Abstract
It has been proved feasible to generate attenuation maps (μ-maps) from cardiac SPECT using deep learning. However, this assumed that the training and testing datasets were acquired using the same scanner, tracer, and protocol. We investigated a robust generation of CT-derived μ-maps from cardiac SPECT acquired by different scanners, tracers, and protocols from the training data. We first pre-trained a network using 120 studies injected with 99mTc-tetrofosmin acquired from a GE 850 SPECT/CT with 360-degree gantry rotation, which was then fine-tuned and tested using 80 studies injected with 99mTc-sestamibi acquired from a Philips BrightView SPECT/CT with 180-degree gantry rotation. The error between ground-truth and predicted μ-maps by transfer learning was 5.13 ± 7.02%, as compared to 8.24 ± 5.01% by direct transition without fine-tuning and 6.45 ± 5.75% by limited-sample training. The error between ground-truth and reconstructed images with predicted μ-maps by transfer learning was 1.11 ± 1.57%, as compared to 1.72 ± 1.63% by direct transition and 1.68 ± 1.21% by limited-sample training. It is feasible to apply a network pre-trained by a large amount of data from one scanner to data acquired by another scanner using different tracers and protocols, with proper transfer learning.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Myocardial perfusion imaging (MPI) using cardiac single-photon emission computed tomography (SPECT) is a widely performed cardiology imaging technique for the detection, localization, and risk stratification of ischemic heart diseases.1,2,3 The attenuation of emitted photons within the patient body is a major obstacle of accurate qualitative and quantitative image analysis for cardiac SPECT.4,5 In clinical practice, even though multiple techniques can be used to estimate attenuation maps (μ-maps), the μ-maps derived from computed tomography (CT) are commonly used for the SPECT attenuation correction (AC)5,6 in clinical practice. CT-based AC can significantly increase “true-positive” and reduce “false-negative” diagnosis.7 However, stand-alone SPECT without CT transmission scanning is still the mainstream and dominating over 80% of the market share.8 Even when hybrid SPECT/CT scanning is available, issues of radiation damage, hardware expense, and SPECT-CT misalignment are still challenging.9,10
Deep-learning-based μ-map generation showed promising performance in a recent study.11 Although this approach proved the feasibility of predicting μ-maps from SPECT images, it was based upon the assumption that the training and testing datasets were acquired from the same scanner using the same tracer and acquisition protocol. Thus, when datasets are acquired using different scanners, tracers, or protocols, ideally a large amount of new data should be collected to train new networks. This time-consuming and expensive process would hinder fast clinical adoption of this approach. Transfer learning is a promising deep learning strategy that re-uses a pre-trained network for a new related problem through a fine-tuning strategy.12,13 It has shown good performance in many medical-imaging-related fields such as denoising and image transformation.14,15 Thus, in this study, we implemented transfer learning with different fine-tuning modes on the μ-map generation across different scanners. The μ-maps used in our study are those with the quality derived from CT images. Recently, a deep learning algorithm, Dual Squeeze-and-Excitation Residual Dense Network (DuRDN), showed superior performance to conventional U-Net in terms of image transformation and multi-channel information extraction.16 In this study, we implemented both widely-used U-Net17 and DuRDN and compared their performance on the transfer learning study.
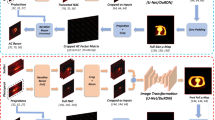
We utilized a previous network trained with 120 anonymized patient stress MPI studies acquired from a GE 850 SPECT/CT scanner. Then, part or all of the parameters of the network were fine-tuned using 10 studies acquired from a Philips BrightView SPECT/CT scanner. The performance of the network with transfer learning was compared to networks only trained by GE dataset or a small Philips dataset without fine-tuning. The ground-truth used in our study is 1) the CT-derived μ-maps; and 2) the AC SPECT images reconstructed using the CT-derived attenuation maps from the Philips dataset.
Materials and Methods
Study Datasets
The pre-training and fine-tuning datasets used in this study are listed in Table 1. The pre-training dataset was acquired from a GE NM/CT 850 SPECT/CT scanner with a 360-degree gantry rotation with heads in H-mode (two heads at 180 degrees) and the injection of 99mTc-tetrofosmin. The fine-tuning dataset was acquired from a Philips BrightView XCT scanner with a 180-degree gantry rotation with heads in L-mode (two heads at 90 degrees) in list-mode and the injection of 99mTc-sestamibi. The GE scanner utilizes a helical CT, while the Philips scanner utilizes a flat-panel cone-beam CT, resulting in μ-maps of different styles. The pre-training and validation datasets included 150 studies scanned using the GE scanner at Yale New Haven Hospital, in which 120 studies were used for training and the other 30 studies were used for validation. The fine-tuning and testing datasets included 80 studies scanned using the Philips scanner at the University of Massachusetts Medical School, in which 10 studies were used for fine-tuning and the other 70 studies were used for testing.
Deep Convolutional Neural Networks
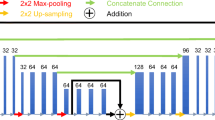
U-Net and DuRDN were implemented in this study. The detailed descriptions of DuRDN are provided in a previous study16 and Section 1 of Supplementary Material. Previous study found that DuRDN outperformed U-Net due to the densely-connected blocks18 and the dual squeeze-and-excitation blocks19. For each task in this study, the performance of U-Net and DuRDN were compared to determine the optimal deep learning algorithm for this transfer learning study.
Image Preprocessing
In the pre-training dataset, the projection data with a matrix size of 64×64×60 and a voxel size of 6.8×6.8×6.8 mm3 was acquired in a 360-degree range with a 6-degree gap between every two angles. SPECT images in photopeak (126.5-154.5 keV) and scatter (114-126 keV) windows were reconstructed with a matrix size of 64×64×64 without AC using Maximum-Likelihood Expectation-Maximization Algorithm20 (MLEM, 30 iterations, no scatter correction).
In the fine-tuning dataset, the projection data with a matrix size of 128 × 128 × 60 and a voxel size of 4.7 × 4.7 × 4.7 mm3 was acquired in a 180-degree range with a 3-degree gap between every two angles. To reduce discrepancies between the two datasets, the original projection data was bilinearly interpolated in each angle and then centrally cropped to the same size (64 × 64 × 60, 6.8 × 6.8 × 6.8 mm3) as that of the pre-training dataset. SPECT images in photopeak (126.5-154.5 keV) and scatter (92.7-125.5 keV) windows were reconstructed without AC (MLEM, 30 iterations). Sample images shown in Figure 1 illustrates the differences in the SPECT images and μ-maps between the two scanners.
Mean normalization that normalizes each image volume by its mean intensity has been proved to be an effective operation to limit the image dynamic range and improve prediction accuracy.21 Thus, all SPECT photopeak and scatter images were mean-normalized before input to the network. Previous studies also showed that concatenating both photopeak and scatter images can improve network performance.11,16 Thus, photopeak and scatter images were concatenated as two-channel inputs to the network in this study.
Strategies of Transfer Learning
The training, fine-tuning, and testing strategies are detailed in Table 2. Net A, B, C referred to testing groups of direct transition, limited-sample training, and transfer learning. Net A was trained with 120 studies and validated with 30 studies in the GE dataset, and then tested directly with 70 studies in the Philips dataset. Net B was trained with only 10 studies and tested with the other 70 studies in the Philips dataset. Net C inherited the parameters of Net A and fine-tuned with 10 studies in the Philips dataset. Then, Net C was tested with the other 70 studies in the Philips dataset. Net A, B, and C were tested with the same 70 Philips studies.
Figure 2 shows the four modes of transfer learning using DuRDN. During the fine-tuning process, partial or all parameters of the pre-trained network were frozen, while other parameters were updated. In Mode 1, only the first input (i1) and the last output (o1) layers were updated. In Mode 2, the first input (i1) and the last output upsampling (o1-o3) layers were updated. In Mode 3, the first downsampling (i1-i3) and the last output upsampling (o1-o3) layers were updated. In Mode 4, all layers (Lall) were updated.
Network Parameters
All the following network parameters were the optimal parameters after repeated tests based on Pytorch.22 Net A was pre-trained for 200 epochs with a learning rate of 2 × 10−3, a batch size of 4, and an Adam optimizer (\({\upbeta }_{1}=0.5, {\upbeta }_{2}=0.999\)). The patch size was 64 × 64 × 16 with 4 patches randomly sampled from each study. A learning rate decay policy with a step size of 1 and a decay rate of 0.99 was employed to avoid overfitting.23,24 Then, Net A was tested with the 70 studies in the Philips dataset.
Net B was trained with 10 studies in the Philips dataset for 200 epochs with the same parameters as above. Then, Net B was tested with the other 70 studies in the Philips dataset. Net B shares the same training parameters with Net A.
Net C inherited parameters from Net A and fine-tuned with 10 studies in the Philips dataset for 50 epochs using the same parameters as above. Then, Net C was tested with the other 70 studies in the Philips dataset. Net C was fine-tuned with 50 epochs. The other hyperparameters for fine-tuning Net C are the same as those for the training of Net A.
Quantitative Evaluations
The voxel-wise quantitative evaluation metrics included normalized mean square error (NMSE), normalized mean absolute error (NMAE), structural similarity index measure (SSIM), and peak signal-to-noise ratio (PSNR). 17-segment polar maps25,26 were generated for quantitative and qualitative evaluations of the SPECT AC images using Carimas.27 The mean intensity of each segment was output to compute the segment-wise absolute percent error (APE) and percent error (PE). In addition, correlation coefficients and coefficients of determination (R2) for the total 70 × 17 = 1190 segments between the predicted and ground-truth polar maps were computed to evaluate the consistency. NMSE, NMAE, PSNR are defined as:
where \({X}_{i}\) and \({Y}_{i}\) are the ith voxels of the predicted image and ground-truth image. \(N\) is the total number of voxels of the image volume. \(Max(Y)\) is the maximum voxel value of the ground-truth image. SSIM is defined as:
where \({C}_{1}={{(K}_{1}\times R)}^{2}\) and \({C}_{2}={{(K}_{2}\times R)}^{2}\) are constants to stabilize the ratios. \(R\) stands for dynamic range of pixel values. Usually, we set \({K}_{1}=0.01\) and \({K}_{2}=0.03\). \({\mu }_{X}\), \({\mu }_{Y}\), \({\sigma }_{Y}^{2}\), \({\sigma }_{X}^{2}\) are the means of \(Y\) and \(X\), deviations of \(Y\) and \(X\), and the correlation between \(Y\) and \(X\) respectively. APE and PE are defined as
where \({\text{Pred}}_{i}\) and \({\text{True}}_{i}\) represent the mean values of the ith segment in the polar maps of the predicted and ground-truth images.
An FDA-510(k)-cleared commercial software package, Wackers-Liu CQTM (WLCQ),28,29,30 was applied in this study for the quantifications of myocardial perfusion defect sizes. We quantified the defect sizes of predicted and ground-truth AC images and computed the errors of defect size at apical, mid-ventricle, basal, and apex respectively. The patterns of MPI defects compared to normal database were visualized by 2D extent iso-contours and 3D topographical maps.
Statistical Analysis
Two-tailed paired t test was performed based on the quantitative metrics above between two groups. A value of 0.05 was set as the significance level for evaluating the statistical significance.
Results
Predicted μ-Maps by Transfer Learning
Figure 3 shows sample predicted μ-maps by Net A, B, and C. It was observed that Net C predicted a more accurate μ-map than Net A and B. Net A didn’t incorporate information from the Philips scanner. Thus, the μ-map predicted by Net A showed lower resolution and fewer details which were like the μ-maps acquired from the GE scanner. The μ-maps predicted by Net B showed many artifacts due to only 10 training studies. Figure 4 shows the voxel-wise correlation maps between the predicted and ground-truth μ-maps based on 70 testing cases. The distributions of segments in Net C (Mode 4) were more concentrated around the line of identity than Net A and B, while the distributions in Net A were the most dispersed. Also, the correlation coefficients and R2 of Net C were the highest.
Table 3 shows detailed quantitative evaluations on the predicted μ-maps by Net A, B, and C based on 70 testing cases. DuRDN showed better performance than U-Net. Besides, transfer learning of Mode 4 showed the highest accuracy compared to the other 3 Modes, and Mode 1 outputted the poorest performance. Net C (Mode 4) showed better performance than Net A and B. The NMSE of Net C (Mode 4) using DuRDN was significantly lower than Net A (5.13% versus 8.24%, P < 0.001) and Net B (5.13% versus 6.45%, P < 0.001).
SPECT AC Images
Figure 5 shows sample SPECT AC images reconstructed with the predicted μ-maps by Net A, B, and C (Mode 4). Net A under-estimated the MPI intensities at the inferior and septal wall, and Net B over-corrected the MPI intensities at the lateral and anterior wall. Net C generated more accurate AC images than Net A and B. Additional sample AC images are shown in Section 2 of Supplementary Material. The voxel-wise correlation maps based on 70 testing cases and image profiles of the SPECT AC images are shown in Section 3 and 4 of Supplementary Material.
Sample SPECT AC images reconstructed with the predicted μ-maps by Net A, B, and C with both photopeak and scatter-window images as input. The AC image from Net A under-estimated the MPI intensities at the inferior and septal (white arrows). The AC image from Net C over-estimated the MPI intensities at the lateral and anterior (yellow arrows) and thus over-corrected the true defects
Table 4 shows the voxel-wise quantitative evaluations of the AC images based on 70 testing cases. The testing group of non-attenuation-correction (NAC) SPECT images was listed as the baseline. DuRDN outperformed U-Net in Net A, B, and C. Besides, transfer learning of Mode 4 showed the highest reconstruction accuracy compared to the other 3 modes, and Mode 1 output the poorest performance. Net C (Mode 4) showed better performance than Net A and B. The NMSE of Net C (Mode 4) using DuRDN was significantly lower than Net A (1.11% versus 1.72%, P < 0.001) and Net B (1.11% versus 1.68%, P < 0.001).
Quantitative Evaluations of Polar Maps
Figure 6 visualizes segment-wise correlation maps and Bland-Altman plots of the ground-truth and predicted polar maps based on 70 testing cases. In the correlation maps of Net C, the point distributions were the most concentrated around the line of identity. Besides, the correlation coefficients and R2 of Net C were higher than those of Net A and B. In the Bland-Altman plots, the point distributions of Net C were more concentrated along the mean line, and the standard deviations were lower than those of Net A and B.
Segment-wise correlation maps of polar maps (top) and Bland-Altman plots of segment PE (bottom) based on 70 testing cases. The correlation coefficients and R2 were listed on the correlation maps. The mean value and the 97.5% confidence interval (1.96 standard deviations) were listed in the Bland-Altman plots
Table 5 lists the segment-wise quantitative evaluations of 17-segment polarmaps of AC images based on 70 testing cases. The testing group of NAC SPECT images was listed as the baseline. Net C generated the lowest APE, the highest correlation coefficient, and the highest fitting R2 compared to other testing groups. The APE of Net C was significantly lower than that of Net A (4.09% versus 4.74%, P < 0.001) and Net B (4.09% versus 5.91%, P < 0.001).
Defect Size Quantifications
Figure 7 shows sample 2D extent iso-contours and 3D topographical maps of defects compared to the normal database. Net A and B under-estimated the defects at the anterior and basal-lateral compared to the ground-truth AC. In contrast, the defect patterns of Net C were closer to those of the ground-truth AC images.
Table 6 lists the MPI defect errors of predicted AC images compared to the ground-truth AC images based on 30 testing cases. It was observed that the defect size errors in the apical, mid-ventricle, basal, apex of Net C were all lower than those of Net A and B. In addition, the total defect size errors of Net C were significantly lower than those of Net A (1.17% versus 1.90%, P = 0.009) and Net B (1.17% versus 2.03%, P < 0.001).
Discussion
It is feasible to apply neural networks trained using data from one scanner to data from another scanner using different tracers and acquisition protocols by transfer learning. Transfer learning (Net C) generated better results than direct transition (Net A) or limited-sample training (Net B). The quantitative evaluations of polar map segments using Carimas and defect size using WLCQ further proved that Net C generated significantly more accurate μ-maps and SPECT AC images than those of Net A and Net B.
Four modes of fine-tuning were tested and compared. It turned out that Mode 4, with all parameters fine-tuned, generated the most accurate results in our case. Previous work on PET denoising showed that fine-tuning only the first and last layers would be a more reliable choice.14 The optimal fine-tuning implementation might be varied by different applications, data-type, and network parameters.
Our work on transfer learning shows the potentials to accelerate clinical adoption of cardiac SPECT μ-map generation for various scanners, tracers, and protocols. Once new scanners, tracers, and protocols are applied in clinical practice, there is no need to collect a large amount of new data using the SPECT/CT counterparts to re-train new networks. Instead, fine-tuning based on a previous well-trained network requires only a few new studies. The fine-tuned networks could show comparable performance with the CT-derived μ-maps for the new scanners, tracers, and protocols.
This transfer learning study is based on the generation of μ-maps as an indirect approach for AC of cardiac SPECT. There are also multiple studies investigating the direct prediction of SPECT AC images from NAC images.16,31 Our recent work made comprehensive comparisons of the direct and indirect deep-learning-based strategies of AC for cardiac SPECT, which demonstrated that indirect approaches are superior to direct approaches.32 In the AC SPECT reconstruction with attenuation maps, the overall attenuation factor of a voxel in one projection direction is the linear integral of all the attenuation coefficients along the projection line. Thus, any inaccurate voxel values of the predicted μ-maps could potentially be compensated by other neighboring voxels in the integration process. In contrast, the direct approaches generate the SPECT AC images directly without any error compensation. This could be a theoretical explanation for why indirect AC approaches outperform direct AC approaches.
The μ-maps in our studies are all derived from CT measurements. The transfer learning on μ-maps derived from other measurements, such as transmission sources, can be another potential research subject in the future.
New Knowledge Gained
For the μ-map prediction task of cardiac SPECT, transfer learning makes it possible to apply a pre-trained neural network to a totally different dataset. The prediction accuracy of transfer learning can be significantly better than just simply transferring networks or limited-sample training. The operations of transfer learning were quite simple and efficient which was promising to be put into clinical practice to accelerate the application of the μ-map prediction approach on new scanners.
Conclusions
We proposed the application of transfer learning on the μ-map prediction from SPECT images. Based on 150 GE and 80 Philips MPI studies, the voxel-wise quantitative evaluations of the predicted μ-maps and predicted AC images proved the superior performance of transfer learning to simple direct transition or limited-sample training. The DuRDN showed superior performance than conventional U-Net in terms of predicted μ-maps, and subsequently reconstructed SPECT AC images of higher accuracy.
Abbreviations
- MPI:
-
Myocardial perfusion imaging
- SPECT:
-
Single-photon emission computed tomography
- μ-Map:
-
Attenuation maps
- CT:
-
Computed tomography
- AC:
-
Attenuation correction
- DuRDN:
-
Dual squeeze-and-excitation residual dense network
- MLEM:
-
Maximum-likelihood expectation-maximization
References
Danad I, Raijmakers PG, Driessen RS, Leipsic J, Raju R, Naoum C. Comparison of coronary CT angiography, SPECT, PET, and hybrid imaging for diagnosis of ischemic heart disease determined by fractional flow reserve. JAMA Cardiol 2017;2:1100‐7.
Gimelli A, Rossi G, Landi P, Marzullo P, Iervasi G, L’abbate A, et al. Stress/rest myocardial perfusion abnormalities by gated SPECT: still the best predictor of cardiac events in stable ischemic heart disease. J Nucl Med 2009;50:546‐53.
Nishimura T, Nakajima K, Kusuoka H, Yamashina A, Nishimura S. Prognostic study of risk stratification among Japanese patients with ischemic heart disease using gated myocardial perfusion SPECT: J-ACCESS study. Eur J Nucl Med Mol Imaging 2008;35:319‐28.
Patton JA, Turkington TG. SPECT/CT physical principles and attenuation correction. J Nucl Med Technol 2008;36:1‐10.
Tavakoli M, Naij M. Quantitative evaluation of the effect of attenuation correction in SPECT images with CT-derived attenuation. Med Imaging 2019;109485U.
Blankespoor S, Xu X, Kaiki K, Brown J, Tang H, Cann C, et al. Attenuation correction of SPECT using X-ray CT on an emission-transmission CT system: myocardial perfusion assessment. IEEE Trans Nucl Sci 1996;43:2263‐74.
Patchett N, Pawar S, Sverdlov A, Miller E. Does improved technology in spect myocardial perfusion imaging reduce downstream costs? An observational study. Int J Radiol Imaging Technol 2017;3:023.
Rahman MA, Zhu Y, Clarkson E, Kupinski MA, Frey EC, Jha AK. Fisher information analysis of list-mode SPECT emission data for joint estimation of activity and attenuation distribution. Inverse Prob 2020;36:084002.
Sakoshi M, Matsutomo N, Yamamoto T, Sato E. Effect of misregistration between SPECT and CT images on attenuation correction for quantitative bone SPECT imaging. Nihon Hoshasen Gijutsu Gakkai Zasshi 2018;74:452‐8.
Saleki L, Ghafarian P, Bitarafan-Rajabi A, Yaghoobi N, Fallahi B, Ay MR. The influence of misregistration between CT and SPECT images on the accuracy of CT-based attenuation correction of cardiac SPECT/CT imaging: Phantom and clinical studies. Iran J Nucl Med 2019;27:63‐72.
Shi L, Onofrey JA, Liu H, Liu Y-H, Liu C. Deep learning-based attenuation map generation for myocardial perfusion SPECT. Eur J Nucl Med Mol Imaging 2020;47:2383‐95.
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285‐98.
Torrey L, Shavlik J, et al. Transfer learning. In: Olivas ES, et al. editors. Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI Global: Hershey; 2010. p. 242‐64.
Liu H, Wu J, Lu W, Onofrey JA, Liu Y-H, Liu C. Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose PET. Phys Med Biol 2020;65:185006.
Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: Understanding transfer learning for medical imaging. http://arxiv.org/abs/190207208 2019.
Chen X, Zhou B, Shi L, Liu H, Pang Y, Wang R, et al. CT-free attenuation correction for dedicated cardiac SPECT using a 3D dual squeeze-and-excitation residual dense network. J Nucl Cardiol 2021. https://doi.org/10.1007/s12350-021-02672-0.
Ronneberger O, Fischer P, Brox T, et al. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, et al. editors. International conference on medical image computing and computer-assisted intervention. Springern Cham; 2015. p. 234‐41.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 4700-8.
Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. p. 7132-41.
Wieczorek H. The image quality of FBP and MLEM reconstruction. Phys Med Biol 2010;55:3161.
Onofrey JA, Casetti-Dinescu DI, Lauritzen AD, Sarkar S, Venkataraman R, Fan RE et al. Generalizable multi-site training and testing of deep neural networks using image normalization. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019); 2019. p. 348-51.
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Pytorch: An imperative style, high-performance deep learning library. Adv Neural Inf Process Syst 2019;32:8026‐37.
Smith LN. A disciplined approach to neural network hyper-parameters: Part 1—learning rate, batch size, momentum, and weight decay. http://arxiv.org/abs/180309820 2018.
You K, Long M, Wang J, Jordan MI. How does learning rate decay help modern neural networks? http://arxiv.org/abs/190801878 2019.
Zoccarato O, Marcassa C, Lizio D, Leva L, Lucignani G, Savi A, et al. Differences in polar-map patterns using the novel technologies for myocardial perfusion imaging. J Nucl Cardiol 2017;24:1626‐36.
Gutierrez M, Furuie S, Rebelo M, Moura L, Moro C, Meio C et al. A polar map representation of myocardial kinetic energy from Gated SPECT. In: Proceedings of 18th annual international conference of the IEEE Engineering in Medicine and Biology Society; 1996. p. 660-1.
Nesterov SV, Han C, Mäki M, Kajander S, Naum AG, Helenius H, et al. Myocardial perfusion quantitation with 15 O-labelled water PET: high reproducibility of the new cardiac analysis software (Carimas™). Eur J Nucl Med Mol Imaging 2009;36:1594‐602.
Liu Y-H, Sinusas AJ, Khaimov D, Gebuza BI, Frans JT. New hybrid count-and geometry-based method for quantification of left ventricular volumes and ejection fraction from ECG-gated SPECT: methodology and validation. J Nucl Cardiol 2005;12:55‐65.
Liu Y-H, Sinusas AJ, DeMan P, Zaret BL, Frans J. Quantification of SPECT myocardial perfusion images: methodology and validation of the Yale-CQ method. J Nucl Cardiol 1999;6:190‐203.
Liu Y-H. Quantification of nuclear cardiac images: the Yale approach. J Nucl Cardiol 2007;14:483‐91.
Yang J, Shi L, Wang R, Miller EJ, Sinusas AJ, Liu CJ, et al. Direct attenuation correction using deep learning for cardiac SPECT: A feasibility study. J Nucl Med 2021;62:1645‐52.
Chen X, Zhou B, Xie H, Shi L, Liu H, Holler W, et al. Direct and indirect strategies of deep-learning-based attenuation correction for general purpose and dedicated cardiac SPECT. Eur J Nucl Med Mol Imaging 2022. https://doi.org/10.1007/s00259-022-05718-8.
Acknowledgments
This work is supported by NIH Grants R01HL154345 and R01HL122484.
Author contribution
All authors contribute to the study’s conception and design. Conception and design of this study: XC, BZ, MK, and CL. Data acquisition: XC, HP, KJ, MK, and CL. Network implementation and code writing: XC and BZ. Image reconstruction: XC, HP, and HL. Image analysis: XC, HP, YL, BZ, MK, and CL. Statistical analysis: XC and HL. Clinical evaluations: XC and YL. Writing of the first draft of the manuscript: XC. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets used in the current study are available from the corresponding author upon reasonable request and with permission of Yale University and University of Massachusetts.
Disclosure
CL, HL, XC, and BZ are named inventors of non-provisional patent applications related to this work that Yale University has filed. HP, KJ, YL, MK have no potential conflicts.
Ethics approval
The retrospective use of the anonymized data in this study was approved by the Institutional Review Boards of Yale University and University of Massachusetts Medical School.
Informed consent
Not applicable.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors of this article have provided a PowerPoint file, available for download at SpringerLink, which summarises the contents of the paper and is free for re-use at meetings and presentations. Search for the article DOI on SpringerLink.com.
The authors have also provided an audio summary of the article, which is available to download as ESM, or to listen to via the JNC/ASNC Podcast.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, X., Hendrik Pretorius, P., Zhou, B. et al. Cross-vender, cross-tracer, and cross-protocol deep transfer learning for attenuation map generation of cardiac SPECT. J. Nucl. Cardiol. 29, 3379–3391 (2022). https://doi.org/10.1007/s12350-022-02978-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12350-022-02978-7