Abstract
Background
The performance of left ventricular (LV) functional assessment using gated myocardial perfusion SPECT (MPS) relies on the accuracy of segmentation. Current methods require manual adjustments that are tedious and subjective. We propose a novel machine-learning-based method to automatically segment LV myocardium and measure its volume in gated MPS imaging without human intervention.
Methods
We used an end-to-end fully convolutional neural network to segment LV myocardium by delineating its endocardial and epicardial surface. A novel compound loss function, which encourages similarity and penalizes discrepancy between prediction and training dataset, is utilized in training stage to achieve excellent performance. We retrospectively investigated 32 normal patients and 24 abnormal patients, whose LV myocardial contours automatically segmented by our method were compared with those delineated by physicians as the ground truth.
Results
The results of our method demonstrated very good agreement with the ground truth. The average DSC metrics and Hausdorff distance of the contours delineated by our method are larger than 0.900 and less than 1 cm, respectively, among all 32 + 24 patients of all phases. The correlation coefficient of the LV myocardium volume between ground truth and our results is 0.910 ± 0.061 (P < 0.001), and the mean relative error of LV myocardium volume is − 1.09 ± 3.66%.
Conclusion
These results strongly indicate the feasibility of our method in accurately quantifying LV myocardium volume change over the cardiac cycle. The learning-based segmentation method in gated MPS imaging has great promise for clinical use.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Myocardial perfusion SPECT (MPS) has been one of the most important imaging modalities for the assessment of left ventricular (LV) function.1,2,3,4 Photons emitted by an injected radioactive perfusion tracer taken up by the LV myocardium are detected to reconstruct perfusion images. With electrocardiographic gating, MPS provides 8 or 16 volumetric perfusion image sets corresponding to different phases of the cardiac cycle.5 Evaluations are performed based on these images by visual and quantitative estimation of the variation of LV during the cardiac cycle.6 LV contractile functional indices can then be derived from MPS images for the diagnosis/prognosis of coronary artery disease and patient risk assessment.7,8,9,10,11,12
The fidelity of LV function assessment by MPS directly relates to the quantification accuracy of the LV myocardium volume.13,14 The measurement of LV myocardium volume starts with delineation of epicardial and endocardial boundaries on the perfusion images, and is calculated as the volume bounded by the epicardial and endocardial surface. Manual segmentation is tedious when it involves studies on multiple volumetric phase images of the cardiac cycle, and is dependent on observers’ experience. It is desirable to develop an observer-independent segmentation method to improve efficiency and reproducibility with comparable accuracy.
Current automated methods extract the epicardial and endocardial boundaries based on general assumptions and rules with empirical parameters. For example, commercially available methods estimate the profiles of the myocardium by identifying the maximal myocardial count, then applying Gaussian fitting with empirical standard deviation or threshold to extract endocardial and epicardial boundaries.15,16 This method is easy and fast to implement, though it neglects the anatomical variations and pathology abnormalities among different patients. Studies have shown that LV myocardium volume would be over- or underestimated by this method,17,18 and manual adjustments are usually required.14
In recent years, machine learning methods are being integrated into segmentation studies. They have been shown to feature better results while requiring less time than traditional methods for CT and MR images19,20,21 due to its data-driven approaches toward automatically learning image features and model parameters. Compared with these common imaging modalities, MPS images have advantages for machine learning methods in that image size is greatly reduced and with higher image contrast, which leads to more efficient extraction of global features from the whole image set during the training stage. Thus, the machine learning method is promising in automatic MPS image segmentation.
In this paper, we propose a novel machine-learning-based method to automatically segment LV myocardium by delineating its endocardial and epicardial surface, and measure its volume in gated MPS imaging. Our method uses a multi-class 3D V-Net, which is an end-to-end fully convolutional neural network. A compound loss function, which simultaneously encourages similarity and penalizes discrepancy between prediction and training datasets, was utilized in training stage to achieve excellent performance. To evaluate our proposed method, we retrospectively investigated 32 normal patients and 24 abnormal patients with clinically acquired MPS. The LV myocardium was segmented by our proposed method and compared with ground truth approved by physicians for evaluation on a total of 32 + 24 patients.
Methods and Materials
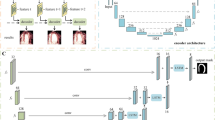
The proposed SPECT LV myocardium segmentation method consists of a training stage and a segmentation stage. For a given SPECT image dataset, the clinically implemented physician-drawn contours of the endocardial and epicardial surface of myocardium are available. These clinical contours were used as the learning-based target of the SPECT image. The region within endocardial surfaces, region within epicardial surfaces, and background region are regarded as training and segmenting classes in our method. The original SPECT images were first automatically cropped into 32 × 32 × 16 voxels to reduce background region: a threshold was used to get rid of background and the centroid of the active heart region was then calculated, based on which a 32 × 32 × 16 voxel region was cropped to cover the active heart region. A volume-based deep learning network was trained based on such extracted SPECT image volume. The 3D multi-class V-Net architecture was used to enable voxel-wise error back-propagation during the training stage, and directly outputting an equal-sized prediction patch with the input patch during the testing procedure.22 By up-scaling low [4 × 4 × 2], modest [8 × 8 × 4], and high-level [16 × 16 × 8] feature volumes at each forwarding path from left to right portions of the hidden network using additional deconvolutional layers, and incorporating last output feature volume, the softmax function was employed on these equal-sized feature volumes to obtain final contour prediction. The Adam gradient decent optimizer was employed to train the V-Net. We used the whole volume as a patch and the batch size (number of patches) is 20. The number of epochs is 180. Compound loss supervision was then integrated into this prediction by considering both binary cross entropy and Dice loss to supervise the back-propagation of gradients for parameter updating in each training epoch. During the segmentation stage, the new arrival 3D patient SPECT was automatically cropped to reduce the background region ([32 × 32 × 16] in this study), and then input to the trained networks. The output volume was a multi-class contour probability maps. Finally, the segmentation was generated by thresholding the probability maps larger than 0.5. Figure 1 outlines the workflow schematic of our segmentation method.
Schematic flow chart of the proposed algorithm for LV segmentation. The upper part of this figure shows the training stage of our proposed method. The upper part also shows the V-Net architecture which has single channel volume input and 3 channels (background, region within endocardium, and region within epicardium) volume output. The lower part (brown) shows the segmentation stage. In segmentation stage, a new SPECT heart image is fed into the well-trained model to get the segmentation
In this study, we propose a compound loss function incorporating both the effectiveness of logistic loss and Dice loss functions to supervise our network. Since the optimization of the prediction task is a binary regression, we first used the voxel-wise binary cross entropy (BCE) loss as the logistic loss function. The BCE loss is defined as follows:
where \( C_{j} \) and \( \hat{C}_{j} \) denote the jth voxel in clinical contour \( C \) and prediction \( \hat{C} \), respectively.
The endocardial surface contour often occupies a small region of the MPS images as compared with epicardial surface contour. This may cause the network to ignore segmented regions and bias network output toward the background. The learning process can be trapped in local minima and unable to obtain accurate results. To address this issue, we additionally incorporated the logistic loss with Dice loss in the final stage as the final objective function. The Dice loss for segmentation was originally proposed in a 3D model defined as
where \( V \) indicates the volume of the region enclosed in the contours.
The compound loss function is a combination of the above BCE and Dice loss functions, which are related to the dissimilarity and similarity between prediction and training dataset, respectively. It is defined as follows:
where \( \mu \) is an empirical parameter, which balances the loss from binary cross entropy and Dice coefficient. In order to fairly compare the performance of the method on different patients, our hyper parameters of the network were fixed before we conducted the leave-one-out experiments. The batch size was set to 20. The number of epochs was set to 180. For the parameter μ, we employed fourfold cross-validation to evaluate its setting. It was shown that the performance is not sensitive when μ is between [0.7, 1.3], and thus we set μ = 1.
To evaluate the performance of the proposed method for LV myocardium segmentation, we compared the difference of contours generated by our method with clinical contours. In this retrospective study, we studied the dataset of 32 patients without hypertension, diabetes, heart dysfunction, family history of heart diseases (mean ± STD age: 63 ± 10, 23 males, 9 females). The cohort of 32 patients were used to evaluate our method using the leave-one-out cross-validation. For one test patient, the model is trained by the rest 31 patients. The model is initialized and re-trained for next test patient by another group of 31 patients. The training dataset and testing dataset are separated and independent during each study. In addition, 24 patients (mean ± STD age: 57 ± 10, 17 males, 7 females) diagnosed with myocardial ischemia ranging from mild, moderate to severe extents were also included to further test the proposed segmentation method with a leave-one-out validation strategy. Institutional review board approval was obtained with no informed consent required for this HIPAA-compliant retrospective analysis. Each patient underwent 8-frame ECG-gated resting SPECT performed 30 minutes after injection of 20-30 mCi Tc-99m sestamibi. The SPECT images were acquired by a dual-headed camera (CardioMD, Philips Medical Systems) using a standard resting protocol. The acquisition parameters were 20% energy window around 140 keV, 180° orbit, 32 steps with 25 seconds per step, 8-bin gating, and 64 projections per gate. Images were reconstructed into transaxial slices by ordered subsets expectation maximization with 3 iterations and 10 subsets, with Butterworth filter of power 10 and cutoff frequency of 0.3 cycles/cm. The reconstructed voxel dimensions of each SPECT image volume was 6.4 × 6.4 × 6.4 mm3.
Each patient had contours of myocardium with endocardial and epicardial surface delineated and approved by physicians, which were treated as the ground truth. Corresponding contours were also generated by our proposed method. The volume between endocardial and epicardial surface was considered as myocardium where the LV myocardium volume is calculated. We first visually checked the similarity of the contours between ground truth and the results of the proposed method. Quantitatively, we characterized the accuracy of the proposed method by calculating two widely used metrics: Dice similarity coefficient (DSC) and Hausdorff distance. The DSC describes the overlapping of the segmented volumes between ground truth and proposed method, which can be calculated by 1-Eq. (2) with \( C \) and \( \hat{C} \) being the contours of ground truth and proposed method results, respectively. A magnitude of DSC closer to 1 indicates higher overlapping with ground truth, thus high accuracy of the proposed method. The DSC metrics were calculated on the contours of endocardial surface and epicardial surface individually, as well as the combined myocardium contour. Hausdorff distance measures the difference of two contours in distance. It is defined as the maximum of the closest distances from each point on one contour to all points on the other contour,23 i.e.,
where \( a \) and \( b \) are points on contours of ground truth \( C \) and proposed method results \( \hat{C} \), respectively. \( a - b \) represents the Euclidean distance between point \( a \) and \( b \). A smaller Hausdorff distance means higher similarity between the two contours. Hausdorff distance metrics were calculated on the contours of endocardial surface and epicardial surface individually. To determine interobserver variability, the DSC and Hausdorff distance metrics were also calculated on the contours delineated by a second observer for three randomly selected patients with all phases from the total dataset.
The accuracy of the LV myocardium volume was evaluated by Pearson Correlation analysis between ground truth and the results of proposed method among all patients for each gating phase (0,1/8,… 7/8), and its correlation coefficient (r) and P value were calculated. A correlation coefficient closer to 1 indicates higher accuracy of proposed method, and a P value of less than 0.05 was considered to be statistically significant. The relative error of measured LV myocardium volume was calculated as the difference between ground truth and the results of proposed method relative to ground truth. A Bland–Altman figure was then plotted to show the absolute systematic bias in error and dependence of error on volume size.
Moreover, we calculated LV ejection fraction (EF) to further evaluate our method. We considered the region within endocardial surface of myocardium as the LV cavity. The maximum and minimum volumes within endocardial surface among all phases were then used as end-diastolic volume (EDV) and end-systolic volume (ESV), respectively, to calculate EF = (EDV − ESV)/EDV. The EF calculation based on the segmentations of our proposed method was compared with that based on the ground truth, as well as compared with results from a commercial software package (Emory Cardiac Toolbox 4.0, Atlanta, USA).
Results
Normal Group
In Figure 2, the segmentations by the proposed method are compared side-by-side with the clinical ground truth at different slices of gating phase 0 from patient #1 (normal) as an example. In this case, the LV myocardium volume measured by the proposed method was 191.5 cc, underestimated 1.74% from ground truth 194.9 cc.
Figure 3 shows a side-by-side comparison between the segmentations by the proposed method and ground truth from the same patient as Figure 2, but across different gating phases of the same slice. The mean and standard deviation (STD) of DSC of endocardial and epicardial surface, and myocardium, and Hausdorff distance of endocardial and epicardial surface among all 32 normal patients are plotted in Figure 4 for each phase and summarized in Table 1. The average DSC metrics and Hausdorff distance are all larger than 0.900 and less than 1 cm, respectively. The minimum DSC on myocardium is 0.783 which is the only case with DCS less than 0.800. These results quantitatively demonstrate high accuracy of contours delineated by the proposed method. No case shows unreasonable result among the 32 patients and 8 phases using our method.
The LV myocardium volumes changing with phases are demonstrated in Fig. 5 for patients #1 to #4. Our method accurately quantified the variation of LV myocardium volume during the cardiac cycle. Note that we present the results of four patients in figures as examples, but similar results can be seen on the other patients.
The correlation analysis of LV myocardium volume between ground truth and the proposed method is shown in Figure 6. Mean (and STD) of r among all phases is 0.910 ± 0.061, and all P values are less than 0.001, which indicates statistically significant linear correlation between LV myocardium volumes measured by proposed method and ground truth. The relative error in LV myocardium volume measurement for each phase is presented in Figure 7 as a Bland–Altman plot. The mean (and STD) relative error among all patients and all phases is − 1.09± 3.66%. The average linear correlation coefficient between volume error and volume size for all phases is − 0.222 (P = 0.238).
The EF results on normal patients are calculated based on our results and ground truth, and obtained from commercial software which are shown in Figure 8 as Bland–Altman plots. Good correlation on EF is shown between our results and ground truth (r = 0.893, P < 0.001). Correlation between our results and commercial software is fair (r = 0.644, P < 0.001). Similar studies on ESV and EDV are shown in Figure 9. Excellent correlations are found between our method and ground truth for both ESV and EDV, and between our method and commercial software for EDV. The correlation of ESV between our method and commercial software is fair.
Abnormal Group
Figure 10 demonstrates a side-by-side comparison between our results and the clinical ground truth at different slices of gating phase 0 from patient #33 (diagnosed with moderate ischemia) as an example. In this case, the LV myocardium volume measured by the proposed method was 212.0 cc, overestimated 1.49% from ground truth 208.9 cc. The mean and standard deviation (STD) of DSC of endocardial and epicardial surface, and myocardium, and Hausdorff distance of endocardial and epicardial surface among all 24 abnormal patients are plotted in Figure 11 for each phase and summarized in Table 2. Overall, the results on abnormal patients are very similar to those of normal patients, with mean DSC larger than 0.9 and Hausdorff distance less than 1 cm. The correlation coefficient of the LV myocardium volume between ground truth and our results is 0.939 ± 0.103 (P ≤ 0.001), and the mean relative error of LV myocardium volume is − 0.567 ± 3.47%.
Interobserver Study
The interobserver study results on normal patients are shown in Figure 12. The average DSC and Hausdorff distance between contours from two observers are 0.890 and 10.99 mm, respectively, which is higher but still comparable to the discrepancy of our method.
Discussion
In this study, we proposed a novel machine-learning-based method to segment LV and measure LV myocardium volume in gated MPS imaging. The average DSC metrics and Hausdorff distance of the contours delineated by our method are > 0.9 and < 1 cm, respectively. The results on abnormal patients are very similar to those on normal patients using our proposed method. The correlation coefficient of the LV myocardium volume between ground truth and results by the proposed method is 0.910 ± 0.061 with statistical significance, and the mean relative error of LV myocardium volume is − 1.09 ± 3.66 %. These results strongly indicate the feasibility of the proposed method in accurately quantifying the changing LV myocardium volume during the cardiac cycle. It also demonstrates the great potential of learning-based segmentation method in gated MPS imaging for clinical use.
Segmentation of LV in MPS imaging studied in this paper is a critical step in clinical evaluations for quantifying multiple LV contractile functional indices. In this study, we validated the accuracy of LV myocardium volume measured by the proposed method with ground truth. An accurate LV myocardium volume measurement can predict adverse cardiovascular events and premature death based on a well-established model,24 and provides prognostic information beyond traditional cardiovascular disease risk factors.25 The endocardium contours segmented for all gating phases would also be tracked to calculate regional endocardial wall motion by computing the distance of the endocardial surface between end-diastole and end-systole. Thus, a segmentation method with high performance is essential to avoid introducing error from the beginning of MPS imaging practice.
Manual contours rely on observer’s experience, and are reported to have substantial intraobserver and interobserver variability and less reproducibility.26 The manual contour from different observers may have systematic errors and random errors. Our learning-based method can mitigate random errors, but cannot correct systematic errors induced by the observers. In this study, we find that current contours for clinical use are represented as unsmooth curvature (see Figures 2 and 3). The contours segmented by our method have better refinement, which is more physically plausible when considering real anatomical structures. Secondly, our method provides comparable results but spends significantly shorter time. With a trained model, it takes around 10 seconds to accurately delineate contours and measure volumes for all phases of MPS imaging on a NVIDIA TITAN XP GPU. Moreover, our method requires no manual input parameters, correction, or intervention. Its speed and reproducibility allow it to be promising for clinical use.
Overestimation of LV myocardium volume for small heart and underestimation for large heart is commonly seen in current existing methods,17,18 which may lead to systematic errors on patient groups with small or large hearts. In our result, such correlation between volume measurement error and volume size is not observed from Figure 6. The linear correlation coefficient between volume error and volume size for all phases is − 0.222 (P = 0.238) on average, which indicates very low correlation without statistical significance. Thus, our method is able to work equally well regardless of the size of LV myocardium volume.
We compared the EF calculated based on our segmentation results with that obtained from a commercial software package, and found a fair correlation. Compared with correlation on EDV which is good (r = 0.914), ESV has larger discrepancy from commercial software (r = 0.818). Thus, the difference in the EF is mainly contributed from the difference in ESV. It may be explained by the different methodology used to determine ESV in the commercial software with various post-processing steps, which leads to larger discrepancy in results from our method. Studies showed that the correlation of EFs between two commercial softwares using different methods could be around 0.800.27 Thus, the commercial software results should be considered as a benchmark instead of a gold standard in this study.
Note that this study does not aim to demonstrate the absolute accuracy of the output contours of the proposed method by comparing with patients’ true myocardial contours which are always unavailable. Instead, we showed the high correlation of the output contours with its training dataset, which is manual contour from one observer in this study. Such high correlation would still exist if the training contours are from another experienced observer since the method is not designed for a specific observer. Thus, if the training dataset is closer to the true patient contour (e.g., consensus contours by multiple observers), the result of our method would also be closer to the true contour. In other words, our method generates contours with similar quality as training contours, and the quality of training dataset directly determines the quality of the output results.
In this study, we proposed a novel method for MPS automatic segmentation, and demonstrated its feasibility with 32 normal patients and 24 abnormal patients. This training/testing dataset has intermediate number of patients with anatomical variations and pathology abnormalities. Future study would involve a comprehensive evaluation with a larger population of patients with diverse demographics and pathological abnormalities. Different testing and training datasets (including normal and abnormal cases) from different observers and institutes would be valuable to further evaluate the clinical utility of our method. Moreover, this study validated the proposed method by quantifying the shape similarity of contours. Small differences from ground truth are observed, and its potential clinical impact (e.g., on functional indices) needs to be understood. Thus, a further investigation in diagnostic accuracy of the proposed method in detection and localization of coronary artery disease would be of great interest for clinical use.
Conclusion
We proposed a learning-based method to automatically segment LV and measure LV myocardium volume in gated MPS imaging. This method would benefit the gated MPS imaging in providing high-quality automatic quantification on multiple LV contractile functional indices without manual intervention. The proposed method was evaluated among 32 normal patients and 24 abnormal patients. The results demonstrate the feasibility of the proposed method in contouring with comparable accuracy as that based on physician experience.
New Knowledge Gained
A learning-based method has been proposed to automatically segment LV and measure myocardium volume in gated MPS imaging. Results show that the proposed method has the feasibility in contouring with comparable accuracy as that based on physician experience.
Abbreviations
- SPECT:
-
Single-photon emission computed tomography
- MPS:
-
Myocardial perfusion SPECT
- LV:
-
Left ventricular
- CT:
-
Computed tomography
- MRI:
-
Magnetic resonance imaging
- BCE:
-
Binary cross entropy
- DSC:
-
Dice similarity coefficient
- EF:
-
Ejection fraction
- EDV:
-
End-diastolic volume
- ESV:
-
End-systolic volume
References
Dvorak RA, Brown RKJ, Corbett JR. Interpretation of SPECT/CT myocardial perfusion images: common artifacts and quality control techniques. RadioGraphics. 2011;31:2041–57.
Holder L, Lewis S, Abrames E, Wolin EA. Review of SPECT myocardial perfusion imaging. J Am Osteopath Coll Radiol. 2016;5:5–13.
Zhou W, Garcia EV. Nuclear image-guided approaches for cardiac resynchronization therapy (CRT). Curr Cardiol Rep. 2016;18:7.
Garcia EV, Cooke CD, Van Train KF, Folks R, Peifer J, DePuey EG, et al. Technical aspects of myocardial SPECT imaging with technetium-99m sestamibi. Am J Cardiol. 1990;66:23e–31e.
Collom SJ, Case JA, Bateman TM. Electrocardiographically gated myocardial perfusion SPECT: technical principles and quality control considerations. J Nucl Cardiol. 1998;5:418–25.
Paul AK, Nabi HA. Gated myocardial perfusion SPECT: basic principles, technical aspects, and clinical applications. J Nucl Med Technol. 2004;32:179–87 quiz 88-9.
Shah PK, Pichler M, Berman DS, Singh BN, Swan HJ. Left ventricular ejection fraction determined by radionuclide ventriculography in early stages of first transmural myocardial infarction. Relation to short-term prognosis. Am J Cardiol. 1980;45:542–6.
Slomka P, Xu Y, Berman D, Germano G. Quantitative analysis of perfusion studies: strengths and pitfalls. J Nucl Cardiol. 2012;19:338–46.
Sharir T, Germano G, Kang X, Lewin HC, Miranda R, Cohen I, et al. Prediction of myocardial infarction versus cardiac death by gated myocardial perfusion SPECT: risk stratification by the amount of stress-induced ischemia and the poststress ejection fraction. J Nucl Med. 2001;42:831–7.
Sharir T, Berman DS, Waechter PB, Areeda J, Kavanagh PB, Gerlach J, et al. Quantitative analysis of regional motion and thickening by gated myocardial perfusion SPECT: normal heterogeneity and criteria for abnormality. J Nucl Med. 2001;42:1630–8.
Shaw LJ, Iskandrian AE. Prognostic value of gated myocardial perfusion SPECT. J Nucl Med. 2004;11:171–85.
Friehling M, Chen J, Saba S, Bazaz R, Schwartzman D, Adelstein EC, et al. A prospective pilot study to evaluate the relationship between acute change in left ventricular synchrony after cardiac resynchronization therapy and patient outcome using a single-injection gated SPECT protocol. Circ Cardiovasc Imaging. 2011;4:532–9.
Faber TL, Cooke CD, Folks RD, Vansant JP, Nichols KJ, DePuey EG, et al. Left ventricular function and perfusion from gated SPECT perfusion images: an integrated method. J Nucl Med. 1999;40:650–9.
Xu Y, Kavanagh P, Fish M, Gerlach J, Ramesh A, Lemley M, et al. Automated quality control for segmentation of myocardial perfusion SPECT. J Nucl Med. 2009;50:1418–26.
Germano G, Kavanagh PB, Waechter P, Areeda J, Van Kriekinge S, Sharir T, et al. A new algorithm for the quantitation of myocardial perfusion SPECT. I: technical principles and reproducibility. J Nucl Med. 2000;41:712–9.
Germano G, Kiat H, Kavanagh PB, Moriel M, Mazzanti M, Su HT, et al. Automatic quantification of ejection fraction from gated myocardial perfusion SPECT. J Nucl Med. 1995;36:2138–47.
Eva P, Marcus C, John P, Olle P, Håkan A. Evaluation of left ventricular volumes and ejection fraction by automated gated myocardial SPECT versus cardiovascular magnetic resonance. Clin Physiol Funct Imaging. 2005;25:135–41.
Soneson H, Ubachs JF, Ugander M, Arheden H, Heiberg E. An improved method for automatic segmentation of the left ventricle in myocardial perfusion SPECT. J Nucl Med. 2009;50:205–13.
Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng P, et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging. 2018;37:2514–25.
Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal. 2017;35:159–71.
Betancur J, Rubeaux M, Fuchs TA, Otaki Y, Arnson Y, Slipczuk L, et al. Automatic Valve Plane Localization in Myocardial Perfusion SPECT/CT by Machine Learning: Anatomic and Clinical Validation. J Nucl Med. 2017;58:961–7.
Milletari F, Navab N, Ahmadi S. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV); 2016. p. 565-71.
Varadhan R, Karangelis G, Krishnan K, Hui S. A framework for deformable image registration validation in radiotherapy clinical applications. J Appl Clin. 2013;14:4066.
Koren MJ, Devereux RB, Casale PN, Savage DD, Laragh JH. Relation of left ventricular mass and geometry to morbidity and mortality in uncomplicated essential hypertension. Ann Intern Med. 1991;114:345–52.
Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Left ventricular mass and incidence of coronary heart disease in an elderly cohort. The Framingham Heart Study. Ann intern med. 1989;110:101.
Trobaugh GB, Wackers FJ, Sokole EB, DeRouen TA, Ritchie JL, Hamilton GW. Thallium-201 myocardial imaging: an interinstitutional study of observer variability. J Nucl Med. 1978;19:359–63.
Nakajima K, Higuchi T, Taki J, Kawano M, Tonami N. Accuracy of ventricular volume and ejection fraction measured by gated myocardial SPECT: comparison of 4 software programs. J Nucl Med. 2001;42:1571–8.
Acknowledgements
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 and Emory Winship Cancer Institute pilot Grant. This research is also supported by the American Heart Association under Award Number 17AIREA33700016.
Disclosures
Dr. Kristin Higgins is consulting for Astra Zeneca, Varian, on advisory board for Genentech, and receiving research funding from RefleXion Medical. Tonghe Wang, Yang Lei, Haipeng Tang, Zhuo He, Richard Castillo, Cheng Wang, Dianfu Li, Tian Liu, Walter J. Curran, Weihua Zhou, and Xiaofeng Yang declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors of this article have provided a PowerPoint file, available for download at SpringerLink, which summarizes the contents of the paper and is free for re-use at meetings and presentations. Search for the article DOI on SpringerLink.com.
Funding
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 and Emory Winship Cancer Institute pilot Grant. This research is also supported by the American Heart Association under Award Number 17AIREA33700016.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, T., Lei, Y., Tang, H. et al. A learning-based automatic segmentation and quantification method on left ventricle in gated myocardial perfusion SPECT imaging: A feasibility study. J. Nucl. Cardiol. 27, 976–987 (2020). https://doi.org/10.1007/s12350-019-01594-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12350-019-01594-2