Abstract
Objectives
Cardiac magnetic resonance (CMR) first-pass perfusion is an established noninvasive diagnostic imaging modality for detecting myocardial ischemia. A CMR perfusion sequence provides a time series of 2D images for dynamic contrast enhancement of the heart. Accurate myocardial segmentation of the perfusion images is essential for quantitative analysis and it can facilitate automated pixel-wise myocardial perfusion quantification.
Methods
In this study, we compared different deep learning methodologies for CMR perfusion image segmentation. We evaluated the performance of several image segmentation methods using convolutional neural networks, such as the U-Net in 2D and 3D (2D plus time) implementations, with and without additional motion correction image processing step. We also present a modified U-Net architecture with a novel type of temporal pooling layer which results in improved performance.
Results
The best DICE scores were 0.86 and 0.90 for LV myocardium and LV cavity, while the best Hausdorff distances were 2.3 and 2.1 pixels for LV myocardium and LV cavity using 5-fold cross-validation. The methods were corroborated in a second independent test set of 20 patients with similar performance (best DICE scores 0.84 for LV myocardium).
Conclusions
Our results showed that the LV myocardial segmentation of CMR perfusion images is best performed using a combination of motion correction and 3D convolutional networks which significantly outperformed all tested 2D approaches. Reliable frame-by-frame segmentation will facilitate new and improved quantification methods for CMR perfusion imaging.
Key Points
• Reliable segmentation of the myocardium offers the potential to perform pixel level perfusion assessment.
• A deep learning approach in combination with motion correction, 3D (2D + time) methods, and a deep temporal connection module produced reliable segmentation results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cardiac magnetic resonance (CMR) is an established noninvasive diagnostic imaging modality for evaluating cardiac disease. A frequent task is CMR first-pass perfusion imaging, where a bolus of gadolinium is injected and a time series of 2D images is acquired as the contrast agent enters the myocardium. This enables the detection of perfusion deficits with high accuracy [1]. Interpretation of these images can be difficult, can be time-consuming, and requires expert knowledge. To improve and simplify analysis, several methods have been proposed including frame-to-frame motion correction [2] and pixel-wise quantitative assessment [3]. An integral part of automatic or semiautomatic CMR perfusion analysis is the segmentation of the left ventricular myocardium. While there are methods for LV segmentation relying on hand-crafted features [4], deep learning methods have more recently outperformed traditional methods in various medical segmentation tasks [5], including CMR image segmentation [6, 7]. The task of segmenting a 2D structure on a sequence of frames—which may have significant frame-to-frame motion and large dynamic contrast changes [6] (see Fig. 1)—can be approached from various angles. We hypothesized that 3D deep learning methods would outperform 2D methods. We compare results of state-of-the-art 2D and 3D deep learning methods with and without motion correction. In addition, we introduce a neural network layer that performs temporal pooling and evaluate impact on performance.
Effects of motion correction: to visualize motion, the part of the image indicated by the orange line on the left panel is shown over time on the middle panel (analogous to echocardiography M-mode). The upper half shows the original non-motion-corrected series and the lower half the motion-corrected series. The right panel shows the corresponding ground truth segmentations of the myocardium. It can be clearly seen that the motion correction makes the borders of the myocardium less jagged (see arrows)
Methods
Data
We used CMR images obtained at the NHLBI imaging laboratory under a research protocol. All individuals provided written informed consent. Data were fully anonymized before analysis started. From the non-motion-corrected cardiac perfusion sequence scanner output, additional motion-corrected images were generated. The method for motion correction is based on optical flow analysis and has been previously published in detail [2].
Scans of a total of 70 patients were included in the study. Data from the initial 50 patients were used in a 5-fold cross-validation approach with a train/validation/test split of 60%/20%/20% with strict sorting of individual patients in the same fold. Then, to further inform generalizability, data from an additional 20 patients was collected and segmented by another observer. Data from these 20 patients were used as an additional test set.
From the non-motion-corrected raw perfusion image series, we used an optical flow method that was published previously to generate separate motion-corrected image series [2]. For the convolutional neural network training, validation, and testing, stress perfusion images of 50 patients (in 3 slice positions, each having 60 images, for a total of 9000 images) were used. Here, 2 proton density images in each image series were not used for segmentation training and testing. Labels for LV myocardium (area between epicardial and endocardial border) and LV cavity (area inside the endocardial border) were manually generated (performed by VS). Papillary muscles were considered to be part of the LV cavity. These manual labels were then transformed to the corresponding non-motion-corrected image series using freely available software tools (elastix 4.9 [8]).
After completing all training and optimization, a separate dataset of 20 patients (3 slice positions, stress perfusion, for a total of 3600 images) with manually generated labels (performed by M.J.) was used for additional final testing.
CMR acquisition
We retrospectively analyzed anonymized images. The initial imaging procedures were described previously [9]. Briefly, perfusion imaging was performed using a 1.5-T scanner (Siemens Healthcare). As vasodilator regadenoson was used and as contrast agent a dose of 0.05 mmol/kg gadolinium was injected intravenously at 2 to 5 ml/s. Typical imaging parameters for the myocardial series included a non-slice-selective 90°composite saturation preparation pulse, 90-ms inversion time, 1.2-ms echo time, 2.3-ms repetition time, 50° flip angle, 8-mm slice thickness, 360 × 270-mm field of view, and a 256 × 192 image matrix [10].
Networks and training
We used a modified U-net architecture [11] with residual connections [12], group normalization [13], and Leaky ReLU as non-linearity. Convolution kernels were 3 × 3 × 3 and 3 × 3 × 1 for 3D and 2D networks, respectively. The initial feature number was 32 and was doubled at each level for a total of 4 levels. Due to the 3D nature of our data and the limited amount of labeled data available, we applied heavy data augmentation including flips, cropping, and non-rigid spatial deformation with following specifics: mirroring (along all axes, x,y,t), switching of axis (x,y), cropping (along x- and y-axis, remove 0–30% along each border randomly, then resize to original dimensions), and nonrigid deformation (using sitk.BSplineTransform, 10 × 10 control point mesh, control points are moved by random distance in x and y direction [distance drawn from normal distribution and multiplied by 2]).
Modified Dice score was used as loss function with s = 1 for training (for improved differentiability). For evaluation, classic Dice score was used:
Training was performed for 10,000 iterations with a batch size of 4 on 4-GPU HPC cluster nodes (4×NVIDIA P100). The Adam optimizer was used for training. Training time was approximately 6 h for all models. Inference time was < 5 s for all models (using 1 GPU). The initial learning rate was 0.0001 and was reduced by a factor of 2 if there was no improvement in validation performance in 30 epochs. The model with the best validation Dice score was used for testing.
Deeply temporally connected pooling layer
The data at hand has specific properties which make it different from spatial 3D data and traditional video content. It is highly anisotropic by definition because the time axis is fundamentally different from the spatial axes. This is problematic for conventional 3D U-nets [14]. All frames of a series show the same object (a cross-section of the heart) with some spatial variation (translation and deformation) and intense brightness variation due to contrast media passage in different tissues. It is therefore likely desirable to have the information of the complete time series available at all time points. Neural networks with a kernel size of 3 need many convolutions to reach a field of view to capture this information. In addition, the effective receptive field of view of imaging neural networks has been shown to be much smaller than the theoretical size [15].
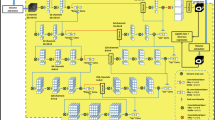
Therefore, we propose a novel deeply temporally connected pooling layer (DTC, see Fig. 2)—where at the deepest level of the U-net, presumably with the most abstract representation of the input—a maximum pooling module is added. This module performs max pooling of the complete time axis down to 1 and 4 frames and then re-expands the feature maps to the full temporal resolution again. This was inspired by the pyramid scene parsing network which employs a similar method to achieved state-of-the art accuracy by incorporating larger context information in 2D image segmentation [16]. These 2 pooling results are concatenated with the original deepest feature map and followed by a 3 × 3 × 3 convolution (for details, see code in the Appendix). Other than the addition of this layer, no changes were made. For comparison, we also applied this layer to the 3D U-net.
Schematic of the U-net architecture in panel a. In a subset of experiments, an additional module was added at the deepest layer of the U-net. This deep temporal connection module DTC is shown in more detail in panel b. In this module, at a relatively abstract feature representation, the complete time series undergoes a maximum pooling to a single frame and to 4 frames. These pooled stacks are then again up-sampled to the full temporal resolution. Then these up-sampled feature maps are concatenated with the module input stack, and a 3 × 3 × 3 convolution is applied. The effect of this module is that information from the complete time series is combined with the feature map of this single frame feature map, thereby—domain specific—forcing a greatly increased receptive field at this location. This is a variation of a concept which was shown to be highly effective in general image segmentation in the competition winning PSPnet (PASCAL VOC)
Evaluation
All experiments on the initial 50 included patients were performed in 5-fold cross-validation while always assuring that all sequences of each patient were assigned in the same fold. Cross-validation test set results were pooled and then submitted to statistical analysis. We used the Dice score (Eq. 1) and Hausdorff distance (in pixels [17], applicability to cardiac MRI discussed in [18]) for the evaluation of the segmentations. If the automatic segmentation was empty, the Hausdorff distance is undefined and we imputed the worst distance that occurred in the dataset. We evaluated each slice separately and averaged the score metrics of all frames of a series. In addition, we measured the time an expert needed to correct the resulting automatic segmentations to an acceptable quality (a series with MRI and segmentation data was loaded in ImageJ and contours were manually corrected).
Statistical analysis
For all statistical calculations, R version 3.5.2 was used. Because we compare multiple observations performed on the same images, we used the Friedman test followed by a pairwise post hoc test (Nemenyi test). The level for statistical significance was p = 0.05.
Results
Patient characteristics
Table 1 shows basic patient demographics for all included patients. Of note, a significant number of patients had abnormal imaging test results (53% with abnormal perfusion and 34% with subendocardial LGE).
Dice scores
The Dice results for LV myocardium and LV cavity segmentations are shown in Table 2 on the left side and in boxplots in Fig. 3. For each image series, a mean Dice score of all frames was calculated. Regarding the LV myocardium, for motion-corrected images, the 3D U-net showed the best result with a Dice score of 0.86 for mean scores. For non-motion-corrected images, the 2D U-net with deep temporal pooling showed the best result (0.85). For the LV cavity segmentations, the best Dice score was 0.90 in the motion-corrected data using the 3D U-net with deep temporal connection (although the performance difference to the 3D U-net without DTC was not statistically significant). For statistical analysis, see supplemental tables.
If using segmentations for pixel precise perfusion quantification, even a single problematic segmentation can affect results. We used an empirical threshold to determine an unusable LV myocardium segmentation as Dice < 0.6 and calculated the percentage of series that contain at least one image with unusable segmentation. The results are shown in Table 3. This data show that up to 31.3% of series contain a problematic LV myocardial segmentation when using non-motion-corrected data and a 2D U-net while the best result is only 1.3% for motion-corrected data and a 3D U-net with deeply connected temporal pooling layer.
Dice scores by slice position
Depending on the MR slice position (basal, mid, or apical), the amount of myocardium present on the slice can vary significantly. If a low amount of myocardium is visualized, segmentation intuitively becomes more challenging with additional issues such as partial volume effects or residual motion. Figure 4 shows the Dice scores for all models and motion correction conditions stratified by slice position. It can be clearly seen that the apical slice segmentations have the lowest Dice scores (dark bars). This is consistently seen in all models.
Box plots of the LV myocardial segmentation Dice scores analogous to Fig. 3 but stratified for slice position (basal, mid, apical). The apical slice position shows the worst segmentation performance. This can be explained by the lower amount of myocardium, LV cavity, and associated structures like the RV present on the apical images—which makes segmentation intuitively more challenging. Partial volume effects and residual motion which are more prominent at the apical slice location can also explain these findings. DTC: deeply connected temporal pooling layer
Hausdorff distance
Technically, if the automatic segmentation was empty, the Hausdorff distance is undefined. In these cases, we imputed the largest distance that occurred in the dataset which was 85.6 pixels, but of note, this only occurred in 5 frames in the combined datasets and therefore had negligible effects on the final results.
The right side of Table 2 shows Hausdorff distance scores (for which a lower value indicates better performance; see also Fig. 3 lower half).
For the LV myocardium, the 3D U-net with DTC performed best in motion-corrected images while the 2D U-net with DTC showed the second best performance. Interestingly, in non-motion-corrected images, the same order of performance was seen as in the Dice scores, with 3D U-net DTC and 2D U-net DTC showing the best results.
Similar results are seen for the mean LV cavity segmentation performance, where the 3D U-net with deep temporal connection again achieved the best results in motion-corrected images while the 2D U-net with deep temporal connection showed the best results in non-motion-corrected images.
Example images
In Fig. 5, typical examples of the proposed 3D U-net DTC segmentation with motion correction are shown. These examples indicate that our methods successfully segment the perfusion image series at both low and high luminal contrast enhancement phases. These qualitative results are in line with our quantitative assessment.
In Fig. 6 example, segmentations of different networks are shown, illustrating difficult cases. The columns show the 4 tested models, expert segmentation, and MRI without segmentation. In the first row (A), a typical failure of the 2D U-net is seen. At this point in the perfusion cycle, there is not sufficient information present in the frame to perform a valid segmentation. Therefore, the 2D U-net, which is unable to access information from other frames, fails to perform a complete segmentation. The 2D U-net with deep temporal connection (DTC), on the other hand, is able to use the information from other slices and performs well. In the second row (B), an unusual but intense failure of the 2D U-nets is shown. A second circular structure (bowel) is present on this frame. As a U-net is a pixel classifier, which has no concept of segmentation shape, the 2D U-net segments the spurious structure in addition to the myocardium. The 3D U-nets have the information of the complete perfusion cycle available and it can be suspected that on other frames the heart is unambiguously visible (e.g., when the contrast agent is in the LV cavity, the bowel loop will still be dark), and this may lead to a correct segmentation on this slice. Panels C and D show a case without motion correction in two different views. In C, an example frame is shown, while in D a single line of the 2D image in C is projected over time as y-axis to visualize the motion that occurs. This is similar to images commonly recorded in echocardiography (M-mode). In panel C, it can be seen that there is a failure to correctly segment the LV in the 3D U-nets while the 2D U-nets perform well (see white arrows). Panel D better visualizes the motion and two episodes of erratic movement can be seen on these images (likely due to breathing motion). It is again clear that the 2D U-nets correctly capture the abrupt motion (see white arrows in panel D) while the 3D U-nets assume as smooth but faulty delineation.
Independent test set
We used the previously trained models to segment an additional independent test set. Of note, the segmentations in this test set were generated by a different researcher and the transformation from motion-corrected to non-motion-corrected images was performed in a different manner.
The results in Table 4 show again that for non-motion-corrected sequences, the 2D U-net with DTC performs best. The result is very similar to the original data set (0.84 vs. 0.85 for Dice score in non-MOCO images and 0.84 vs. 0.86 in MOCO images) and the overall best performing models again were the 3D U-nets with motion correction. These results thus further validate the robustness of our approach.
Manual segmentation correction effort
In a subsample of segmentations from the 2D U-net without motion correction and from the 3D U-net with motion correction and DTC, we performed timing of the effort of manual contour correction. For the 3D U-net MOCO DTC, the effort was significantly and meaningfully less compared to a traditional 2D U-net approach (median correction times per patient of 0 s [range 0–61 s] versus 78 s [range 0–229 s], respectively, p = 0.002, n = 20).
Discussion
CMR perfusion imaging is an important and increasingly used modality for detecting myocardial ischemia clinically [1]. Quantitative image analysis of perfusion MRI has been proven to be equal or superior to visual assessment [1, 3]. Segmentation of the left ventricular myocardium is an essential step in quantitative image analysis for research and in clinical practice in CMR. Perfusion CMR images have some unique properties like dramatic changes of signal intensity due to contrast passage in different tissues, lack of myocardial signal on some individual frames, and erratic frame-to-frame body and cardiac motion that make segmentation using conventional methods difficult. We hypothesized that using 3D deep learning methods with time as the third axis and with applied motion correction will overcome these difficulties and may outperform 2D methods. In addition, we tested the utility of a modification of the U-net architecture that enables a deep temporal connection between all frames of a sequence (DTC layer).
We systematically evaluated four models and two preprocessing variations for LV segmentation in CMR perfusion images. The results show that the combination of classic advanced motion correction and a 3D U-net yields the best results in Dice score and Hausdorff distance. Our novel deeply temporally connected (DTC) pooling layer in many cases improved the network performance further (the difference was statistically significant for the 2D U-net but not for the 3D U-net) (Fig. 3).
There are numerous publications in regard to deep learning LV segmentation on cine MR [2], but to our knowledge, relatively few publications exist on deep learning–based segmentation of first-pass myocardial perfusion CMR. A general overview of traditional methods can be found in [19]. A recent work by Scannell et al [6] used a 2D deep learning method without motion correction and achieved a Dice similarity coefficient of 0.8 for LV segmentation while our method with motion correction and 3D analysis resulted in a Dice coefficient of 0.86 in our data. A study by Kim et al took a different approach and selected the frame of the perfusion sequence with the lowest predicted segmentation uncertainty and segmented this frame [20] which also improved performance (DICE 0.81). These studies emphasize that traditional 2D methods do not yield optimal results and that there is ongoing work to improve on this unique segmentation task.
Example segmentations: these example shows the difficulties in segmenting CMR perfusion series. The first 4 columns show the different tested LV segmentation models. The last two columns show the expert segmentation and the MRI without segmentation. a Typical failure of 2D U-net when there is little contrast present on a frame (motion-corrected series), (b) example failure of mainly of the 2D U-net and less prominent of the 2D U-net DTC segmenting additional structures because of a lack of temporal context (motion-corrected series), (c, d) showing the same dataset: failure of the 3D U-nets on a non-motion-corrected sequence with erratic movement (3rd and 4th column, white arrows). Images in row d show time in the Y-axis (similar to M-mode echocardiography images)
The specific challenges of CMR perfusion data are variable information content of frames, large changes of contrast and brightness, and frame-to-frame motion. The variable information content makes pure 2D methods perform less favorably because of the need to access information across multiple frames which 3D methods can do (this can be seen in the examples shown in Fig. 6). On the other hand, frame-to-frame motion with rapid and erratic movements actually adversely affect 3D methods (see Fig. 6, panel D). This is likely because 3D U-nets work best for isotropic data and implicitly assume that voxels adjacent to a voxel are directly related. This assumption is frequently violated in a temporal 2D stack of images with rapid frame-to-frame motion where often—due to frame shift—there is no direct relation of a voxel to a neighboring voxel in an adjacent frame.
We chose to alleviate this issue with motion correction and this showed significant improvement as can be seen in Fig. 3 with significantly improved Dice scores and Hausdorff distance in motion-corrected data compared with non-motion-corrected data in the 3D networks.
We also used a second approach to cope with the frame-to-frame motion issue. We used a U-net which in the initial stages has only 2D convolutions but processes a complete time stack of frames at the same time. In the lowest level of the U-net, the resolution of the feature maps is greatly reduced and intuitively more abstract features are represented. On this deep level, we perform a temporal pooling step (Fig. 2; for details, see the Methods section) that performs a maximum pooling function over the complete time series in a similar fashion as imaging physicians frequently use maximum intensity projections when evaluating cardiovascular images. This enables the 2D stacked network to be relatively resistant to frame-to-frame motion but at the same time allows for drawing information from the complete time cycle. This deeply connected temporal pooling layer resulted in the best performance on non-motion-corrected images (see Table 2) and also improved performance in the 3D U-net numerically (although the difference did not meet criteria for statistical significance).
The full extent of differences in consistency of the tested segmentation methods becomes apparent if evaluating the percentage of series containing at least one frame with failed segmentation (Dice < 0.6) where the worst performance was 31.3% for non-motion-corrected data and 2D U-net while the best performance was an error rate of only 1.3% for motion-corrected data and a 3D U-net with deeply temporally connected layer (Table 3). In addition, manual correction of the automatic contours was needed significantly less frequently and to a lesser extent when using 3D segmentation methods compared to 2D methods.
Perfusion imaging is usually performed at multiple short-axis slice locations which have different properties. Stratified results by slice location (Fig. 4) show that the apical slice is the most difficult location. This can be explained by the small size of the apex compared to the basal myocardium and the lack of additional visual cues at this location.
We also evaluated an additional independently created test data set. The results shown in Table 4 corroborate the previously mentioned results. Of note, in this separate test set, the top performing method (Dice) was the 3D U-net with deeply temporally connected pooling layer and motion correction.
Of note, we believe that the concept of a deeply temporally connected pooling layer may benefit many other medical imaging applications wherever there is a time sequence of images showing a relatively stationary scene but which may contain only partial information on each of the frames or may show changes in contrast and image characteristics frame to frame. Other examples with these properties in medical imaging are X-ray angiography, MRI time-resolved angiography, CT perfusion imaging of the heart and brain, and phase contrast MRI.
This study has several limitations. The analysis is based on images from a single center and a single MRI scanner type. It would be preferable to use a wider range of scanner technologies and patient populations. This would, in turn, necessitate a significantly larger sample size. Yet, we believe that given an appropriate sample size, the described algorithms will perform similarly because the issue of breathing motion is universal and we did not add any additional scanner specific modifications to the architecture. In addition, we developed and tested our method for stress perfusion images only because this is the more clinically relevant part of the exam; therefore, we cannot comment on performance in rest perfusion images.
Conclusion
We showed that, of the tested methods, the optimal approach for LV myocardial segmentation of CMR perfusion images was to use motion correction in combination with a 3D U-net. Addition of a deeply temporally connected pooling module did result in numerically slightly higher performance, but this difference was not statistically significant in this setting. If motion correction is not available or feasible, the best performance was achieved using a 2D approach with deeply temporally connected pooling module. We believe that the concept of a deep temporal connection may apply to many other medical imaging tasks.
Abbreviations
- CMR:
-
Cardiac magnetic resonance imaging
- DTC:
-
Deeply temporally connected pooling layer
- GPU:
-
Graphics processing unit
- LV:
-
Left ventricle
- MOCO:
-
Motion corrected
References
Danad I, Szymonifka J, Twisk et al (2016) Diagnostic performance of cardiac imaging methods to diagnose ischaemia-causing coronary artery disease when directly compared with fractional flow reserve as a reference standard: a meta-analysis. Eur Heart J 095. https://doi.org/10.1093/eurheartj/ehw095
Benovoy M, Jacobs M, Cheriet F, Dahdah N, Arai AE, Hsu LY (2017) Robust universal nonrigid motion correction framework for first-pass cardiac MR perfusion imaging. J Magn Reson Imaging 46(4):1060–1072. https://doi.org/10.1002/jmri.25659
Hsu L-Y, Jacobs M, Benovoy M et al (2018) Diagnostic performance of fully automated pixel-wise quantitative myocardial perfusion imaging by cardiovascular magnetic resonance. JACC Cardiovasc Imaging 11(5):697–707. https://doi.org/10.1016/j.jcmg.2018.01.005
Huang H-H, Huang C-Y, Chen C-N, Wang Y-W, Huang T-Y (2017) Automatic regional analysis of myocardial native t1 values: left ventricle segmentation and AHA parcellations. Int J Cardiovasc Imaging 34(1):131–140. https://doi.org/10.1007/s10554-017-1216-x
Sahiner B, Pezeshk A, Hadjiiski et al (2018) Deep learning in medical imaging and radiation therapy. Med Phys 46(1):1–36. https://doi.org/10.1002/mp.13264
Scannell CM, Veta M, Villa ADM et al (2019) Deep-learning-based preprocessing for quantitative myocardial perfusion MRI. J Magn Reson Imaging. https://doi.org/10.1002/jmri.26983
Fahmy AS, El-Rewaidy H, Nezafat M, Nakamori S, Nezafat R (2019) Automated analysis of cardiovascular magnetic resonance myocardial native t1 mapping images using fully convolutional neural networks. J Cardiovasc Magn Reson 21(1). https://doi.org/10.1186/s12968-0180516-1
Klein S, Staring M, Murphy K, Viergever MA, Pluim J (2010) elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205. https://doi.org/10.1109/tmi.2009.2035616
Jacobs M, Benovoy M, Chang L-C, Arai AE, Hsu L-Y (2016) Evaluation of an automated method for arterial input function detection for first pass myocardial perfusion cardiovascular magnetic resonance. J Cardiovasc Magn Reson 18(1). https://doi.org/10.1186/s12968-0160239-0
Kellman P, Arai AE (2007) Imaging sequences for first pass perfusion - a review. J Cardiovasc Magn Reson 9(3):525–537. https://doi.org/10.1080/10976640601187604
Cicek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. ArXiv e-prints. 1606.06650
Kayalibay B, Jensen G, van der Smagt P (2017) CNN-based Segmentation of medical imaging data. ArXiv e-prints. 1701.03056
Wu Y, He K (2018) Group normalization. ArXiv e-prints. 1803.08494
Liu S, Xu D, Zhou SK et al (2018) 3d anisotropic hybrid network: Transferring convolutional features from 2d images to 3d anisotropic volumes. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-Lo’pez C, Fichtinger G (eds) Medical image computing and computer assisted intervention – MICCAI 2018. Springer, Cham, pp 851–858
Luo W, Li Y, Urtasun R, Zemel R (2016) Understanding the effective receptive field in deep convolutional neural networks. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R (eds) Advances in Neural Information Processing Systems 29. Curran Associates, Inc., New York, pp 4898–4906
Zhao H, Shi J, Qi X, Wang X, Jia J (2016) Pyramid scene parsing network. arXiv:1612.01105
MedPy package by Maier, O.: https://pypi.org/project/MedPy/ Accessed 8/10/2020
Petitjean C, Zuluaga MA, Bai W et al (2015) Right ventricle segmentation from cardiac MRI: a collation study. Med Image Anal 19(1):187–202. https://doi.org/10.1016/j.media.2014.10.004
Gupta V, Kirişli HA, Hendriks EA et al (2012) Cardiac MR perfusion image processing techniques: a survey. Med Image Anal 16(4):767–785. https://doi.org/10.1016/j.media.2011.12.005
Kim YC, Kim KR, Choe YH (2020) Automatic myocardial segmentation in dynamic contrast enhanced perfusion MRI using Monte Carlo dropout in an encoder-decoder convolutional neural network. Comput Methods Programs Biomed 185. https://doi.org/10.1016/j.cmpb.2019.105150
Acknowledgements
This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). We are grateful for the support of the NIH Biowulf team.
Funding
This work was supported by the NIH intramural research program of the National Heart, Lung and Blood Institute (ZIA HL006137-08).
Author information
Authors and Affiliations
Contributions
VS planned and performed experiments, analyzed data, and drafted the manuscript. LH and AEA conceived experiments, evaluated data, and edited manuscript. MJ contributed to data, experiments, and manuscript.
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Veit Sandfort, MD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in “Evaluation of an Automated Method for Arterial Input Function Detection for First-Pass Myocardial Perfusion Cardiovascular Magnetic Resonance” Matthew Jacobs, Mitchel Benovoy, Lin-Ching Chang, Andrew E Arai, Li-Yueh Hsu, 10.1186/s12968-016-0239-0.
Methodology
• Retrospective, performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 19.7 kb)
Rights and permissions
About this article
Cite this article
Sandfort, V., Jacobs, M., Arai, A.E. et al. Reliable segmentation of 2D cardiac magnetic resonance perfusion image sequences using time as the 3rd dimension. Eur Radiol 31, 3941–3950 (2021). https://doi.org/10.1007/s00330-020-07474-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07474-5