Abstract
Purpose
We aimed to evaluate the performance of deep learning-based generalization of ultra-low-count amyloid PET/MRI enhancement when applied to studies acquired with different scanning hardware and protocols.
Methods
Eighty simultaneous [18F]florbetaben PET/MRI studies were acquired, split equally between two sites (site 1: Signa PET/MRI, GE Healthcare, 39 participants, 67 ± 8 years, 23 females; site 2: mMR, Siemens Healthineers, 64 ± 11 years, 23 females) with different MRI protocols. Twenty minutes of list-mode PET data (90–110 min post-injection) were reconstructed as ground-truth. Ultra-low-count data obtained from undersampling by a factor of 100 (site 1) or the first minute of PET acquisition (site 2) were reconstructed for ultra-low-dose/ultra-short-time (1% dose and 5% time, respectively) PET images. A deep convolution neural network was pre-trained with site 1 data and either (A) directly applied or (B) trained further on site 2 data using transfer learning. Networks were also trained from scratch based on (C) site 2 data or (D) all data. Certified physicians determined amyloid uptake (+/−) status for accuracy and scored the image quality. The peak signal-to-noise ratio, structural similarity, and root-mean-squared error were calculated between images and their ground-truth counterparts. Mean regional standardized uptake value ratios (SUVR, reference region: cerebellar cortex) from 37 successful site 2 FreeSurfer segmentations were analyzed.
Results
All network-synthesized images had reduced noise than their ultra-low-count reconstructions. Quantitatively, image metrics improved the most using method B, where SUVRs had the least variability from the ground-truth and the highest effect size to differentiate between positive and negative images. Method A images had lower accuracy and image quality than other methods; images synthesized from methods B–D scored similarly or better than the ground-truth images.
Conclusions
Deep learning can successfully produce diagnostic amyloid PET images from short frame reconstructions. Data bias should be considered when applying pre-trained deep ultra-low-count amyloid PET/MRI networks for generalization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
With the development of the imaging-based “AT(N) framework [1]” for neurodegenerative research, simultaneous amyloid positron emission tomography/magnetic resonance imaging (PET/MRI) provides a potential “one-stop shop” imaging exam for dementia research, diagnosis, and clinical trials [2,3,4]. PET allows the acquisition of the amyloid (A) and tau (T) biomarkers, hallmarks of Alzheimer’s disease neuropathology [3, 5,6,7,8], while MRI with its exquisite soft tissue contrast allows for imaging cortical atrophy, representative of neurodegeneration (N) [9].
However, multiple factors will affect the utility of PET. The scan time, the cost of the tracer, and the radiation given for the PET imaging might all be limiting factors, affecting respectively the logistics, economics, and the scanned subjects. Since PET imaging quality is highly dependent upon the number of detected events (counts), reduced dose or reduced scan time typically results in lower signal-to-noise images. Previous work to tackle this issue involves direct interpretation of the low-count images [10,11,12] or with machine learning-based methods [13, 14] but using few subjects collected at a single site. To increase the utility of this hybrid modality in ultra-low-dose imaging, we have previously trained deep learning (DL) networks using a U-net structure with residual learning [15, 16] to generate diagnostic amyloid PET images from PET/MRI scans with simulated ultra-low injected radiotracer dose [17].
To move single-site studies to multi-center studies, traditionally for multi-center machine learning applications, a DL network would be trained centrally, on data collected using a harmonized protocol from multiple sites. However, privacy issues such as sharing patient information and data ownership often limit the ability to collect a large number of medical images from multiple institutions [18,19,20]. Moreover, when a pre-trained network is applied to data acquired at other sites, performance of the network may decrease [21]. To overcome this data bias, a sequential training approach may be considered for network generalization, a method sometimes known as “transfer learning” [22]. Under this approach, the network may be applied to data acquired on different scanner models, with different scan protocols, and reconstructed with different methods or parameters. Most previous machine learning work using data from different sites assume that the same image types exist for input to the network [18,19,20, 23]. However, it is more realistic that different sites with different scanners also employ different scan protocols that may not include all the inputs required to directly apply an algorithm trained elsewhere. Local populations with different disease prevalence might also affect the results, and there is evidence that for optimal performance, networks should be trained depending on the target study population [24].
In this project, we investigated various approaches to apply a pre-trained convolutional neural network (CNN) originally meant to denoise ultra-low-dose amyloid PET/MRI on new cases from a separate institution, collected on a different PET/MRI scanner, and with different reconstruction parameters and MR sequences. Moreover, we focused on whether these approaches can improve ultra-low-count PET data obtained from severely reduced imaging duration (1 min, far lower counts than previous work in literature [10, 14]). A better understanding of how to best apply a pre-trained network to a new population should enable the optimal performance for generalizing DL-based image synthesis tasks.
Methods
This study was approved by the local institutional review boards. Written informed consent for imaging was obtained from all participants or an authorized surrogate decision-maker.
PET/MRI data acquisition: Site 1
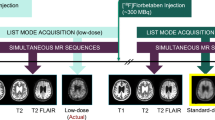
Forty datasets from 39 participants (23 female, 67 ± 8 years; one female participant was scanned twice, 9 months apart) with MRI and PET data were simultaneously acquired on scanner 1: an integrated PET/MRI scanner with time-of-flight capabilities (SIGNA PET/MR, GE Healthcare). T1-weighted, T2-weighted, and T2 FLAIR morphological MR images were acquired, with the parameters listed in Chen et al. [17].
330 ± 30 MBq of the amyloid radiotracer [18F]florbetaben (Life Molecular Imaging, Berlin, Germany) was injected intravenously with PET acquired 90–110 min post-injection. The list-mode PET data were reconstructed for the ground-truth (i.e., reconstructed from 20-min full-dose PET acquisitions) image as well as a random subset containing 1/100th of the events (also taking the different randoms rate into account) to produce a low-dose PET image [25]. Time-of-flight ordered-subsets expectation-maximization (OSEM), with two iterations and 28 subsets, accounting for randoms, scatter, dead-time, and attenuation, and a 4 mm full-width at half-maximum post-reconstruction Gaussian filter was used for all PET images. MR attenuation correction was performed using the vendor’s atlas-based method [26].
PET/MRI data acquisition: site 2
Analysis was performed on 40 participants (23 female, 64 ± 11 years) who were scanned on scanner 2 (mMR, Siemens Healthineers). Only the T1-weighted and T2-weighted (no T2-FLAIR-weighted) MR images (parameters in Table S1) were acquired. [18F]florbetaben (283 ± 10 MBq) was injected with PET and MRI acquired 90–110 min after injection. The 20-min list-mode PET data was reconstructed for the ground-truth image. The first minute of PET acquisition was reconstructed to produce low-count, short-time (5% of the original) PET images. OSEM, with 8 iterations and 21 subsets, accounting for randoms, scatter, dead-time, and attenuation, and a 3 mm full-width at half-maximum post-reconstruction Gaussian filter was used for all PET images, based on the standard protocol at site 2. MR attenuation correction was performed using RESOLUTE [27].
Image preprocessing
The site 2 ground-truth PET images were resliced based on the site 1 PET volumes: 89 2.78-mm-thick slices with 256-by-256 matrix size (1.17 × 1.17 mm2 in-plane voxel size); to compensate for any residual motion between the modalities and sequences, all other images from site 2 were co-registered to the resliced ground-truth PET image following the pipeline outlined in Chen et al. [17].
CNN training and testing
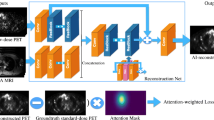
We trained a CNN (using a “U-net” structure [16]) with the structure, hyperparameters, and training algorithm described in Chen et al., using site 1 data (32 training datasets, chosen randomly) [17]. The network inputs are multi-contrast MR (T1-, T2-, and T2 FLAIR-weighted) and the ultra-low-dose PET images. The network was trained using residual learning, based on the ground-truth PET image [15] (Fig. 1). The remaining datasets were used as the test set. In the previous work, 5-fold cross-validation was used, resulting in five trained networks; one was randomly selected for this study. Network training details and network selection can be found in the Supplementary Materials.
Schematic of the U-Net used in this work and its inputs and outputs. The arrows denote computational operations, and the tensors are denoted by boxes with the number of channels indicated above each box. Note that for the site 2 data in which T2-FLAIR was not available, this input was replaced with the T1-weighted image. BN batch normalization, Conv convolution, ReLU rectified linear unit activation, tanh hyperbolic tangent
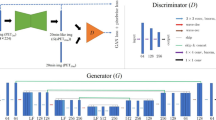
To apply this trained network to site 2 data, two preliminary studies were conducted: for the missing T2-FLAIR channel, T1-weighted images were used as inputs. Site 2 1-min images were chosen as inputs to the network. The choices for time reduction and contrast replacement was made based on the peak signal-to-noise ratio (PSNR) between the low-count and its corresponding ground-truth image (details outlined in the Supplementary Materials [28]). Four approaches were investigated (schematically shown in Fig. 2): in the first (method A), the site 2 data were input directly into the network trained at site 1, with no attempts to account for site differences. In the second (method B), network weights were initialized with the final weights from site 1 data, then further tuned for 100 epochs with a learning rate of 0.0001. 5-fold cross-validation (i.e., 32 datasets for training, 8 for testing per network trained) was used during transfer learning. For method C, a new network was trained from random initialization on site 2 data only. Finally, method D was trained from random initialization on all data from both scanners (32 cases from site 1 and 32 cases from site 2, with testing on 8 cases from site 2). The network inputs for methods C and D are the multi-contrast MR images (T1- and T2-weighted images only) and the low-count PET image. The hyperparameters and training follow that implemented in Chen et al. [17].
The various methods used in this work for network generalization. Five-fold cross-validation was used to utilize all datasets (when coming from the same site) for testing and training. The training and testing data for method A were from different sites; therefore, all site 2 data could be applied to one network
Assessment of image quality
Dataset-specific FreeSurfer-based [29, 30] T1-derived brain masks were used for voxel-based analyses. For each axial slice, the image quality of the synthesized PET images and the original low-count PET images within the brain mask were assessed using peak signal-to-noise ratio (PSNR), structural similarity (SSIM) [31], and root mean square error (RMSE), where:
and IGT denotes the ground-truth image (mean μGT, variance σGT2, maximum pixel value MAX(IGT)), Ix denotes the image to be tested, σGT,x denotes the covariance of IGT and Ix, C1 and C2 are the square of 0.01 and 0.03 times the pixel value range of IGT, respectively, MSE denotes the mean squared error, and || . || denotes the Frobenius norm.
The respective metrics for each slice were then averaged (weighted by the number of voxels in the brain mask). A repeated measures analysis of variance (ANOVA) test followed by pair-wise paired t-tests were conducted at the p = 0.05/3 level with Bonferroni correction for multiple (three) comparisons, to compare the values of the image quality metrics across the different image processing methods. Image metric improvement from the low-count to the synthesized image was also calculated for data from both sites; this comparison was conducted using the one-tailed two-sample unequal variance t-test (also at the p = 0.05/3 level).
Region-based analyses
Region-based analyses were carried out to assess the agreement of the tracer uptake between images as well as differentiating between amyloid positive and negative images. FreeSurfer-based cortical parcellations and cerebral segmentations based on the Desikan-Killiany Atlas [32] were created, yielding a maximum of 111 regions per dataset. Mean standard uptake value ratios (SUVR, normalized to the cerebellar cortex) in 4054 total regions from 37 successful segmentations were calculated and compared between methods and evaluated by Bland-Altman plots. Next, a composite ROI was derived from the frontal, parietal, lateral temporal, occipital, anterior and posterior cingulate cortices and the mean composite SUVR was calculated by Hermes BRASS software for all datasets, again with the cerebellar cortex as reference. Using the clinical readers’ majority ground-truth reads, receiver operator characteristic (ROC) analysis was carried out using different SUVRs as cutoff values for amyloid positive vs. negative and the area under the ROC curve (AUC) was calculated for each image type; the AUCs were analyzed according to DeLong et al. [33] for significance and a non-inferiority threshold of 5% was set to compare the DL-based AUCs and the ground-truth AUC. Cohen’s d [34] was also calculated for the composite SUVRs between amyloid positive and negative groups of each image type.
Clinical readings
All PET images of each dataset were anonymized, their series numbers were randomized, then presented to four readers (H.B., O.S., G.Z.: board-certified physicians with 10+ years’ experience of reading amyloid images; M.E.I.K.: resident with 4 years’ experience) for independent reading. The amyloid uptake status (positive, negative, uninterpretable) of each image was determined; the ground-truth amyloid status was based on the majority read from the ground-truth images. A fifth reader (G.D.: board-certified physician with 10 years’ experience) was a tiebreaker for a single case of a 2–2 positive-negative reading. Reader agreement was assessed using the Krippendorff’s alpha test. The accuracy, sensitivity, and specificity were calculated for the readings of the short-time and synthesized images. Symmetry tests were also carried out to examine whether the readings produced an equal number of false positives and negatives.
For each PET image, the four physicians also assigned an image quality score on a five-point scale: 1 = uninterpretable, 2 = poor, 3 = adequate, 4 = good, 5 = excellent. Also, these scores were dichotomized into 1–3 vs. 4–5 to analyze the percentage of images with high scores.
Results
Assessment of image quality
Visually, all synthesized images showed marked noise reduction (Fig. 3). For site 2 data, the ANOVA test showed that the four methods yielded different results than the low-count images and from each other (Table 1), indicating that image quality improved from the low-count images but improved the least with method A (i.e., simply applying the site 1 model to site 2 data). Pair-wise t tests showed that image quality improved the most using method B (Fig. 4; p < 0.05/3 for all metric comparisons). Comparing the metric difference (improvement from the low-dose/short-time images to the images output from different trained/tuned networks) across sites, all methods showed more improvement (p < 0.05/3) in SSIM than that in site 1 data. For RMSE, methods B, C, and D results showed more improvement (p < 0.05/3) than the site 1 results, while for PSNR, methods C and D showed similar improvement (p > 0.05/3) and method B showed more improvement (p < 0.05/3) than the site 1 results. Method A results showed less improvement (p < 0.05/3) compared with the site 1 results in both PSNR and RMSE.
Representative amyloid positive (top)/negative (bottom) images, with T1-weighted MRI and the corresponding PET images. Difference images between the ground-truth and the other images are also shown. All synthesized images show marked noise reduction. However, method A images are blurrier than the other synthesized images. Network training methods: A, direct application of pre-trained network; B, transfer learning starting with pre-trained network; C, training new network from scratch; D, training new network with combined datasets
The peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and root mean square error (RMSE) of the synthesized and low-count images compared to the ground-truth image. Site 1 data (used to train the original network in Chen et al. [17]) are also shown for comparison. Network training methods: A, direct application of pre-trained network; B, transfer learning starting with pre-trained network; C, training new network from scratch; D, training new network with combined datasets
Region-based analyses
SUVRs derived from method B had the least variability from the ground-truth SUVRs (Fig. 5). Out of all image types, images generated by method B also yielded the highest AUC (Fig. 6) and the largest Cohen’s d values to distinguish positive and negative amyloid status (Table 2). Comparing the four DL method-based AUCs with the low-count AUC and the ground-truth AUC yielded p values of 0.46 and 0.70, respectively, and the 95% confidence interval of the DL-based AUCs fell within the non-inferiority threshold of the ground-truth AUC.
Bland-Altman 2-D histograms of regional standardized uptake value ratios (SUVRs) compared between methods (ground-truth to low-count and methods A, B, C, and D) across all datasets with FreeSurfer segmentations (n = 37). The scale bar denotes the number of data points in each pixel; the solid and dashed lines denote the mean and 95% confidence interval of the SUVR differences respectively. GT ground-truth. Network training methods: A, direct application of pre-trained network; B, transfer learning starting with pre-trained network; C, training new network from scratch; D, training new network with combined datasets
The receiver operating characteristic (ROC) curves of the standardized uptake value ratios (SUVRs) from the various image types used to differentiate between amyloid positive and negative readings. Network training methods: A, direct application of pre-trained network; B, transfer learning starting with pre-trained network; C, training new network from scratch; D, training new network with combined datasets
Clinical readings
Inter-reader agreement on amyloid uptake status was high (Krippendorff’s alpha > 0.7) for all methods except for method A (Krippendorff’s alpha = 0.5) and the readings from all four readers were pooled. Seventy-six of 160 (47.5%) total reads of the ground-truth images were amyloid positive.
When comparing the accuracy, sensitivity, and specificity of the readings between the synthesized images and the ground-truth images, methods B, C, and D produced higher values than those from method A (Table 3). The accuracy of the readings from images synthesized using methods B, C, and D was high, though method B produced more false positives than false negatives (p = 0.031 for the symmetry test). For the short-time images that were interpretable (only 56% of them), the accuracy, sensitivity, and specificity of the clinical assessments were also high (Confusion matrices in Table S2).
The mean image quality scores assigned by each reader to all PET volumes are shown in Table 4 and Table S3. The results showed relevant inter-reader variability and limited agreement, and thus, no statistics were done on the readings. However, for all readers, methods B, C, and D had similar (greater than − 10%) or even higher proportions of high-scoring (i.e., 4 or 5) readings compared to those of the ground-truth images. In contrast, readings of images from method A performed worse than the other deep-learning-based methods and the ground-truth images.
Discussion
When conducting retrospective multi-center imaging studies or applying models trained on one site to another, differences such as scanner hardware, acquisition protocol, and reconstruction parameters will pose challenges in the generalization of these trained models. In this work, we were able to apply a pre-trained network to ultra-short-time duration amyloid PET/MRI data from another institution, overcoming the differences in acquisition protocol. Through further training iterations, the pre-trained network adjusted for data bias stemming from the differences in acquisition and reconstruction between institutions. Furthermore, we showed that the network could still be used in the event of missing input data; providing another structurally similar MRI contrast (the approach used in this work) as an input for the missing channel preserved the functionality of the network [28]. From this and the previous study [17], we have shown that DL-assisted extreme time-shortening and dose-reducing methods for amyloid PET/MRI can potentially increase the utility of PET imaging.
Certainly, the hyperparameter space for network tuning is vast, and methods for data/network sharing across institutions are many. However, we believe the methods investigated in this study represent four main DL-based approaches in multi-site studies: the first (method A), naïvely applying the network trained on data from one site to another. However, network tuning is needed to account for data bias from each site, evidenced by the weak performance of this method. The second method (method B) requires passing the network between sites, using pre-training from site 1 to act as the initialization of a model that is further trained on site 2 data (i.e., transfer learning). Method B represents the most extreme case of optimizing the test set results, where the network is first trained on site 1 data, then tuned with site 2 training data (tuning all layers of the network), and directly applied on the site 2 test set. The final two methods are extreme examples in which institutions approach data sharing: method C where each site keeps its own data and trains its own network for the institution’s own use, and the traditional “data-sharing” approach (method D) for multi-center studies where all data are collected and stored in a central repository for training. To simulate this approach for method D, where all data acquisition protocols are harmonized across sites, we did not include the T2 FLAIR channel during training.
Based on the evaluation metrics, selection of the optimal network training/tuning method is application-specific. In this specific study, while training an institution’s own network (method C) produces good results quantitatively and qualitatively as expected (the network is specifically trained on the image quality of the institution), for the network generalization and data sharing methods, using the pre-trained network (method B), an efficient way to allow each institution to keep its own data, provides better quantification results overall, possibly due to the image quality difference from the scanner and image processing protocols between the two sites. The U-Net architecture, which emphasizes low spatial frequencies in the input and results in a blurrier output [35], also contributes to a slightly blurrier image when using methods C and D. On the other hand, for applications involving expert readers, with methods B, C, and D performing similarly in the clinicians’ image quality preference and amyloid status readings, any of the methods would be sufficient as long as the data bias from each site is accounted for. This is not surprising since previous studies have shown that clinicians can confidently read PET images that have lower counts than that routinely acquired, though not as extremely reduced as in the current study [11, 12].
There are several limitations to this study. First, the network training and tuning methods evaluated are not exhaustive. Second, with the approach of method B, the problem of “forgetting [19, 36, 37]” how to produce an optimal image for site 1 data is unavoidable, since the pre-trained network is now tuned for site 2 data instead. However, in this project, our focus was on sharing the pre-trained network to other sites and thus in actual practice the newer networks will not be applied back on site 1 data. Finally, in this study, there are potential sources of bias such as the site 1 training dataset (there are two datasets from one participant, though the two scans are independent, with the head positioned differently, and took place 9 months apart); the readers’ experience or institution may also lead to bias. For example, we enlisted multiple readers so that a consensus reading by majority vote could be used as the ground-truth, but in terms of image quality readers 3 and 4 (belonging to the same institution) showed a clear preference for the synthesized images while readers 1 and 2 (belonging to the same institution) preferred the site 2 ground-truth images. This preference may be due to many reasons, such as the PET image quality at the readers’ original sites, or experience with reading lower count images [11]. However, variability in the image quality scores also prevented pooling scores for further statistical analyses. This reader bias also demonstrated the need for multiple readers from different sites when conducting reader studies.
Conclusion
To perform deep-learning-based ultra-short-time amyloid PET/MRI imaging using transfer learning methods, further tuning of pre-trained networks or training new networks including data acquired from the new scanner is required to overcome data bias. Sharing the network parameters between sites rather than the images themselves can be a potential way for collaboration across multiple amyloid PET/MRI sites.
References
Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–62. https://doi.org/10.1016/j.jalz.2018.02.018.
Barthel H, Schroeter ML, Hoffmann KT, Sabri O. PET/MR in dementia and other neurodegenerative diseases. Semin Nucl Med. 2015;45:224–33. https://doi.org/10.1053/j.semnuclmed.2014.12.003.
Catana C, Drzezga A, Heiss WD, Rosen BR. PET/MRI for neurologic applications. J Nucl Med. 2012;53:1916–25. https://doi.org/10.2967/jnumed.112.105346.
Drzezga A, Barthel H, Minoshima S, Sabri O. Potential clinical applications of PET/MR imaging in neurodegenerative diseases. J Nucl Med. 2014;55:47S–55S. https://doi.org/10.2967/jnumed.113.129254.
Rowe CC, Villemagne VL. Brain amyloid imaging. J Nucl Med. 2011;52:1733–40. https://doi.org/10.2967/jnumed.110.076315.
Sperling RA, Mormino EC, Schultz AP, Betensky RA, Papp KV, Amariglio RE, et al. The impact of amyloid-beta and tau on prospective cognitive decline in older individuals. Ann Neurol. 2019;85:181–93. https://doi.org/10.1002/ana.25395.
Barthel H, Gertz HJ, Dresel S, Peters O, Bartenstein P, Buerger K, et al. Cerebral amyloid-beta PET with florbetaben (18F) in patients with Alzheimer’s disease and healthy controls: a multicentre phase 2 diagnostic study. Lancet Neurol. 2011;10:424–35. https://doi.org/10.1016/S1474-4422(11)70077-1.
Villemagne VL. Selective Tau Imaging: Der Stand der Dinge. J Nucl Med. 2018;59:175–6. https://doi.org/10.2967/jnumed.117.198325.
Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, et al. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. https://doi.org/10.1093/cercor/bhn113.
Herholz K, Evans R, Anton-Rodriguez J, Hinz R, Matthews JC. The effect of 18F-florbetapir dose reduction on region-based classification of cortical amyloid deposition. Eur J Nucl Med Mol Imaging. 2014;41:2144–9. https://doi.org/10.1007/s00259-014-2842-3.
Tiepolt S, Barthel H, Butzke D, Hesse S, Patt M, Gertz HJ, et al. Influence of scan duration on the accuracy of beta-amyloid PET with florbetaben in patients with Alzheimer's disease and healthy volunteers. Eur J Nucl Med Mol Imaging. 2013;40:238–44. https://doi.org/10.1007/s00259-012-2268-8.
Schiller F, Frings L, Thurow J, Meyer PT, Mix M. Limits for reduction of acquisition time and administered activity in (18)F-FDG PET studies of Alzheimer dementia and frontotemporal dementia. J Nucl Med. 2019;60:1764–70. https://doi.org/10.2967/jnumed.119.227132.
Bland J, Mehranian A, Belzunce MA, Ellis S, McGinnity CJ, Hammers A, et al. MR-guided kernel EM reconstruction for reduced dose PET imaging. IEEE Trans Radiat Plasma Med Sci. 2018;2:235–43. https://doi.org/10.1109/TRPMS.2017.2771490.
Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep auto-context convolutional neural networks for standard-Dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017;267:406–16. https://doi.org/10.1016/j.neucom.2017.06.048.
Chen H Low-dose CT with a residual encoder-decoder convolutional neural network (RED-CNN). arXiv: arXiv. 2017.
Ronneberger O, Fischer P, Brox T U-Net: convolutional networks for biomedical image segmentation. arXiv. 2015.
Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, et al. Ultra-low-dose (18)F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290:649–56. https://doi.org/10.1148/radiol.2018180940.
Chang K, Balachandar N, Lam C, Yi D, Brown J, Beers A, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc. 2018;25:945–54. https://doi.org/10.1093/jamia/ocy017.
Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. Brainlesion. 2019;11383:92–104. https://doi.org/10.1007/978-3-030-11723-8_9.
McClure P, Zheng CY, Kaczmarzyk JR, Lee JA, Ghosh SS, Nielson D, et al. Distributed weight consolidation: a brain segmentation case study. In Proceedings of the 32nd International Conference on Neural Information Processing Systems (NIPS’18). Curran Associates Inc.: Red Hook, NY. 2018: 4097–4107.
Yune S, Lee H, Pomerantz SR, Romero JM, Kamalian S, Gonzalez RG, et al. Real-world performance of deep-learning-based automated detection system for intracranial hemorrhage. 2018 SIIM Conference on Machine Intelligence in Medical Imaging: San Francisco; 2018.
Pan SJ, Yang QA. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–59. https://doi.org/10.1109/Tkde.2009.191.
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2015;34:1993–2024. https://doi.org/10.1109/TMI.2014.2377694.
Guo J, Gong E, Fan AP, Goubran M, Khalighi MM, Zaharchuk G. Predicting (15)O-Water PET cerebral blood flow maps from multi-contrast MRI using a deep convolutional neural network with evaluation of training cohort bias. J Cereb Blood Flow Metab. 2019:271678X19888123. https://doi.org/10.1177/0271678X19888123.
Gatidis S, Wurslin C, Seith F, Schafer JF, la Fougere C, Nikolaou K, et al. Towards tracer dose reduction in PET studies: simulation of dose reduction by retrospective randomized undersampling of list-mode data. Hell J Nucl Med. 2016;19:15–8. https://doi.org/10.1967/s002449910333.
Iagaru A, Mittra E, Minamimoto R, Jamali M, Levin C, Quon A, et al. Simultaneous whole-body time-of-flight 18F-FDG PET/MRI: a pilot study comparing SUVmax with PET/CT and assessment of MR image quality. Clin Nucl Med. 2015;40:1–8. https://doi.org/10.1097/RLU.0000000000000611.
Ladefoged CN, Benoit D, Law I, Holm S, Kjaer A, Hojgaard L, et al. Region specific optimization of continuous linear attenuation coefficients based on UTE (RESOLUTE): application to PET/MR brain imaging. Phys Med Biol. 2015;60:8047–65. https://doi.org/10.1088/0031-9155/60/20/8047.
Chen KT, Schürer M, Ouyang J, Gong E, Tiepolt S, Sabri O, et al. How to generalize a deep learning model to new data lacking appropriate MR inputs? An Exploration using Ultra-low-dose Amyloid PET/MRI. Montreal: Annual Meeting ISMRM; 2019.
Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. https://doi.org/10.1006/nimg.1998.0396.
Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I Segmentation and surface reconstruction. Neuroimage. 1999;9:179–94. https://doi.org/10.1006/nimg.1998.0395.
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–12.
Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80. https://doi.org/10.1016/j.neuroimage.2006.01.021.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45.
Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Hillsdale: L. Erlbaum Associates; 1988.
Ye JC, Han Y, Cha E. Deep convolutional framelets: a general deep learning framework for inverse problems. SIAM J Imaging Sci. 2018;11:991–1048. https://doi.org/10.1137/17M1141771.
French RM. Catastrophic forgetting in connectionist networks. Trends Cogn Sci. 1999;3:128–35.
Kirkpatrick J, Pascanu R, Rabinowitz N, Veness J, Desjardins G, Rusu AA, et al. Overcoming catastrophic forgetting in neural networks. Proc Natl Acad Sci U S A. 2017;114:3521–6. https://doi.org/10.1073/pnas.1611835114.
Acknowledgments
This project was made possible by the NIH grants P41-EB015891 and P50-AG047366 (Stanford Alzheimer’s Disease Research Center), GE Healthcare, the Michael J. Fox Foundation for Parkinson’s Disease Research, the Foundation of the ASNR, and Life Molecular Imaging. The authors would also like to thank Tie Liang, EdD, for the statistical analysis.
Availability of data and material
Data was collected at the authors’ institutions and is available when requested for review.
Funding
This project was made possible by the NIH grants P41-EB015891 and P50-AG047366 (Stanford Alzheimer’s Disease Research Center), GE Healthcare, the Michael J. Fox Foundation for Parkinson’s Disease Research, the Foundation of the ASNR, and Life Molecular Imaging.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Outside submitted work: GZ-Subtle Medical Inc., co-founder and equity relationship. No other potential conflicts of interest relevant to this article exist.
Ethics approval
All procedures involving human participants were in accordance with the ethical standards of the Stanford University Institutional Review Board and the Leipzig University Ethics Committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate
Written informed consent for imaging was obtained from all participants or an authorized surrogate decision-maker.
Code availability
Custom code was used for this project.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Electronic supplementary material
ESM 1
(DOCX 1.50 mb)
Rights and permissions
About this article
Cite this article
Chen, K.T., Schürer, M., Ouyang, J. et al. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur J Nucl Med Mol Imaging 47, 2998–3007 (2020). https://doi.org/10.1007/s00259-020-04897-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-020-04897-6