JS, Chung CH, Ohlrogge JB, Suh MC (2006) Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′UTR intron. Mol Genet Genomics 276:351–368
Kinney AJ (1994) Genetic modification of the storage lipids of plants. Curr Opin Biotechnol 5:144–151
Kinney AJ (1996) Development of genetically engineered soybeans for food applications. J Food Lipids 3:273–292
Kinney AJ, Cahoon EB, Hitz WD (2002) Manipulating desaturase activities in transgenic crop plants. Biochem Soc Trans 30:1099–1103
Klein TM, Fromm M, Weissinger A, Tomas D, Schaaf S, Sletten K, Sanford JC (1988) Transfer of foreign genes into intact maize cells with high-velocity microprojectiles. Proc Natl Acad Sci USA 85:4305–4309
Ledent V, Vervoort M (2001) The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res 11:754–770
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184
Littlewood TD, Evan GI (1998) Basic helix-loop-helix transcription factors. Oxford University Press, Oxford
Liu Q, Singh S, Green A (2000) Genetic modification of cotton seed oil using inverted-repeat gene-silencing techniques. Biochem Soc Trans 28:927–929
Ludwig SR, Habera LF, Dellaporta SL, Wessler SR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc homology region. Proc Natl Acad Sci USA 86:7092–7096
Ma PCM, Rould MA, Weintraub H, Pabo CO (1994) Crystal structure MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77:451–450
Maruyama I, Rakow TL, Maruyama HI (1995) cRACE: a simple method for identification of the 5′-end of mRNAs. Nucleic Acids Res 23:3796–3797
Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol 20:429–440
Moon H, Smith MA, Kunst L (2001) A condensing enzyme from the seeds of Lesquerella fendleri that specifically elongates hydroxy fatty acids. Plant Physiol 127:1635–1643
Murre C, McCaw PS, Baltimore D (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783
Nair SK, Burley SK (2000) Functional genomics: recognizing DNA in the library. Nature 404:715–718
Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6:147–158
Pirtle IL, Kongcharoensuntorn W, Nampaisansuk M, Knesek JD, Chapman KD, Pirtle RM (2001) Molecular cloning and functional expression of the gene for a cotton Δ-12 fatty acid desaturase (FAD2). Biochim Biophys Acta 1522:122–129
Radicella JP, Turks D, Chandler VL (1991) Cloning and nucleotide sequence of cDNA encoding B-Peru, a regulatory protein of the anthocyanin pathway in maize. Plant Mol Biol 17:127–130
Regier JL, Shen F, Triezenberg SJ (1993) Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA 90:883–887
Rossak M, Smith M, Kunst L (2001) Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol Biol 46:717–725
Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol 49:611–641
Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T (1997) Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J 16:4689–4697
Slocombe SP, Cummins I, Paul Jarvis R, Murphy DJ (1992) Nucleotide sequence and temporal regulation of a seed-specific Brassica napus cDNA encoding a stearoyl-acyl carrier protein (ACP) desaturase. Plant Mol Biol 20:151–155
Somerville C, Browse J, Jaworski JG, Ohlrogge J (2000) Lipids. In: Buchanan B, Gruissem W, Jones R (eds) Biochemistry & molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 456–527
Suh MC, Schultz DJ, Ohlrogge JB (1999) Isoforms of acyl carrier protein involved in seed-specific fatty acid synthesis. Plant J 17:679–688
Suh MC, Kim MJ, Hur C-G, Bae JM, Park YI, Kang C-W, Ohlrogge JB (2003) Comparative analysis of expressed sequence tags from Sesamum indicum and Arabidopsis thaliana developing seeds. Plant Mol Biol 52:1107–1123
Tang GQ, Novitzky WP, Carol Griffin H, Huber SC, Dewey RE (2005) Oleate desaturase enzymes of soybean: evidence of regulation through differential stability and phosphorylation. Plant J 44:433–446
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770
Triezenberg SJ (1995) Structure and function of transcriptional activation domains. Curr Opin Genetics Dev 5:190–196
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Acknowledgements
The authors would like to thank Kyung Hee Paek for providing the pCAMBIA 1305.1 vector containing the minimal CaMV35S promoter and the NOS terminator and Sung Han Ok for helpful discussion about yeast one-hybrid screening. This work was supported by grants from the Agricultural Plant Stress Research Center (R11-2001-0920301-0) and the Interdisciplinary Research Program (R01-2006-000-11056-0) of the Korea Science and Engineering Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence of the SebHLH cDNA from Sesamum indicum reported here has been registered in the GenBankTM/EBI Data Bank under accession number EF397568.
Rights and permissions
About this article
Cite this article
Kim, M.J., Kim, JK., Shin, J.S. et al. The SebHLH transcription factor mediates trans-activation of the SeFAD2 gene promoter through binding to E- and G-box elements. Plant Mol Biol 64, 453–466 (2007). https://doi.org/10.1007/s11103-007-9165-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9165-8