Abstract
Spinocerebellar ataxia subtypes 1, 3, and 6 (SCA1, MJD/SCA3, and SCA6) are among the most prevalent autosomal dominant cerebellar ataxias worldwide, but their relative frequencies in Peru are low. Frequency of large normal (LN) alleles at spinocerebellar ataxia-causative genes has been proposed to be associated with disease prevalence. To investigate the allelic distribution of the CAG repeat in ATXN1, ATXN3, and CACNA1A genes in a Peruvian mestizo population and examine their association with the relative frequency of SCA1, MJD/SCA3, and SCA6 across populations. We genotyped 213 healthy mestizo individuals from Northern Lima, Peru, for ATXN1, ATXN3, and CACNA1A using polymerase chain reaction (PCR) and polyacrylamide gel electrophoresis (PAGE). We compared the frequency of LN alleles and relative disease frequency between populations. We also tested 40 samples for CAT repeat interruptions within the CAG tract of ATXN1. We found no association between disease frequency and population frequency of LN alleles at ATXN1 and ATXN3. All 40 ATXN1 samples tested for CAT interruptions were positive. Frequency of LN alleles at CACNA1A correlates with SCA6 frequency across several populations, but this effect was largely driven by data from a single population. Low frequency of SCA1 and MJD/SCA3 in Peru is not explained by frequency of LN alleles at ATXN1 and ATXN3, respectively. The observed correlation between CACNA1A LN alleles and SCA6 frequency requires further assessment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spinocerebellar ataxias (SCAs) are a group of genetically heterogeneous neurodegenerative disorders characterized by the deterioration of the cerebellum and its afferent and efferent pathways. Some of these are caused by expansion of polyglutamine-encoding CAG trinucleotides, like SCA1, SCA2, Machado–Joseph disease (MJD/SCA3), SCA6, SCA7, SCA12, SCA17, and dentatorubro-pallidoluysian atrophy [1].
The overall prevalence of SCAs is ~ 3 per 100,000 individuals, with MJD/SCA3, SCA6, and SCA1 among the most common worldwide [2, 3]. Nevertheless, there are local differences in SCA subtypes prevalence and relative frequency. SCA1 represents approximately 6% of patients with autosomal dominant cerebellar ataxia (ADCA) worldwide; however, its relative frequency among ADCAs ranges between 0% in Mexico and South Korea [4, 5] and 88% in Siberian Yakuts [6]. SCA3 is considered the most frequent ADCA in Japan, Taiwan, Portugal, and Brazil with a relative frequency between 43 and 85% [7,8,9]. In contrast, no cases of SCA3 have been reported in Czech and Finns, and its frequency is very low in Southern India (0.8%) [10,11,12]. The overall prevalence of SCA6 is approximately 0.02 to 0.31 per 100,000 individuals and represents approximately 15% from ADCAs worldwide [3, 13]. The highest frequencies of SCA6 were reported in Japan, Australia, and Germany with 25.5%, 30%, and 42%, respectively [7, 14, 15]. However, there are no cases in Mexicans and Northern Indians [5, 16].
Frequency of LN alleles can influence repeat disease prevalence by altering the microsatellite mutation rate. Disease-causing microsatellites have higher mutation rates than non-disease microsatellites [17], which stem from their propensity to form secondary DNA structures that promote defective DNA synthesis mediated by repair complexes [18, 19]. Microsatellite mutation rate increases with repeat size, even within non-pathogenic ranges [20,21,22]; thus, a high population frequency of LN alleles is expected to increase the probability that at least one non-expanded allele will reach a pathogenic size. There is a well-documented correlation between Huntington disease prevalence and frequency of large alleles within non-pathogenic range (LN alleles) [23, 24]. Likewise, in the case of SCA2, the high frequency of LN alleles has been related to the high prevalence of this disease in Cubans [25]. While an association between frequency of LN alleles and disease prevalence has also been proposed for SCA1, MJD/SCA3, and SCA6, other studies did not validate this association [5].
Lack of interruptions in the CAG repeat tract is also associated with microsatellite instability. A pure CAG repeat tract is prone to form hairpin structures that promote faulty DNA synthesis mediated by repair complexes targeting the DNA hairpins [18, 19]. Thus, DNA sequences interrupting CAG repeat tracts destabilize secondary DNA structures and impair hairpin formation [18, 26]. The ATXN1 microsatellite presents up to three instances of the CAT trinucleotide within the CAG tract, with varying frequency across populations. Studies performed in North American, Indian, Chinese, and Siberian Yakuts people suggest that the higher the population frequency of alleles carrying a single CAT interruption, the higher the relative frequency of SCA1 among SCAs [27,28,29]. Furthermore, except for very rare cases, most expanded alleles are uninterrupted by CAT trinucleotides [30]. A study of a SCA17-causing microsatellite shows similar results, with mutation frequency of pure CAG tracts at least twice as high as in alleles bearing CAA interruptions [31].
The distribution of normal alleles at ATXN1, ATXN3, and CACNA1A has been described in several ethnic groups from Asia, Europe, and Africa, but studies in Latin American populations other than Brazilians and Mexicans are lacking [5, 9, 11, 32,33,34,35,36]. Furthermore, the frequency or presence of CAT interruptions at ATXN1 gene has been described in some populations but not in Latin America [27, 28, 36]. Studying the population frequency of LN alleles and microsatellite repeat interruptions could therefore help inform the epidemiology of these diseases in Latin America.
The Neurogenetics Research Center has implemented the genotyping of ATXN1, ATXN3, and CACNA1A CAG microsatellites, which has allowed us to provide the diagnosis for SCA1, SCA3, and SCA6, as well as to study the epidemiology of these diseases in Peruvian population. Little is known about the factors that drive the prevalence and incidence of these diseases in Peruvian population. In this study, we sought to test whether allele size distribution correlates with frequency of SCA1, MJD/SCA3, and SCA6 in Peru. To this end, we PCR-genotyped 213 individuals of self-reported mestizo ancestry from Northern Lima, Peru, and compared the allele distribution of ATXN1, ATXN, and CACNA1A unstable microsatellites with the frequency of SCA1, MJD/SCA3, and SCA6 across populations. We also tested 40 individuals for the presence of CAT interruptions within the ATXN1 microsatellite.
Methods
Subjects
Using the Minsage software, we estimated that we required 213 subjects to obtain a 95% probability to detect an allele with 1.4% population frequency under no Hardy–Weinberg equilibrium [37]. We targeted for such allele because it was the rarest allele observed, in Brazilians, in any of the studied genes that has a high-enough frequency not to be considered a mutation (< 1%) [32]. Therefore, we recruited 213 healthy unrelated individuals from Northern Lima during a 6-month period of 2013. All participants were self-declared mestizo adults (admixed Peruvian individuals of mostly Amerindian and European ancestry), having Peruvian ancestors for at least two generations. Lima’s population has admixed ancestry, mostly from native Americans and Europeans [38]. Participants were 26.3% male, with ages ranging between 18 and 84 years (median 36, interquartile range 29.25). A standardized neurological examination by trained neurologists was performed with all participants to exclude any current gait instability and any other neurodegenerative disorder. The study was approved by the Institutional Review Board at the Instituto Nacional de Ciencias Neurológicas and informed consent was obtained from all individuals.
Samples
Genomic DNA was extracted from peripheral blood samples by the salting out method and were stored at − 20 °C until analysis [39]. DNA concentration and quality were assessed by EPOCH spectrophotometer (Biotek ®, Winnoski, USA).
Genotyping
Samples were genotyped for CAG repeat size at ATXN1, ATXN3, and CACNA1A. All three loci were amplified by PCR in different reactions using an ABI Prism Thermal cycler. Primers for (CAG)n in ATXN1 gene were Rep1: 5′-GTACGTCCACATTTCCAGTT-3′ and Rep2: 3′-CAACATGGGCAGTCTGAG-5′ [40]; for (CAG)n in ATXN3 were MJD52 5′-CCAGTGACTACTTTGATTCG-3′ and MJD25 3′-AACCCTCACTAGATCCATTC-5′ [41]; and for CACNA1A were S-5-F1 5′-CACGTGTCCTATTCCCCTGTGATCC-3′ and S-5-R1 5′-CACCAGCGGCCCTCGGAGGTACCCA-3′ [42]. PCR products were genotyped by 6% polyacrylamide gel electrophoresis. Briefly, we used samples of known genotype (previously analyzed by capillary electrophoresis) to perform a linear regression of repeat size versus distance migrated on polyacrylamide gel electrophoresis. Next, we used the linear model to predict repeat size of unknown samples based on their migrated distance.
Given that the mutability of the ATXN1 CAG microsatellite is influenced not only by the repeat tract length, but also by the presence of CAT interruptions, we also analyzed the frequency of this interruption in the normal population. We selected the forty individuals with the largest ATXN1 normal alleles and tested them for the presence of CAT interruptions by RFLP. The PCR products were enzymatically digested by LweI (SfaNI) 10 U/μL (Fermentas Life Sciences), for 5 h at 37 °C, and subsequently were electrophoresed in 8% PAGE.
Statistical Analysis
Hardy–Weinberg equilibrium was tested with the Genepop v.4.5.1 [43]. For each gene analyzed, alleles were grouped in normal and large normal according to previously reported thresholds [44]. Specifically, we considered LN alleles those with over 31, 27 and 13 CAG repeats for ATXN1, ATXN3, and CACNA1A, respectively [44]. Normality of each distribution was assessed by Shapiro–Wilk test (S-W). Frequency of LN alleles across populations was compared by a chi-squared test (α = 0.05) and Fisher’s exact test performed with the Stata v.12. We used the Bonferroni correction to adjust for multiple testing. We divided 0.05 by the number of total comparisons to find the new cutoff value to declare that a statistical test was significant. We estimated the 95% confidence interval of the frequencies of large normal alleles using the Klopper–Pearson method [45] implemented in the binom package [46] for the R v.3.5 [47].
Relationship between frequency of LN alleles and relative SCA frequency was tested by Spearman’s correlation coefficient. All figures were created using the R v.3.5.
Results
Distribution of CAG Repeat Size of ATXN1, ATXN3, and CACNA1A Genes in Peruvian Population
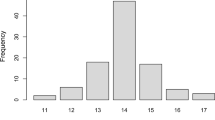
ATXN1 showed a normal distribution (S-W, p value > 0.05) slightly asymmetric, while ATXN3 and CACNA1A presented a prominently asymmetric and non-normal distribution (S-W, p value < 0.05) (Fig. 1). ATXN1 presented 12 different alleles ranging between 24 and 35 CAG repeats, with 29 and 30 CAG repeats as the most frequent alleles. DNA digestion with LweI restriction enzyme showed that all forty individuals carried at least one CAT interruption within the ATXN1 CAG repeat tract in both chromosomes. ATXN3 showed 28 different alleles in the range between 9 and 38 CAG repeats; the most frequent ones were 14, 23, and 24 CAG repeats. CACNA1A displayed 12 different alleles ranging from 3 to 17 CAG repeats. The most frequent ones were 11, 12, and 13 CAG repeats. All three loci deviated from Hardy–Weinberg equilibrium with p values 0.014 (ATXN1), < 0.001 (ATXN3), and < 0.001 (CACNA1A) and presented heterozygotes deficit, as shown by the moderate FIS statistics (0.114, 0.154, and 0.227 for ATXN1, ATXN3, and CACNA1A, respectively).
Frequency of LN Alleles
We found 68 (16.0%), 40 (9.4%), and 31 (7.3%) LN alleles at ATXN1, ATXN3, and CACNA1A genes, respectively. Differences in LN allele frequency between Peru and other populations did not allow us to conclude any association with relative frequency of SCA1, MJD/SCA3, and SCA6 (Table 1). There was no correlation between frequency of LN alleles and frequency of SCA1 or MJD/SCA3 cases (p value > 0.05). However, there was a significant correlation between frequency of LN alleles at CACNA1A and relative SCA6 frequency (Fig. 2).
Discussion
Frequency of LN alleles at disease-causing unstable microsatellites has been proposed to be associated with prevalence of the corresponding diseases including SCAs [23, 24, 44], presumably because mutation rate increases with microsatellite length, even within non-pathogenic range [22, 50]. In this study, we sought to test whether allele size distribution correlates with frequency of SCA1, SCA3, and SCA6 in Peru. For this purpose, we PCR-genotyped 213 self-reported mestizo individuals from Northern Lima, Peru, and compared the allele distribution of ATXN1, ATXN3, and CACNA1A unstable microsatellites across populations with differing frequency of SCA1, MJD/SCA3, and SCA6. The frequency of LN alleles in the studied population was 16%, 9.4%, and 7.3% for ATXN1, ATXN3, and CACNA1A genes, respectively, and was not associated with relative frequencies of SCA1, MJD/SCA3, and SCA6 in the Peruvian population.
Observed allele size ranges were similar to those reported in other populations (Fig. 1). ATXN1 allele size range (24–35 CAG repeats) in Peruvians was similar to that of Mexican (26–38) [5], Chinese (26–35) [28], and African (21–36) [51] populations. The allele size range of ATXN3 (9–38) was similar to that of individuals in other Latin Americans countries such as Mexicans (14–34) [5], Brazilians (12–34) [32], and Portugueses (14–36) [52], as well as to that of other populations across the globe [14, 16, 44]. CACNA1A repeat range (3–17) was similar to previous reports worldwide [5, 44, 53, 54]. We did not find any individual carrying triplet expansions or mutation-prone, intermediate alleles on any of the genes analyzed.
Frequency of ATXN1 LN alleles in Peru (16%) was moderate compared with other populations (Table 1). Interestingly, frequency of LN alleles in Peruvians was similar to that of Caucasians (16%) even though SCA1 frequency in this population is much higher (15%) than that in Peru (2.6%) [44, 49]. Given the intermediate frequency of ATXN1 LN alleles in Peruvians (Table 1), a moderate frequency of SCA1 cases could be expected in Peru according to the hypothesis that LN alleles are a source of de novo repeat expansions [23, 24, 44]. Moreover, frequency of LN alleles of ATXN1 in Mexican population (27.5%) is among the highest worldwide, although there are no reported SCA1 cases [5]. Furthermore, populations with a varying frequency of SCA1 cases, such as Brazilian (Paraná) [32], Northern Indian [16], Caucasian [44], and Portuguese [9], do not show statistically significant differences in frequency of LN alleles. Particularly, the Siberian Yakuts show a markedly high frequency of SCA1 (88.1%), but their frequency of LN alleles (11.2%) is similar to that of Peru (16.0%, Table 1). These results seem to challenge the role of LN alleles on influencing the SCA1 prevalence; however, we note that, if this effect exists, it may be stronger in populations with high frequency of low number of CAT interruptions. For instance, the frequency of singly interrupted CAG tracts in Siberian Yakuts (65%) is much greater than that of other populations where SCA1 has a low prevalence, such as Chinese Han (8% singly interrupted alleles) [28]. The frequency of LN alleles in India is similar to that of Caucasian populations, but their SCA1 prevalence is higher; these results have been attributed to the difference in the frequency of singly interrupted ATXN1 alleles (2% in Polish and 16% in Indians) [12, 36].
All 40 samples analyzed tested positive for the presence of CAT interruptions within the CAG tract at both chromosomes. DNA sequences interrupting (CAG)n tracts destabilize repeat-induced secondary DNA structures and impair formation of hairpins that promote expansions during DNA synthesis [18, 19, 26]. Thus, the lack of agreement between the frequency of LN alleles and SCA1 frequency in Peruvians might be explained by the high frequency of alleles that have at least two CAT interruptions in this population. However, further analyses quantifying the number of CAT interruptions in the CAG tracts are required to test this hypothesis.
The frequency of LN alleles at the ATXN3 locus was also intermediate in Peru (9%) compared with countries with the most extreme values, such as Southern India (1%) and Japan (21%) [12, 44]. Frequency of LN alleles in Peruvian people was similar to that of Spanish [53], Northern Indian [16], and Caucasian [44] populations, where the frequency of SCA3 ranges between 5 and 30%. However, frequency of LN alleles in Peru was greater than in Mexican [5], Southern Brazilian [32], and Australian [14] which feature low frequency of LN alleles (1.75–5%) but high frequency of SCA3 (12–74%) compared with Peru (5.26%) [49]. Likewise, Czechs and Finnish populations have a similar frequency of LN alleles to that of Peruvians but no cases of SCA3 [10, 11]. These seemingly random differences and similarities in allele frequencies between divergent populations could be explained by genetic drift [55]. Therefore, we could not identify a clear-cut correlation between frequency of LN alleles and relative SCA3 frequency as was suggested in a previous study [44]. This is in agreement with the putatively few inferred origins of the SCA3 mutations according to the association of two common haplotypes with the SCA3 mutation in different worldwide populations and lack of associations of these haplotypes with large normal alleles in healthy population [56, 57].
Frequency of LN alleles at CACNA1A in Peru was moderate (7.28%) compared with other locations such as Northern India (1.7%) and Japan (20%, see Table 1) [16, 44]. Interestingly, populations with higher frequency of CACNA1A LN alleles tend to have a greater SCA6 frequency (Fig. 2), which is consistent with previous studies [44]. Nevertheless, this correlation was strongly driven by a single point corresponding to Japanese population, since statistical significance was lost by removing this single point (p = 0.0745). Further studies are necessary to confirm the observed correlation.
Overall, we found no evidence of association between SCA1 and SCA3 frequency with frequency of LN alleles at their corresponding causal genes. CACNA1A LN alleles showed a statistically significant correlation with SCA6 frequency, but doubts regarding its robustness remain. Thus, our results disagree with a previous study that proposed that frequency of LN alleles is associated with SCA prevalence [44].
We propose that further studies are necessary to discern the role of the frequency of LN alleles, especially for ATXN1 and CACNA1A. Further studies of ATXN1 allele distribution should quantify the number of CAT interruptions, not performed in this study, since this genetic feature seems to play a major role in ATXN1 repeat instability. Correlation between CACNA1A LN alleles and SCA6 frequency should be confirmed by collecting additional data from other populations. Future studies should replace relative disease frequency with disease prevalence, because the relative frequency of any particular SCA will be affected by the epidemiology of other disorders.
References
López-Flores I, Garrido-Ramos MA. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 2010;7:1–28.
Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9:885–94.
Schöls L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304.
Jin DK, Oh MR, Song SM, Koh SW, Lee M, Kim GM, et al. Frequency of spinocerebellar ataxia types 1,2,3,6,7 and dentatorubral pallidoluysian atrophy mutations in Korean patients with spinocerebellar ataxia. J Neurol. 1999;246:207–10.
Magaña JJ, Tapia-Guerrero YS, Velázquez-Pérez L, Cerecedo-Zapata CM, Maldonado-Rodríguez M, Jano-Ito JS, et al. Analysis of CAG repeats in five SCA loci in Mexican population: epidemiological evidence of a SCA7 founder effect. Clin Genet. 2014;85:159–65.
Puzyrev VP, Maximova NP. Hereditary diseases among Yakuts. Russ J Genet. 2008;44:1141–7.
Maruyama H, Izumi Y, Morino H, Oda M, Toji H, Nakamura S, et al. Difference in disease-free survival curve and regional distribution according to subtype of spinocerebellar ataxia: a study of 1,286 Japanese patients. Am J Med Genet. 2002;114:578–83.
Soong B, Lu Y, Choo K, Lee H. Frequency analysis of autosomal dominant cerebellar ataxias in Taiwanese patients and clinical and molecular characterization of spinocerebellar ataxia type 6. Arch Neurol. 2001;58:1105–9.
Silveira I, Miranda C, Guimaraes L, Moreira M-C, Alonso I, Mendonca P, et al. Trinucleotide repeats in 202 families with ataxia: a small expanded (CAG) n allele at the SCA17 locus. Arch Neurol. 2002;59:623–9.
Bauer PO, Zumrova A, Matoska V, Marikova T, Krilova S, Boday A, et al. Absence of spinocerebellar ataxia type 3/Machado–Joseph disease within ataxic patients in the Czech population. Eur J Neurol. 2005;12:851–7.
Juvonen V, Hietala M, Kairisto V, Savontaus M-L. The occurrence of dominant spinocerebellar ataxias among 251 Finnish ataxia patients and the role of predisposing large normal alleles in a genetically isolated population. Acta Neurol Scand. 2005;111:154–62.
Alluri RV, Komandur S, Wagheray A, Chaudhuri JR, Null S, Meena AK, et al. Molecular analysis of CAG repeats at five different spinocerebellar ataxia loci: correlation and alternative explanations for disease pathogenesis. Mol Cells. 2007;24:338–42.
Gomez CM. Spinocerebellar Ataxia Type 6. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Stephens K, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993 [cited 2018 Oct 26]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1140/.
Storey E, du Sart D, Shaw JH, Lorentzos P, Kelly L, McKinley Gardner RJ, et al. Frequency of spinocerebellar ataxia types 1, 2, 3, 6, and 7 in Australian patients with spinocerebellar ataxia. Am J Med Genet. 2000;95:351–8.
Schöls L, Amoiridis G, Büttner T, Przuntek H, Epplen JT, Riess O. Autosomal dominant cerebellar ataxia: phenotypic differences in genetically defined subtypes? Ann Neurol. 1997;42:924–32.
Saleem Q, Choudhry S, Mukerji M, Bashyam L, Padma MV, Chakravarthy A, et al. Molecular analysis of autosomal dominant hereditary ataxias in the Indian population: high frequency of SCA2 and evidence for a common founder mutation. Hum Genet. 2000;106:179–87.
Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Sci. 1997;94:1041–6.
Gacy AM, Goellner G, Juranić N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–40.
Guo J, Gu L, Leffak M, Li G-M. MutSβ promotes trinucleotide repeat expansion by recruiting DNA polymerase β to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis. Cell Res. 2016;26:775–86.
Ellegren H. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 2000;16:551–8.
Fu Y-H, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–58.
Falush D, Iwasa Y. Size-dependent mutability and microsatellite constraints. Mol Biol Evol. 1999;16:960.
Kay C, Collins JA, Wright GEB, Baine F, Miedzybrodzka Z, Aminkeng F, et al. The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am J Med Genet Part B Neuropsychiatr Genet. 2018;177:346–57.
Squitieri F, Andrew SE, Goldberg YP, Kremer B, Spence N, Zelsler J, et al. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum Mol Genet. 1994;3:2103–14.
Laffita-Mesa JM, Velázquez-Pérez LC, Santos Falcón N, Cruz-Mariño T, González Zaldívar Y, Vázquez Mojena Y, et al. Unexpanded and intermediate CAG polymorphisms at the SCA2 locus (ATXN2) in the Cuban population: evidence about the origin of expanded SCA2 alleles. Eur J Hum Genet. 2012;20:41–9.
Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, Nelson DL, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–8.
Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993;5:254–8.
Zhou YX, Qiao WH, Gu WH, Xie H, Tang BS, Zhou LS, et al. Spinocerebellar ataxia type 1 in China: molecular analysis and genotype-phenotype correlation in 5 families. Arch Neurol. 2001;58:789–94.
Mittal U, Sharma S, Chopra R, Dheeraj K, Pal P, Srivastava A, et al. Insights into the mutational history and prevalence of SCA1 in the Indian population through anchored polymorphisms. Hum Genet. 2005;118:107–14.
Spinocerebellar Ataxia Type 1 - GeneReviews™ - NCBI Bookshelf [Internet]. [cited 2012 Sep 17]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1184/.
Gao R, Matsuura T, Coolbaugh M, Zühlke C, Nakamura K, Rasmussen A, et al. Instability of expanded CAG/CAA repeats in spinocerebellar ataxia type 17. Eur J Hum Genet. 2008;16:215–22.
Freund AA, Scola RH, Teive HAG, Arndt RC, da Costa-Ribeiro MCV, Alle LF, et al. Spinocerebellar ataxias: microsatellite and allele frequency in unaffected and affected individuals. Arq Neuropsiquiatr. 2009;67:1124–32.
Popova SN, Slominsky PA, Pocheshnova EA, Balanovskaya EV, Tarskaya LA, Bebyakova NA, et al. Polymorphism of trinucleotide repeats in loci DM, DRPLA and SCA1 in East European populations. Eur J Hum Genet. 2001;9:829.
Limprasert P, Nouri N, Heyman RA, Nopparatana C, Kamonsilp M, Deininger PL, et al. Analysis of CAG repeat of the Machado-Joseph gene in human, chimpanzee and monkey populations: a variant nucleotide is associated with the number of CAG repeats. Hum Mol Genet. 1996;5:207–13.
Limprasert P, Nouri N, Nopparatana C, Deininger PL, Keats BJ. Comparative studies of the CAG repeats in the spinocerebellar ataxia type 1 (SCA1) gene. Am J Med Genet. 1997;74:488–93.
Sobczak K, Krzyzosiak WJ. Patterns of CAG repeat interruptions in SCA1 and SCA2 genes in relation to repeat instability. Hum Mutat. 2004;24:236–47.
Details: IMBS [Internet]. [cited 2019 Mar 24]. Available from: https://www.imbs.uni-luebeck.de/forschung/software/details.html.
Harris DN, Song W, Shetty AC, Levano KS, Cáceres O, Padilla C, et al. Evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire. Proc Natl Acad Sci. 2018;115:E6526–35.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215.
Orr HT, Chung MY, Banfi S, Kwiatkowski TJ Jr, Servadio A, Beaudet AL, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–6.
Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–8.
Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the α1A-voltage-dependent calcium channel. Nat Genet. 1997;15:62–9.
Rousset F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–6.
Takano H, Cancel G, Ikeuchi T, Lorenzetti D, Mawad R, Stevanin G, et al. Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am J Hum Genet. 1998;63:1060–6.
Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–13.
Dorai-Raj S. binom: binomial confidence intervals for several parameterizations [Internet]. 2014 [cited 2019 May 8]. Available from: https://CRAN.R-project.org/package=binom.
R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. p. 2014.
Gan S-R, Shi S-S, Wu J-J, Wang N, Zhao G-X, Weng S-T, et al. High frequency of Machado-Joseph disease identified in southeastern Chinese kindreds with spinocerebellar ataxia. BMC Med Genet. 2010;11:47.
Cornejo-Olivas M, Inca-Martinez M, Castilhos R, Vasata Furtado G, Preusser Mattos E, Bavia Bampi G, et al. Hereditary ataxias in Peru: genetic analysis of familial and sporadic cases. The Cerebellum. 2019;In press.
Semaka A, Kay C, Doty C, Collins JA, Bijlsma EK, Richards F, et al. CAG size-specific risk estimates for intermediate allele repeat instability in Huntington disease. J Med Genet. 2013;50:696–703.
Andrés AM, Lao O, Soldevila M, Calafell F, Bertranpetit J. Dynamics of CAG repeat loci revealed by the analysis of their variability. Hum Mutat. 2003;21:61–70.
Lima M, Costa M d C, Montiel R, Ferro A, Santos C, Silva C, et al. Population genetics of wild-type CAG repeats in the Machado-Joseph disease gene in Portugal. Hum Hered. 2005;60:156–63.
Pujana MA, Corral J, Gratacòs M, Combarros O, Berciano J, Genís D, et al. Spinocerebellar ataxias in Spanish patients: genetic analysis of familial and sporadic cases. Hum Genet. 1999;104:516–22.
Jiang H, Tang B, Xia K, Zhou Y, Xu B, Zhao G, et al. Spinocerebellar ataxia type 6 in Mainland China: molecular and clinical features in four families. J Neurol Sci. 2005;236:25–9.
Templeton AR. Population genetics and microevolutionary theory: John Wiley & Sons; 2006.
Gaspar C, Lopes-Cendes I, Hayes S, Goto J, Arvidsson K, Dias A, et al. Ancestral origins of the Machado-Joseph disease mutation: a worldwide haplotype study. Am J Hum Genet. 2001;68:523–8.
Maciel P, Gaspar C, Guimarães L, Goto J, Lopes-Cendes I, Hayes S, et al. Study of three intragenic polymorphisms in the Machado-Joseph disease gene (MJD1) in relation to genetic instability of the (CAG)n tract. Eur J Hum Genet. 1999;7:147–56.
Acknowledgments
CGS and CCR contributed equally to this study, and are both to be considered first authors. Results of this study are derived from CGS and CCR Master’s thesis’ work.
We are very grateful to the participants of the community of Huaura, Santa María, Hualmay, and Huacho for contributing samples for this study. We kindly thank the Universidad de Huacho for the logistical support on the recruitment of participants. We thank Saul Lindo Samanamud for his cooperation in the recruitment of participants. We thank Kevin Boehnke, Miguel Inca Martínez, Hugo Sarapura Castro, and Erick Figueroa Ildefonso for their contribution on critical review and genotyping.
Funding
Research reported in this publication was supported by the Fogarty International Center (FIC) of the National Institutes of Health and the National Institute of Neurological Disorders and Stroke (NINDS) under grant #D43TW009345 awarded to the Northern Pacific Global Health Fellows Program and grant #D43TW009137 awarded to the Interdisciplinary Cerebrovascular Diseases Training Program in South America.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures performed in this study involving human participants were in accordance with the standards of the Ethical Research Committee of the Instituto Nacional de Ciencias Neurológicas and with the 1964 Helsinki declaration and its later amendments.
Conflict of Interest
The authors declare that they have no conflict of interest.
Disclosure
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gonzales-Sáenz, C., Cruz-Rodriguez, C., Espinoza-Huertas, K. et al. Distribution of the CAG Triplet Repeat in ATXN1, ATXN3, and CACNA1A Loci in Peruvian Population. Cerebellum 19, 527–535 (2020). https://doi.org/10.1007/s12311-020-01129-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12311-020-01129-3