Abstract
Chlorophytum borivilianum is a traditional medicinal plant distributed throughout the tropics and subtropics. In the present investigation, AFLP analysis was used to assess the genetic similarity among 34 accessions. Nine primer sets of AFLP amplified 612 fragments, of which 246 fragments were found to be polymorphic. The average number of polymorphic bands per AFLP primer pair was 27.33. The amplified fragments ranged from 50 base pairs to 600 base pairs. Significant correlation was observed between total number of amplified fragments and polymorphic bands (p > 0.05) per primers. Cluster analysis based on AFLP data revealed limited genetic variation within the thirty four accessions collected from various parts of Central Indian forests.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorophytum borivilianum commonly known as ‘Safed Musli’ has many therapeutic applications in Ayurvedic, Unani, and Homeopathic and Allopathic system of medicine. The Chlorophytum genus having about 300 species are found throughout the tropical and subtropical parts of the world (Vijaya and Chavan 2009). In India, Safed musli is found in Southern Rajasthan, Northern Gujarat and Western Madhya Pradesh (Maiti and Geetha 2005). C. borivilianum roots (tubers) are rich source of over 25 alkaloids, vitamins, proteins, carbohydrates, steroids, saponins, potassium, calcium, magnesium, phenol, resins, mucilage, and polysaccharides and also contains high quantity of simple sugars, mainly sucrose, glucose, fructose, galactose, mannose and xylose (Thakur et al. 2009). Safed musli is considered to be very effective for enhancing the general body immunity. It has been listed as an endangered species in “Red data book of Indian plants” by the Botanical Survey of India (Nayar and Shastry 1988) and it is predicted that if steps for conservation are not taken, the Indian forests will lose this valuable plant (Oudhia 2001).
The characterization of C. borivilianum germplasm has been based mainly on phenotypic characteristics. C. borivilianum shows wide morphological and agronomic variations for roots (Geetha and Maiti 2002) and leaves (Kothari and Singh 2001; Kumar et al. 2008). However, morphological characteristics can vary under different agro-climatic conditions. Divergence studies made on the basis of molecular markers provide precise information about phylogenetic descriptive and hierarchical account of individuals or groups of individuals. PCR technology has promoted the development of a range of molecular assay systems which detect polymorphism at the DNA level. In this study, we used the most widely adopted AFLPs for characterizing the natural variations of C. borivilianum. AFLP’s have been used to estimate genetic relationships and authentication of many medicinal plants species e.g. Aloe vera (Tripathi et al. 2011), Swertia (Misra et al. 2010), Echinacea species (Russi et al. 2009), Tribulus terrestris (Sarwat et al. 2008) and Allium sativum (Ipek et al. 2003). Comparative studies of RAPD, RFLP, AFLP and microsatellites have shown that AFLP is the most-efficient method to estimate genetic diversity because of high reproducibility and multiplex ratio (Powell et al. 1996; Pejic et al. 1998). Knowledge on the genetic diversity and relationships among plant varieties is important to recognize gene pools, for intellectual property rights (IPR) purposes and to develop effective conservation and management strategies. The objective of this study was to evaluate genetic diversity among C. borivilianum accessions collected from Central India using AFLP markers.
Materials and methods
Plant material and DNA extraction
Thirty four C. borivilianum accessions collected from different parts of Central India and maintained at Department of Plant Physiology, J. N. Agricultural University, Jabalpur, India were used in this study. The fresh young leaves were excised from single plant of each accession, and used for DNA extraction by the CTAB method (Doyle and Doyle 1990).
AFLP fingerprinting
The AFLP procedure was performed according to the protocol of Vos et al. (1995) with minor modifications. Approximately 350 ng DNA of each of the accession was digested simultaneously with EcoRI and MseI at 37 °C for 2 h. A small aliquot of the digested DNA was run on a 1.5 % (w/v) agarose gel to check if the DNA digestion was complete. The digested samples were incubated at 70 °C for 15 min to inactivate the restriction endonucleases. EcoRI and MseRI adaptors were ligated to the digested DNA samples to generate template DNA for amplification. The ligation products were diluted 10 fold and 5 μl added to preamplification reaction. Preamplification was carried out with one selective nucleotide MseI complementary primer which had a 3’-C and the EcoRI complementary primer had a 3’-A, in a thermocycler for 20 cycles set at 94 °C denaturation (30 s), 56 °C annealing (60 s) and 72 °C extensions (60 s). The amplification products were diluted 50 folds and stored at −20 °C.
Selective AFLP amplification was carried out with EcoRI and MseI primers each carrying extra three and two selective nucleotides respectively. The PCR selective amplification temperature profile was as follows: one cycle at 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 60 s; followed by 12 cycles of touchdown PCR in which the annealing temperature decreased by 1.0 °C every cycle until a ‘touchdown’ annealing temperature of 56 °C was reached. Once reached, another 23 cycles were conducted as described above for preamplification. Ten μl of the reaction product was mixed with an equal volume of 3X STR loading buffer (95 % [v/v] formamide, 0.005 % [v/v] of each of xylene cyanol and bromophenol blue), denatured by incubating at 90 °C for 5 min and quickly cooled on ice. The products were analyzed on 4 % (w/v) denaturing polyacrylamide gels. The gel was run at constant power (50 ± 2 W) until the xylene cyanol was about two-thirds down the length of the gel. The gels were silver stained and scanned with scanner for analysis and documentation.
Data analysis
To analyze the AFLP markers data, the amplified fragments were scored qualitatively as a dominant allele at a unique locus. Polymorphic amplified fragments were scored manually as ‘1’ for the presence and ‘0’ for the absence of an allele at a particular locus across all the 34 genotypes for each primer combination. Only clearly distinct bands were scored for data analysis. Binary data obtained for the AFLP primer combinations was used for assessing the discriminatory power of AFLP primer combinations. PIC was calculated via the formula PICi = 1—Σ P2ij where, Pij is the frequency of the jth allele for ith locus summed across all alleles for the locus. Cluster analysis was based on Jaccard’s similarity coefficient (Jaccard 1908) by using the unweighted pair group method using the arithmetic averages (UPGMA) method in NTSYS-pc version 2.1 (Rohlf 2000) software package.
Results and discussion
Fundamental goal of conservation strategy is to preserve genetic diversity within and among the natural and wild populations of potential economic values. Currently, biotechnological approaches are an essential component of plant genetic resources management. C. borivilianum is one of the potential ayurvedic Indian medicinal herb of global interest with variety of medicinally valuable compounds. Identification of elite C. borivilianum accessions with high concentrations of valuable compounds is important in ayurvedic and pharmaceutical industries, since it can directly affect the quality of pharma products. One of the important ways by which these can be identified is to screen the available germplasm for extent of existing genetic diversity using molecular markers. During the present investigation, 34 accessions collected from Central India were analyzed for molecular diversity using AFLP.
Cultivar identification using AFLP markers
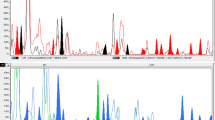
A total of 16 EcoRI and MseI primer combinations (Table 1) were tested and nine were selected on the basis of good banding profile. A total of 612 electromorphs were amplified with an average of 68 electromorphs per combination. Out of 612 electromorphs, 246 amplified as polymorphic alleles with an average of 27.33 ± 4.670 per primer combination. Polymorphic alleles are present in one or more but not in all the individuals and unique ones are present at least in one individual but not in others (Mehetre et al. 2004). During the present investigation, AFLPs analysis indicated the presence of small amount of genetic diversity amongst the cultivars in spite of the presence of high number of polymorphic bands. This situation arose as most of these polymorphic bands appeared at a high frequency only in some of the genotypes evaluated. The combination EcoRI GCA+MseI AG amplified maximum electromorphs (91), while EcoRI ATC+MseI AT amplified minimum (47). The primer combination EcoRI GCA+MseI GA exhibited highest (65.43) polymorphism among all the primer combinations. An example of AFLP banding patterns has been presented in Fig. 1a and b. The size of amplified fragments ranged from 50 to 600 bp. There was a significant correlation (p > 0.05) between total number of amplified fragments and polymorphic bands per primer. The low PIC values ranging from 0.125 to 0.379 with an average 0.257 ± 0.031 indicated the presence of less genetic diversity amongst the accessions under study.
AFLP profiles of Chlorophytum borivilianum as amplified by (a) ECoRI GCA+MseI GA primer (b) ECoRI ATC+MseI AT primer. M=Marker (100 bp), Lane 1-CBI 1, Lane 2-CBI 2, Lane 3-CBI 3, Lane 4-CBI 4, Lane 5-CBI 5, Lane 6-CBI 6, Lane 7-CBI 7, Lane 8-CBI 8, Lane 9-CBI 9, Lane 10-CBI 10, Lane 11-CBI 11, Lane 12-CBI 12, Lane 13-CBI 13, Lane 14-CBI 15, Lane 15-CBI 16, Lane 16-CBI 18, Lane 17-CBI 19, Lane 18-CBI 20, Lane 19-CBI 21, Lane 20-CBI 22, Lane 21-CBI 23, Lane 22-CBI 24, Lane 23-CBI 25, Lane 24-CBI 26, Lane 25-CBI 31, Lane 26-CBI 32, Lane 27-CBI 33, Lane 28-CBI 34, Lane 29-CBI 35, Lane 30-CBI 36, Lane 31-CBI 37, Lane 32-CBI 39, Lane 33-CBI 40, Lane 34-MCB 405
The primer combinations amplified specific alleles enabling differentiation of specific accessions from others. The primer combination EcoRI ATC+MseI GG amplified maximum number of specific alleles while EcoRI GCA+MseI AT amplified only one specific allele whereas, primer combination EcoRI ATC+MseI GC failed to amplify any specific allele. Among the 34 accessions, only 16 accessions possessed specific alleles (Table 2). Seven accessions namely CBI-3, CBI-4, CBI-7, CBI-9, CBI-25, CBI-32 and CBI-37 exhibited only one specific allele, four accessions CBI-6, CBI-12, CBI-34 and CBI-39 had two specific alleles wherein four accessions viz. CBI-11, CBI-13, CBI-33 and CBI-40 had 3–4 specific alleles. The accessions MCB-405 had the highest specific alleles (33) and almost all the primers amplified specific allele in this accession. Identification and authentication of genotypes based on the presence of specific alleles have been reported in many medicinal plant species (Hosokawa et al. 2000; Negi et al. 2006; Khan et al. 2009) including Chlorophytum species (Katoch et al. 2010).
Genetic relationships among C. borivilianum genotypes
The similarity matrix generated using Jaccard’s similarity coefficient was used for UPGMA cluster analysis. On the basis of similarity coefficient, all the accessions were found to be very close and had low level of molecular diversity except for MCB-405. Katoch et al. (2010) also reported the low molecular diversity between two Chlorophytum species with 69 % similarity and more than 85 % similarity in five accessions of C. borivilianum using RAPD markers. Jatav et al. (2011) also observed higher genetic similarity (78 %) using RAPD markers among C. borivilianum accessions collected from different places of Central India. Our results are also in agreements with other vegetatively propagated species which exhibit extremely low or no genetic variations such as Ligusticum chuanxiong (Chen et al. 2010), Allium sativum (Paredes et al. 2008), Eichhornia crassipes (Li et al. 2006), Potamogeton maackianus (Li et al. 2004). The presence of a greater morphologic variability for agronomic traits in the collected accessions as compared to the molecular data can be assigned to the different evolutionary and environmental factors. The phenotypic variability reported in previous studies (Kothari and Singh 2001; Geetha and Maiti 2002; Kumar et al. 2008) could be explained by an intense selection pressure on this species during the domestication process. The selected characteristic adapts to the environment, the greater it is the probability to survive in time (Gepts 2004). Several researchers held responsible other forces for contributing to the high phenotypic variations such as photoperiod and temperature (Bradley et al. 1996; Ipek et al. 2003), mutations (Koul et al. 1997) and crossing over (Ordoñez et al. 2002).
On the basis of dendrogram, two groups of accessions were formed with 33 accessions in one group and MCB-405 solely in the second group (Fig. 2). This distant genetic diversity is strongly supported by AFLP electrophoretic patterns (Fig. 1). To ensure this observed genetic distance, the AFLP experiments with this cultivar were repeated several times. Due to this large genetic distance, the accession may serve as a valuable genetic source and a promising parental material in C. borivilianum improvement programmes. This should however be further evaluated and confirmed based upon more divergent varieties from more diversified regions. The presence of only two groups during the present investigation is in correspondence with the studies of Dwivedi and Sharma (2011) with RAPD markers. PCA analyses (Fig. 3), carried out using the similarity matrices for 9 AFLP primer combinations also produced trends similar to the UPGMA cluster analysis. The assessment of genetic diversity provide estimates on the level of genetic variation among materials that can be used in crop improvement programme for getting transgressive segregation (Wuang and Zhou 1995) and also exposes duplicate germplasm collections for better management and conservation. The specific alleles observed during fingerprinting will help in the assessment of purity and stability of genotypes entering in breeding and seed multiplication programmes.
The genetic diversity of a species has great implications for its long-term survival and evolution. Therefore, knowledge of the levels and patterns of genetic diversity is important for designing conservation strategies for threatened and endangered species. C. borivilianum accessions are maintained by vegetative propagation and accurate identification of vegetative materials is crucial for growers. Propagation of this species takes place vegetatively as well as with seeds. In a long domestication process, asexual reproduction must have contributed to the genetic uniformity or monoculture of the species (Gepts 2004). Estimates of genetic diversity are highly influenced by the genome selected for evaluation and by the number of markers assayed. The small sample sizes in this study, however, restrict the relevance of the analysis and the credibility of results for more generalized conclusions. Further studies should therefore be carried out using larger samples derived from diverse regions to elucidate the genetic variations of C. borivilianum and define valuable germplasm for the improvement of this important medicinal crop.
Conclusions
Results from the present study confirm the robustness and the suitability of the AFLP approach for diversity analysis and for the assessment of genetic relationship among individuals of C. borivilianum. The observed narrow genetic base in C. borivilianum might be due to the vegetative propagation and whatsoever variability present could be attributed to the spontaneous mutation. Exploitation of other molecular markers such as ISSR or microsatellites could be useful to reveal in depth genetic relationship among accessions of C. borivilianum. Knowledge of diversity patterns and specific genetic distance estimates might increase the efficiency of C. borivilianum genetic improvement in India among adapted parents used for cultivar development providing information on genetic variance and heterosis. Further studies will also be necessary to further elucidate the genetic basis of morphological and biochemical traits.
Abbreviations
- SSR:
-
Simple sequence repeat
- AFLP:
-
Amplified fragment length polymorphism
- RAPD:
-
Random amplified polymorphic DNA
- ISSR:
-
Inter simple sequence repeats
- RFLP:
-
Restriction fragment length polymorphism
- PIC:
-
Polymorphic information content
- DNA:
-
Deoxyribonucleic acid
- PCR:
-
Polymerase chain reaction
- UPGMA:
-
Unweighted pair group method with the arithmetic averaging algorithm
- PCA:
-
Principal component analysis
References
Bradley KF, Rieger MA, Collins GG (1996) Classification of Australian garlic cultivars by DNA fingerprinting. Aust J Exp Agric 36:613–618
Chen Y, Wang L, Ma Y, Xiao H, Tang L, An Q (2010) Genetic diversity of Ligusticum chuanxiong based on inter simple sequence repeat (ISSR) and amplified fragment length polymorphism (AFLP) analyses. Afr J Biotechnol 9:8290–8295
Doyle LJ, Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–14
Dwivedi R, Sharma DK (2011) Assessment of genetic diversity among collected genotypes of Chlorophytum borivilianum using random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol 10:5268–5272
Geetha K, Maiti S (2002) Biodiversity in Chlorophytum borivilianum Santapau and Fernandes. PGR Newsletter 129:52–53
Gepts P (2004) Crop domestication as a long-term selection experiment. In: Janick J (ed) Plant Breed Rev 24:1–44
Hosokawa K, Minami M, Kawahara K, Nakamura I, Shibata T (2000) Discrimination among three species of medicinal Scutellaria plants using RAPD markers. Planta Med 66:270–272
Ipek M, Ipek A, Simon P (2003) Comparison of AFLPs, RAPD markers, and isozymes for diversity assessment of garlic and detection of putative duplicates in germplasm collections. J Am Soc Hort Sci 128:246–252
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Jatav DS, Tiwari S, Kumar S, Saini N (2011) Study of polymorphism among mother stocks and genetic fidelity of regenerated plants in Chlorophytum spp. using RAPD markers. J Tropic Biodivers 19 (in press).
Katoch M, Kumar R, Pal S, Ahuja A (2010) Identification of Chlorophytum species (C. borivilianum, C. arundinaceum, C. laxum, C. capense and C. comosum) using molecular markers. Indus Crop Prod 32:389–393
Khan S, Mirza KJ, Abdin MZ (2009) Development of RAPD markers for authentication of medicinal plant Cuscuta reflexa. EurAsia J BioSci 4:1–7
Kothari SK, Singh K (2001) Evaluation of Safed musli (Chlorophytum borivilianum Santapau and Fernandes) germplasm. J Spices Arom Crops 10:147–149
Koul AK, Gohil RN, Langer A (1997) Prospects of breeding improved garlic in the light of its genetic and breeding systems. Euphytica 28:457–464
Kumar B, Verma AK, Singh HP, Mishra HO, Lal RK (2008) Genetic divergence and correlations study in Chlorophytum borivilianum. J New Seeds 9:321–329
Li W, Li QX, Jian QL, Guang XW (2004) Genetic diversity of Potamogeton maackianus in the Yangtze River. Aquat Bot 80:227–240
Li WG, Wang BG, Wang JB (2006) Lack of genetic variation of an invasive clonal plant Eichhornia crassipes in China revealed by RAPD and ISSR markers. Aquat Bot 84:176–180
Maiti S, Geetha KA (2005) Characterization, genetic improvement and cultivation of Chlorophytum borivilianum—an important medicinal plant of India. Pl Genet Res 3:264–272
Mehetre SS, Gomes M, Eapen S (2004) RAPD analysis of hybrid nature of the offspring of Gossypium hirsutum x G. raimondii. Curr Sci 84:24–28
Misra A, Shasany AK, Shukla AK, Darokar MP, Singh SC, Sundaresan V, Singh J, Bagchi GD, Jain SP, Saikia D, Khanuja SP (2010) AFLP markers for identification of Swertia species (Gentianaceae). Genet Mol Res 9:1535–1544
Nayar MP, Shastry ARK (1988) Chlorophytum borivilianum. In: Nayar MP, Sastry ARK (eds) Red data book of Indian plants. Botanical Survey of India, Calcutta, India 2:142
Negi MS, Sabharwal V, Wilson N, Lakshmi kumaran MS (2006) Comparative analysis of the efficiency of SAMPL and AFLP in assessing genetic relationships among Withania somnifera genotypes. Curr Sci 91:464–471
Ordoñez A, Torres LE, Hidalgo MG, Muñoz JO (2002) Análisis citológico de una variante genetic somática de ajo (Allium sativum L.) tipo rosado. Agriscientia 19:37–43
Oudhia P (2001) Problem perceived by Safed Musli (Chlorophytum borivilianum) grower of Chhattisgarh (India) region: a study. J Med Arom Plant Sci 22(4A)/23 (1A):396–399
Paredes MC, Becerra VV, María I, González A (2008) Low genetic diversity among garlic (Allium sativum L.) accessions detected using random amplified polymorphic DNA (RAPD). Chilean J Agric Res 68:3–12
Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor Appl Genet 97:1248–1255
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Rohlf FJ (2000) NYSYS-pc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter publications, Setauket
Russi L, Moretti C, Raggi L, Albertini E, Falistocco E (2009) Identifying commercially relevant Echinacea species by AFLP molecular markers. Genome 52:912–918
Sarwat M, Das S, Srivastava PS (2008) Analysis of genetic diversity through AFLP, SAMPL, ISSR and RAPD markers in Tribulus terrestris, a medicinal herb. Plant Cell Rep 27:519–528
Thakur GS, Bag M, Sanodiya BS, Debnath M, Zacharia A, Bhadauriya P, Prasad GB, Bisen PS (2009) Chlorophytum borivilianum: a white gold for biopharmaceuticals and neutraceuticals. Curr Pharm Biotechnol 10:650–666
Tripathi N, Saini N, Tiwari S (2011) Assessment of genetic diversity among Aloe vera accessions using amplified fragment length polymorphism. Int J Med Arom Plants 1:115–121
Vijaya NK, Chavan PD (2009) Chlorophytum borivilianum (Safed musli): a review. Phcog Rev 3:154–169
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wuang YY, Zhou J (1995) Blood relationship analysis and breeding procedures comparison between Chinese and American main tobacco cultivars. Chin Tobacco Sin 3:11–22
Acknowledgements
We are thankful for grants from Madhya Pradesh Minor Forest Produce Cooperative Ltd., Bhopal, MP, India and Department of Plant Physiology, Jawaharlal Nehru Agricultural University, Jabalpur, India for providing accessions of C. borivilianum.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tripathi, N., Saini, N., Nair, P. et al. Lack of genetic diversity of a critically endangered important medicinal plant Chlorophytum borivilianum in Central India revealed by AFLP markers. Physiol Mol Biol Plants 18, 161–167 (2012). https://doi.org/10.1007/s12298-012-0108-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-012-0108-8