Abstract
Cisplatin resistance is the main cause of colorectal cancer (CRC) treatment failure, and the cause has been reported to be related to Fusobacterium nucleatum (Fn) infection. In this study, we explored the role of Fn in regulating cisplatin resistance of CRC cells and its underlying mechanism involved. The mRNA and protein expressions were examined by qRT-PCR and western blot. Cell proliferation and cell apoptosis were assessed using CCK8 and flow cytometry assays, respectively. Dual-luciferase reporter gene assay was adopted to analyze the molecular interactions. Herein, our results revealed that Fn abundance and miR-135b expression were markedly elevated in CRC tissues, with a favorable association between the two. Moreover, Fn infection could increase miR-135b expression via a concentration-dependent manner, and it also enhanced cell proliferation but reduced apoptosis and cisplatin sensitivity by upregulating miR-135b. Moreover, KLF13 was proved as a downstream target of miR-135b, of which overexpression greatly diminished the promoting effect of miR-135b or Fn-mediated cisplatin resistance in CRC cells. In addition, it was observed that upstream 2.5 kb fragment of miR-135b promoter could be interacted by β-catenin/TCF4 complex, which was proved as an effector signaling of Fn. LF3, a blocker of β-catenin/TCF4 complex, was confirmed to diminish the promoting role of Fn on miR-135b expression. Thus, it could be concluded that Fn activated miR-135b expression through TCF4/β-catenin complex, thereby inhibiting KLF13 expression and promoting cisplatin resistance in CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is a very common human malignancy, ranking third in incidence and second in mortality among all cancers worldwide (Bray et al., 2018). Cisplatin is a well-known antitumor agent (Dasari & Tchounwou, 2014). At present, surgery combined with cisplatin-based chemotherapy is a conventional treatment strategy for CRC, but due to treatment failure, the 3-year survival rate of CRC has not been improved (Du et al., 2018; Franke et al., 2018). Cisplatin resistance is the main reason for treatment failure of CRC (Fuertes et al., 2002). Therefore, it’s urgent to find novel treatment strategies for CRC, and understanding the specific molecular mechanism in regulating cisplatin resistance in CRC is the key to achieving this goal.
Massive evidence has determined that CRC chemoresistance is a result of complex interactions between gene regulation and environment. The intestinal microbiota refers to the bacterial population inhabiting the human gastrointestinal tract, which more than thousands of different species (Gill et al., 2006). Previous evidence has revealed that the intestinal microbiota is closely related to CRC initiation and development by regulating intestinal inflammation (Arthur et al., 2012). Notably, several recent animal studies suggested that gut microbiota might influence chemotherapy sensitivity by modulating local immune responses (Iida et al., 2013; Viaud et al., 2013). Fusobacterium nucleatum (Fn) is a Gram-negative, spindle-shaped, non-spore forming oral anaerobe, which is one of the most abundant Gram-negative bacteria (Socransky et al., 1998). Surprisingly, it was observed that Fn abundance was significantly increased in CRC tissues compared with adjacent normal tissues (Kostic et al., 2012), suggesting that Fn might be a risking factor promoting CRC progression. In addition, the increase of Fn abundance in CRC tissue was related to the shorter survival period of patients (Mima et al., 2016). Notably, a previous study demonstrated that the abundance of Fn was significantly increased in the CRC tissue of patients with relapse after chemotherapy, and Fn could improve the resistance of CRC cells to chemotherapy (Yu et al., 2017). Nevertheless, the mechanism of Fn promoting cisplatin resistance in CRC remain larges unknown, which deserves further research.
MicroRNAs (miRNAs) refer to a class of small noncoding RNA molecules of about 20 nucleotides in length. There a growing body of evidences revealed that miRNA is an essential tumor regulator which can be divided into “protocancer miRNA” or “tumor suppressor miRNA” (Saliminejad et al., 2019). In CRC, dysregulated miRNAs taken part in various cancerous phenotypes such as increased proliferative and invasive abilities, immune escape, angiogenesis, and drug resistances by targeting multiple targeted genes, which was an important inducement for the pathogenesis and development of CRC (Huang et al., 2021). For instance, miR-875-3p was significantly upregulated in CRC tissues and cells, and its knockdown inhibited CRC cell proliferation and migration (Li et al., 2020a). Chen et al. (2017) revealed that miR-199a/b upregulation facilitated cisplatin resistance in CRC. Recently, aberrant expression of miR-135b was found to be associated with carcinogenesis in variety of organs (e.g. non-small-cell lung cancer, ovarian cancer and CRC) (Dai et al., 2022; Wang et al., 2022; Zhao et al., 2021). It was previously described that miR-135b induced protective autophagy to promote the drug sensitivity of CRC cells against oxaliplatin (Wang et al., 2021). Likewise, it was also confirmed that enforced miR-135b expression dramatically elevated the chemoresistance of CRC through modulating PI3K/AKT/ ST6GALNAC2 signaling pathway (Liu et al., 2017). Moreover, previous evidences reported that increasing miRNAs were functioned as downstream targets of Fn and participated in the regulating of Fn on CRC malignant phenotypes, such as miR-21, miR-1246, miR-92b-3p, miR-27a-3p, etc (Guo et al., 2021; Yang et al., 2017). Whereas, whether miR-135b could involve in Fn-induced cisplatin resistance in CRC remains need further study.
In the present research, the experimental data uncovered that Fn infection apparently activated miR-135b expression through acting on transcription factor 4 (TCF4)/β-catenin complex, thereby inhibiting Kruppel-like factor 13 (KLF13) expression and promoting cisplatin resistance in CRC. Our research provided novel perspectives for the treatment of CRC in patients with cisplatin resistance.
Materials and Methods
Clinical Samples Collection
The CRC tissues and normal adjacent tissues were collected from 25 diagnosed CRC patients who received surgical resection at Changsha First Hospital. The CRC patients were clearly diagnosed by pathology and had no radiotherapy. This study was passed the review of Ethics Committee of Changsha First Hospital before enrollment of patients and all participants signed informed consent (Approval Number: No. 2022-171).
Cell Culture and Treatment
Human CRC cells (SW620, SW480, HCT116, HT-29 and Caco2 cells) and human colon epithelial cells (NCM460 and HCoEpiC cells) were all purchased from American Type Culture Collection (ATCC) and cultured in DMEM (Gibco) containing 10% fetal bovine serum (FBS, Gibco) with 5% CO2 at 37 °C. HCoEpiC cells were cultured in colonic epithelial cell medium (CoEpiCM, ScienCell Research Laboratory) also containing 10% FBS according to the manufacturer’s instructions. Fn was also obtained from ATCC and was grown in trypticase soy broth (TSB) agar plates (containing 5% defibrinated sheep blood, 5 µg/ml hemin and 0.5% yeast extract and 1 µg/ml vitamin K1) under anaerobic conditions at 37 °C.
For cell infection, SW620 and Caco2 cells were seeded into a 6 well-plate and infected with Fn at a multiplicity of infection (MOI) of 100:1, 500:1 and 1000:1 for 48 h. For cisplatin treatment, CRC cells were treated with cisplatin (5, 10, 15 or 25 µg/ml) for 48 h.
Cell Transfection
The overexpression plasmid of KLF13 and TCF4 (pcDNA3.1-KLF13, pcDNA3.1-TCF4), mimic/inhibitor of miR-135b and their negative controls (pcDNA3.1, mimics NC, inhibitor NC) were purchased from GenePharma. Lipofectamine™ 3000 (Invitrogen) was acquired to transfect the above RNAs into SW620 and Caco2 cells in accordance with the manufacturer’s instructions. The transfection efficiency was assessed at post-transfection 48 h. The sequences of mimics and inhibitors used in this work were provided as follows: inhibitor NC, 5′-GAA UUA AUUAAAGAUGGCCCGUUGUACU-3′; miR-135b inhibitor, 5′- UCACAUAGGAAUGAAAAGCCAUA-3′; mimics NC, (sense) 5′-CAGUACUAUUGUGUAGUACAA − 3′, (antisense) 5′-GUACUUUUGUGUAGUACAAUU-3′; miR-135b mimics, (sense) 5′-UAUGGCUUUUCAUUCCUAUGUGA-3′, (antisense) 5′-GUACUUUUGUGUAGUACAAUU-3′.
DNA Extraction and Bacteria Quantification
Bacteria DNA was extracted using the QIAGEN stool kit (QIAGEN). Fn quantification was performed by quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR regent (Thermo Fisher Scientific). The data was analyzed with − 2ΔΔCT method. Universal Eubacteria 16s was used as internal reference gene. The primers were listed as follows (5′ –3′): Fn (F): CGGGTGAGTAACGCGTAAAG; Fn (R): ACATTGTGCCACGGACATCTTG; 16s (F): CGGCAACGAGCGCAACCC; 16s (R): CCATTGTAGCACGTGTGTAGCC.
RNA Extraction and qRT-PCR
Total RNA was extracted with TRIzol (Thermo Fisher Scientific). For mRNA, the cDNA was synthesized using the Reverse Transcription Kit (Toyobo). For miRNA, the cDNA was synthesized with the first-strand cDNA synthesis kit (Sangon). Then, SYBR (Thermo Fisher Scientific) was employed for the qRT-PCR assay. GAPDH and U6 were used as the reference gene for mRNA or miRNA, respectively. The data was analyzed with 2−ΔΔCT method. The primers were listed as follows (5′–3′): miR-135b (F): CTGTGGCCTATGGCTTTTCAT, miR-135b (R): GCTCGCCCCTCACTGTAG; KLF13 (F): CGGCCTCAGACAAAGGGTC, KLF13 (R): TTCCCGTAAACTTTCTCGCAG; TCF4 (F): TGCAAAGCCGAATTGAAGATCG, TCF4 (R): AGAAGGTCCAATGATTCCATGC; GAPDH (F): ATGACTCTACCCACGGCAAG, GAPDH (R): GGAAGATGGTGATGGGTTTC; U6 (F): CTCGCTTCGGCAGCACA, U6 (R): AACGCTTCACGAATTTGCGT.
Cell Counting Kit-8 (CCK-8) Assay
SW620 and Caco2 cells were plated in a 96 well-plate with 5000 cells/well. After the corresponding interventions for 48 h, the cells were incubated with 10 µl of CCK-8 solution (Dojindo) at 37 °C for 3 h. Absorbance was analyzed at 450 nm with a microplate spectrophotometer (Bioteke).
Cell Apoptosis Assay
For cell apoptosis detection, SW620 and Caco2 cells after different treatment were collected and washed with PBS for three times. Then, cells were resuspended in 500 µl of 1× Annexin-binding buffer (BD), and then incubated with 10 µl Annexin V-FITC and 5 µl PI stain avoiding light for 10 min. After washing, the cells were immediately analyzed using flow cytometry (BD).
Western Blot
The proteins were isolated with RIPA, and the protein concentration was analyzed using the BCA kit (Beyotime). Then, the same amount of protein was separated by 12% SDS-PAGE gel, which further transferred to a PVDF membrane (Millipore). After blocking, the membranes were incubated with primary antibodies against KLF13 (Proteintech, 18352-1-AP), TCF4 (Abcam, ab217668), β-catenin (Abcam, ab32572) and GAPDH (Abcam, ab8245) at 4 °C for overnight, then hybridized with secondary antibody (Abcam, ab7090) for 60 min. Blots were visualized by GEL imaging system (Bio-Rad) and subsequently analyzed with ImageJ software.
Dual-Luciferase Reporter Gene Assay
For validation the regulation between miR-135b and KLF13, the 3’-UTR fragment of KLF13 containing wide-type (wt) and mutation site (mut) miR-135b binding site were amplified by PCR, and inserted into the pmirGLO vector (Promega). Then, CRC cells were co-transfected with recombinant KLF13-wt or KLF13-mut vectors and miR-135b mimics/inhibitor or their negative controls for 48 h. To examine the effects of TCF4/β-catenin complex on miR-135b expression, the full length of miR-135b promoter fragment containing wt- or mut-TCF4 binding site were amplified by PCR, and inserted into the pGL3 vector (Promega). Then, CRC cells were co-transfected with the recombinant pGL3.0 vector and pcDNA3.1 or pcDNA3.1-TCF4 for 48 h. Finally, the luciferase activity was subsequently assessed via a dual luciferase reporter assay system (Promega).
Statistical Analysis
All our data were obtained from three independent experiments. Statistical data was analyzed by SPSS 19.0 (IBM, Armonk, NY) and expressed as means ± SD. Between-group differences and multi-group comparisons were determined using Student’s t test and one-way ANOVA, respectively. The p values less than 0.05 were considered significant.
Results
Fn Infection Gradually Increased miR-135b Expression Via a Concentration-Depend Manner
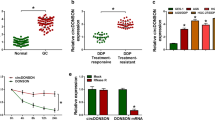
The CRC tissues and normal adjacent tissues were collected from diagnosed CRC patients, and it was found that Fn abundance and miR-135b expression were markedly increased in CRC tissues when compared with those in normal adjacent tissues (Fig. 1A, B). In addition, it was observed that the enrichment of Fn exhibited a positive correlation with miR-135b expression in clinical samples (Fig. 1C). Meanwhile, qRT-PCR assay also revealed that miR-135b expression in CRC cells was examined, and our results revealed that miR-135b was significantly upregulated in CRC cell lines (SW620, SW480, HCT116, HT-29, and Caco2 cells) compared with human colon epithelial cells (NCM460 cells, HCoEpiC cells) (Fig. 1D). To investigate the relationship between Fn and miR-135b in CRC, CRC cells were infected with Fn with different MOI. It was turned out that Fn infection increased miR-135b expression in CRC cells in a MOI dependent manner (Fig. 1E). Collectively, these findings elucidated the potential connection between Fn and miR-13b in CRC.
Fn infection gradually increased miR-135b expression via a concentration-dependent manner. The CRC tissues and normal adjacent tissues were collected from diagnosed CRC patients, and Fn abundance (A) and miR-135b expression (B) in tissues were detected by qRT-PCR (n = 25). C The correlation between Fn abundance and miR-135b expression was analyzed using Pearson correlation analysis. D qRT-PCR was adopted to measure miR-135b expression in CRC cells (SW620, SW480, HCT116, HT-29 and Caco2 cells) and human colon epithelial cells (NCM460 and HCoEpiC cell lines). SW620 and Caco2 cells were infected with Fn (MOI = 0, 100, 500 or 1000) for 48 h, and miR-135b expression in cells was measured by qRT-PCR. Data were expressed as mean ± SD. All our data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Fn Infection Promoted Cisplatin Resistance of CRC Cells by Upregulating miR-135b
After Fn infection, results from CCK8 assay revealed that Fn infection enhanced the cell viability of SW620 and Caco cells in a MOI dependent manner (Fig. 2A). In addition, SW620 and Caco2 cells were subjected to cisplatin, and it was observed that cisplatin treatment suppressed cell viability in a dose dependent manner (Fig. 2B). Following qRT-PCR assay showed that cisplatin treatment gradually increased miR-135b expression in SW620 and Caco cells at concentration of 0–15 µg/ml, while miR-135 expression was no significant difference between 15 µg/ml and 25 µg/ml cisplatin treatment (Fig. 2C). Previous researches have confirmed the evidences highlighting the regulatory roles of Fn infection and miR-135b on drug resistance against CRC (Hong et al., 2023; Liu et al., 2017). Here, we further explored the roles of Fn and miR-135b on cisplatin resistance against CRC cells. Firstly, miR-135b was greatly silenced after miR-135b inhibitor transfection (Fig. 2D). Then, it was demonstrated that Fn infection greatly significantly enhanced cell viability and reduced cell apoptosis in SW620 and Caco2 cells exposed to cisplatin, which dramatically reduced the drug sensitivity of CRC cells to cisplatin, however, these roles could be impeded by miR-135b inhibition (Fig. 2E, F). Taken together, Fn facilitated cisplatin resistance in CRC by increasing miR-135b expression.
Fn infection promoted cisplatin resistance of CRC cells by upregulating miR-135b. A SW620 and Caco2 cells were infected with Fn (MOI = 0, 100, 500 or 1000) for 48 h, and cell proliferation was examined by CCK8 assay. B SW620 and Caco2 cells were treated with cisplatin (5, 10, 15 or 25 µg/ml) for 48 h, and cell proliferation was assessed using CCK8 assay. C The expression of miR-135b in SW620 and Caco2 cells after cisplatin (5, 10, 15 or 25 µg/ml) treatment were detected by qRT-PCR. D miR-135b expression in SW620 and Caco2 cells following inhibitor NC or miR-135b inhibitor transfection was analyzed using qRT-PCR. miR-135b knockdown was induced in CRC cells combined with cisplatin and Fn treatments. E CCK8 assay was adopted to determine cell proliferation. F Cell apoptosis was assessed using flow cytometry. Data were expressed as mean ± SD. All our data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
miR-135b Negatively Modulated KLF13 Expression
Next, the potential targets of miR-135b were further analyzed. Through multiple databases prediction (Starbase, TargetScan and miRDB), it was found that KLF13 was a downstream target gene of miR-135b (Fig. 3A, B). Recently, KLF13 was considered as a tumor suppressor in various types of cancer, including CRC (Yao et al., 2020). Thus, the regulatory relationship between KLF13 and miR-135b was detected. As shown in Fig. 3C, the results from qRT-PCR and/or western blot subsequently revealed that miR-135b mimics transfection resulted in increased miR-135b expression and reduced KLF13 expression in CRC cells, while miR-135b inhibitor transfection presented the opposite effects (Fig. 3C, E). Next, dual luciferase assay also showed that miR-135b mimics transfection evidently downregulated the luciferase activities of wt-KLF13 plasmids expressed cells, and miR-135b inhibitor could obviously enhanced the luciferase activities of cells containing wt-KLF13 plasmids, however, either miR-135b mimics and miR-135b inhibitor had a little effect on CRC cells with mut-KLF13 transfection (Fig. 3F), suggesting that miR-135b could negatively regulate KLF13 expression.
miR-135b negatively regulated KLF13 expression. A Databases (Starbase, TargetScan and miRDB) were employed to predict the downstream target of miR-135b. B The potential binding site between miR-135b and KLF13 was presented. C–E miR-135b and KLF13 expressions in SW620 and Caco2 cells following mimics NC or miR-135b mimics transfection were analyzed using qRT-PCR and/or western blot. F The interaction between miR-135b and KLF13 was analyzed by dual-luciferase reporter gene assay. Data were expressed as mean ± SD. All our data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
KLF13 Overexpression Reversed the Regulatory Effect of Fn/miR-135b-Mediated Cisplatin Resistance
To investigate the functional association between KLF13 and Fn or miR-135b in regulating cisplatin resistance in CRC cells. The KLF13 overexpressing cells were induced by transfection with pcDNA3.1-KLF13. As Fig. 4A showed that KLF13 mRNA level in SW620 and Caco2 cells was significantly increased upon pcDNA3.1-KLF13 transfection. Then, it was further observed that both Fn infection and miR-135b overexpression markedly enhanced the cell viability but decreased cell apoptosis, and all these changes could be restrained when KLF13 was co-overexpressed (Fig. 4B, C). In conclusion, KLF13 functioned as the target of Fn/miR-135b in regulating cisplatin resistance of CRC cells.
Fn activated miR-135b expression through acting on TCF4/β-catenin complex. A KLF13 mRNA level in SW620 and Caco2 cells following pcDNA3.1 or pcDNA3.1-KLF13 transfection was analyzed using qRT-PCR. KLF13 overexpressing vectors were transfected in to cells with Fn infection or miR-135b mimics co-transfection. B CCK8 assay was adopted to determine cell proliferation. C Cell apoptosis was assessed using flow cytometry. Data were expressed as mean ± SD. All our data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Fn Activated miR-135b Expression Through Acting on TCF4/β-catenin Complex
As previously reported, miR-135b upregulation in CRC might be related to transcriptional activation of TCF4/β-catenin (Valeri et al., 2014). Consistent with the previous study, by using database prediction, it was found that TCF4 had a potential binding site at − 2.5 kb upstream of the pre-miR-135b promoter, and the luciferase vector was constructed (Fig. 5A). As shown in Fig. 5B, pcDNA3.1-TCF4 transfection markedly increased TCF4 mRNA level in SW620 and Caco2 cells. The results of dual-luciferase reporter gene assay subsequently displayed that pcDNA3.1-TCF4 transfection increased the luciferase activity of cells expressing recombinant wt-luciferase vectors, while had no significant role on cells transfected with recombinant mut-luciferase vectors (Fig. 5C). Moreover, we also confirmed that Fn infection increased TCF4 and β-Catenin protein levels in SW620 and Caco2 cells (Fig. 5D), and upon LF3 treatment, the promoting effect of Fn on miR-135b expression was diminished (Fig. 5E). Thus, it could be concluded that Fn promoted miR-135b expression in CRC cells via the TCF4/β-catenin complex.
The promotion effect of Fn on miR-135b expression in CRC cells might be related to TCF4/β-Catenin complex. A Database was employed to predict the potential binding site between TCF4 and miR-135b, and the luciferase vector construction diagram was presented. B TCF4 mRNA level in SW620 and Caco2 cells following pcDNA3.1 or pcDNA3.1-TCF4 transfection was analyzed using qRT-PCR. C The interaction between TCF4 and miR-135b was analyzed by dual-luciferase reporter gene assay. D Western blot was adopted to analyze TCF4 and β-Catenin protein levels in SW620 and Caco2 cells following Fn infection. E qRT-PCR was adopted to detect miR-135b expression in SW620 and Caco2 cells following Fn and LF3 treatments. Data were expressed as mean ± SD. All our data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
The intestinal microbiota is made up of a vast array of microbes and is often considered the “forgotten organ” of human disease (Cheng et al., 2020). In recent years, more and more evidence has shown that the ecological imbalance of intestinal microbiota is closely related to the occurrence and development of CRC (Tilg et al., 2018). Several studies have identified microorganisms as CRC candidate pathogens and revealed that these pathogens promote CRC initiation, progression, and metastasis, as well as cisplatin resistance, through a variety of mechanisms including promoting inflammation and secreting virulence factors (Cheng et al., 2020). Therefore, intestinal microbiota is of great clinical value for the prevention and treatment of CRC, and exploring the regulatory mechanism of intestinal microbiota in the occurrence and development of CRC can provide a theoretical basis for the development of novel treatment strategies for CRC.
Fn is an invasive, adhesive, and pro-inflammatory anaerobic bacterium that functions in regulating intestinal immune system (Su et al., 2019). It was previously reported that Fn showed a significant enrichment in the feces and tumor tissues of CRC patients (Xu et al., 2021). Moreover, there were a great body of evidences highlighted that the abnormally enrichment of Fn was could involve in CRC development. Brennan et al showed that Fn affected intestinal immunity by forming a Th17 response in an FFAR2-dependent manner, thus potentiating intestinal tumorigenesis (Kostic et al., 2013). Chen et al. (2022)’ s also proved that Fn driven CRC metastasis through repressing METTL3-medaited m6A modification of KIF26B. Moreover, Fn was also confirmed to act on TLR4 and MYD88 innate immune signaling and specific miRNAs to enhance autophagy, thus promote the drug chemoresistance (Yu et al., 2017). Cisplatin is the most used chemotherapy drug for CRC in clinic (Tan et al., 2022). Although there were several literatures uncovered that Fn could make oesophageal squamous cell carcinoma cells and oral squamous carcinoma cells acquire more stronger resistance against cisplatin (Chen et al., 2022; Da et al., 2021). Nevertheless, the role of Fn in mediating cisplatin resistance in CRC and its underlying mechanism remain clear. Consistent with the previous study (Xu et al., 2021), our results also demonstrated the Fn exhibited an abnormal abundance. Importantly, we also demonstrated that Fn administration greatly facilitated the cell viability and decreased apoptosis of cisplatin-treated CRC cells, supporting that Fn accumulation might be a key cause of cisplatin resistance in CRC.
miRNA has been shown to play an important role in cancer biology, and miRNA dysregulation is an important factor in the development of CRC as well as chemotherapy resistance (Balacescu et al., 2018). Notably, miRNAs acted as the downstream targets of Fn in promoting CRC progression. As proof, Yang et al. (2017) revealed that Fn facilitated CRC cell proliferation by upregulating miR-21. In our paper, it was observed that miR-135b expression was markedly increased in CRC tumor tissues and CRC cells, and its expression was positively correlated with Fn abundance in clinical samples. In addition, Fn infection increased miR-135b expression in CRC cells in a MOI dependent manner. It has been widely reported that miR-135b is markedly upregulated in CRC and acts as an oncogene (Jia et al., 2017; Li et al., 2015). For instance, as revealed by Li et al. (2015) miR-135b upregulation promote CRC cell proliferation and inhibited the apoptosis. Importantly, miR-135b knockdown sensitized CRC cells to oxaliplatin (Qin et al., 2018). However, the function of miR-135b in regulating Fn-mediated cisplatin resistance in CRC hasn’t been fully elucidated. Here, we have proved that miR-135b knockdown greatly reversed the promoting effect of Fn on cisplatin resistance. Subsequently, we attempted to elucidate the pathway by which Fn activated miR-135b. TCF4/β-catenin complexes are major transcriptional regulators of the Wnt signaling pathway. The translocation of β-catenin from cytoplasm to nucleus leads to the formation of the TCF4/β-catenin complex, which activates downstream target genes (Gordon & Nusse, 2006; Schuijers et al., 2014). Much evidence has revealed the correlation between the TCF4/β-catenin complex and CRC progression. For example, Wang et al. (2018b) displayed that TCF4/β-catenin complex activation promoted CRC development by inhibiting tumor-suppressive miR-145 expression. It was well known that Fn could positively promote signal transduction of Wnt/β-catenin signaling to participate in tumor carcinogenesis (Li et al., 2020b; Rubinstein et al., 2019), and it further confirmed in our work, demonstrating that Fn infection markedly elevated TCF4 and β-catenin expressions. Notably, we also found TCF4 targeted miR-135b promoter region and elevated its expression. Upon TCF4/β-catenin complex was blocked, the promotion effect of Fn on miR-135b obviously disappeared.
It is well known that the main biological function of miRNA was depended on binding to the 3′-TUR region of the target gene to degrade RNA or blocking the translation of mRNA into protein, thus involving the regulation of cell process. After multiple bioinformatics analyses, KLF13 was found to be a downstream target of miR-135b, and the relationship was validated by dual luciferase assay. KLF13, a tumor-suppressive member of Kruppel-like factors family, was a newly identified tumor regulator in cancer biology (Yao et al., 2020). Generally, KLF13 also named as BTEB3, FKLF-2, and RFLAT-1, was downregulated in prostate cancer, and its overexpression could promote AKT signaling to suppress tumor growth (Wang et al., 2018a). Newly, Yao et al. reported that KLF13 was also downregulated in CRC tissues and could repress cell proliferation by transcriptionally inhibiting HMGCS1-medaited cholesterol biosynthesis (Yao et al., 2020). Moreover, KLF13 was also found to promote the sensitivity of acute lymphoblastic leukemia cells to dexamethasone by inactivating MYB (Heard et al., 2015). However, the function of KLF13 in regulating cisplatin resistance in CRC is still unclear. In the current paper, it was observed that KLF13 overexpression strikingly reversed the promoting roles of Fn infection and miR-135b overexpression on cisplatin resistance. Therefore, we came to the conclusion that KLF13 functioned as the target of miR-135b in regulating Fn-mediated cisplatin resistance in CRC.
Taken together, our experimental data suggested that Fn infection promoted cisplatin resistance in CRC by regulation of the miR-135b/KLF13 axis. Our study provides a theoretical basis for reserving cisplatin resistance in CRC.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
Arthur, J. C., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T. J., Campbell, B. J., Abujamel, T., Dogan, B., Rogers, A. B., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science,338, 120–123.
Balacescu, O., Sur, D., Cainap, C., Visan, S., Cruceriu, D., et al. (2018). The impact of miRNA in colorectal cancer progression and its liver metastases. International Journal of Molecular Sciences,19, 3711.
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians,68, 394–424.
Chen, B., Zhang, D., Kuai, J., Cheng, M., Fang, X., & Li, G. (2017). Upregulation of miR-199a/b contributes to cisplatin resistance via Wnt/β-catenin-ABCG2 signaling pathway in ALDHA1+ Colorectal cancer stem cells. Tumour Biology,39, 101042831771515. https://doi.org/10.1177/1010428317715155
Chen, S., Zhang, L., Li, M., Zhang, Y., Sun, M., et al. (2022). Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nature Communications,13, 1248.
Cheng, Y., Ling, Z., & Li, L. (2020). The intestinal microbiota and colorectal cancer. Frontiers in Immunology,11, 615056.
Da, J., Wang, X., Li, L., & Xu, Y. (2021). Fusobacterium nucleatum promotes cisplatin-resistance and migration of oral squamous carcinoma cells by up-regulating Wnt5a-mediated NFATc3 expression. The Tohoku Journal of Experimental Medicine,253, 249–259.
Dai, X., Xie, Y., & Dong, M. (2022). Cancer-associated fibroblasts derived extracellular vesicles promote angiogenesis of colorectal adenocarcinoma cells through miR-135b-5p/FOXO1 axis. Cancer Biology & Therapy,23, 76–88.
Dasari, S., & Tchounwou, P. B. (2014). Cisplatin in cancer therapy: molecular mechanisms of action. European Journal of Pharmacology,740, 364–378.
Du, D., Su, Z., Wang, D., Liu, W., & Wei, Z. (2018). Optimal interval to surgery after neoadjuvant chemoradiotherapy in rectal cancer: a systematic review and meta-analysis. Clinical Colorectal Cancer,17, 13–24.
Franke, A. J., Parekh, H., Starr, J. S., Tan, S. A., Iqbal, A., & George, T. J., Jr. (2018). Total neoadjuvant therapy: a shifting paradigm in locally advanced rectal cancer management. Clinical Colorectal Cancer,17, 1–12.
Fuertes, M. A., Castilla, J., Alonso, C., & Pérez, J. M. (2002). Novel concepts in the development of platinum antitumor drugs. Current Medicinal Chemistry - Anti-Cancer Agents,2, 539–551.
Gill, S. R., Pop, M., DeBoy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., Gordon, J. I., Relman, D. A., Fraser-Liggett, C. M., & Nelson, K. E. (2006). Metagenomic analysis of the human distal gut microbiome. Science,312, 1355–1359.
Gordon, M. D., & Nusse, R. (2006). Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. The Journal of Biological Chemistry,281, 22429–22433.
Guo, S., Chen, J., Chen, F., Zeng, Q., Liu, W. L., & Zhang, G. (2021). Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut,70, 1507–1519.
Heard, M. E., Velarde, M. C., Giudice, L. C., Simmen, F. A., & Simmen, R. C. (2015). Krüppel-like factor 13 deficiency in uterine endometrial cells contributes to defective steroid hormone receptor signaling but not lesion establishment in a mouse model of endometriosis. Biology of Reproduction,92, 140.
Hong, X. L., Yu, T. C., Huang, X. W., Wang, J. L., Sun, T. T., Yan, T. T., Zhou, C. B., Chen, H. M., Su, W. Y., Du, W., et al. (2023). Metformin abrogates Fusobacterium nucleatum-induced chemoresistance in colorectal cancer by inhibiting miR-361-5p/sonic hedgehog signaling-regulated stemness. British Journal of Cancer,128, 363–374.
Huang, X., Zhu, X., Yu, Y., Zhu, W., Jin, L., Zhang, X., Li, S., Zou, P., Xie, C., & Cui, R. (2021). Dissecting miRNA signature in colorectal cancer progression and metastasis. Cancer Letters,501, 66–82.
Iida, N., Dzutsev, A., Stewart, C. A., Smith, L., Bouladoux, N., Weingarten, R. A., Molina, D. A., Salcedo, R., Back, T., Cramer, S., et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science,342, 967–970.
Jia, L., Luo, S., Ren, X., Li, Y., Hu, J., Liu, B., Zhao, L., Shan, Y., & Zhou, H. (2017). miR-182 and miR-135b mediate the tumorigenesis and invasiveness of colorectal cancer cells via targeting ST6GALNAC2 and PI3K/AKT pathway. Digestive Diseases and Sciences,62, 3447–3459.
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., Clancy, T. E., Chung, D. C., Lochhead, P., Hold, G. L., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe,14, 207–215.
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., Ojesina, A. I., Jung, J., Bass, A. J., Tabernero, J., et al. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research,22, 292–298.
Li, J., Liang, H., Bai, M., Ning, T., Wang, C., Fan, Q., Wang, Y., Fu, Z., Wang, N., Liu, R., et al. (2015). miR-135b promotes cancer progression by targeting transforming growth factor beta receptor II (TGFBR2) in colorectal cancer. PLoS ONE,10, e0130194.
Li, S. S., Zhu, H. J., Li, J. Y., Tian, L. M., & Lv, D. M. (2020a). MiRNA-875-3p alleviates the progression of colorectal cancer via negatively regulating PLK1 level. European Review for Medical and Pharmacological Sciences,24, 1126–1133.
Li, X., Huang, J., Yu, T., Fang, X., Lou, L., et al. (2020b). Fusobacterium nucleatum promotes the progression of colorectal cancer through Cdk5-activated Wnt/β-catenin signaling. Frontiers in Microbiology,11, 545251.
Liu, B., Liu, Y., Zhao, L., Pan, Y., Shan, Y., Li, Y., & Jia, L. (2017). Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Molecular Carcinogenesis,56, 2669–2680.
Mima, K., Nishihara, R., Qian, Z. R., Cao, Y., Sukawa, Y., Nowak, J. A., Yang, J., Dou, R., Masugi, Y., Song, M., et al. (2016). Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut,65, 1973–1980.
Qin, Y., Li, L., Wang, F., Zhou, X., Liu, Y., Yin, Y., & Qi, X. (2018). Knockdown of Mir-135b sensitizes colorectal cancer cells to oxaliplatin-induced apoptosis through increase of FOXO1. Cellular Physiology and Biochemistry,48, 1628–1637.
Rubinstein, M. R., Baik, J. E., Lagana, S. M., Han, R. P., Raab, W. J., Sahoo, D., Dalerba, P., Wang, T. C., & Han, Y. W. (2019). Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator annexin A1. EMBO Reports,20, e47638.
Saliminejad, K., Khorshid, K., Fard, H. R. S., & Ghaffari, S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. Journal of Cellular Physiology,234, 5451–5465.
Schuijers, J., Mokry, M., Hatzis, P., Cuppen, E., & Clevers, H. (2014). Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. The EMBO Journal,33, 146–156.
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., & Kent, R. L., Jr. (1998). Microbial complexes in subgingival plaque. Journal of Clinical Periodontology,25, 134–144.
Su, S. H., Wu, Y. F., Lin, Q., Wang, D. P., & Hai, J. (2019). URB597 protects against NLRP3 inflammasome activation by inhibiting autophagy dysfunction in a rat model of chronic cerebral hypoperfusion. Journal of Neuroinflammation,16, 260.
Tan, L., Qu, W., Wu, D., Liu, M., Ai, Q., Hu, H., Wang, Q., Chen, W., & Zhou, H. (2022). The interferon regulatory factor 6 promotes cisplatin sensitivity in colorectal cancer. Bioengineered,13, 10504–10517.
Tilg, H., Adolph, T. E., Gerner, R. R., & Moschen, A. R. (2018). The intestinal microbiota in colorectal cancer. Cancer Cell,33, 954–964.
Valeri, N., Braconi, C., Gasparini, P., Murgia, C., Lampis, A., Paulus-Hock, V., Hart, J. R., Ueno, L., Grivennikov, S. I., Lovat, F., et al. (2014). MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell,25, 469–483.
Viaud, S., Saccheri, F., Mignot, G., Yamazaki, T., Daillère, R., Hannani, D., Enot, D. P., Pfirschke, C., Engblom, C., Pittet, M. J., et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science,342, 971–976.
Wang, H., Wang, X., Zhang, H., Deng, T., Liu, R., Liu, Y., Li, H., Bai, M., Ning, T., Wang, J., et al. (2021). The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene,40, 4695–4708.
Wang, J., Zhang, R., Zhang, B., Zhang, L., Jiang, W., Liu, X., & Duan, X. (2022). MiR-135b improves proliferation and regulates chemotherapy resistance in ovarian cancer. Journal of Molecular Histology,53, 699–712.
Wang, Q., Peng, R., Wang, B., Wang, J., Yu, W., Liu, Y., & Shi, G. (2018a). Transcription factor KLF13 inhibits AKT activation and suppresses the growth of prostate carcinoma cells. Cancer Biomarkers,22, 533–541.
Wang, W., Xiao, X., Chen, X., Huo, Y., Xi, W. J., Lin, Z. F., Zhang, D., Li, Y. F., Yang, F., Wen, W. H., et al. (2018b). Tumor-suppressive miR-145 co-repressed by TCF4-β-catenin and PRC2 complexes forms double-negative regulation loops with its negative regulators in colorectal cancer. International Journal of Cancer,142, 308–321.
Xu, C., Fan, L., Lin, Y., Shen, W., Qi, Y., Zhang, Y., Chen, Z., Wang, L., Long, Y., Hou, T., et al. (2021). Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes,13, 1980347.
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., Gao, R., Liu, M., Yin, M., Pan, C., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology,152, 851–866.
Yao, W., Jiao, Y., Zhou, Y., & Luo, X. (2020). KLF13 suppresses the proliferation and growth of Colorectal cancer cells through transcriptionally inhibiting HMGCS1-mediated cholesterol biosynthesis. Cell & Bioscience,10, 76.
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., Qian, Y., Kryczek, I., Sun, D., Nagarsheth, N., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell,170, 548–563.
Zhao, J., Wang, X., Mi, Z., Jiang, X., Sun, L., Zheng, B., Wang, J., Meng, M., Zhang, L., Wang, Z., et al. (2021). STAT3/miR-135b/NF-κB axis confers aggressiveness and unfavorable prognosis in non-small-cell lung cancer. Cell Death & Disease,12, 493.
Acknowledgements
The study was supported by Changsha Natural Science and Technology Foundation (kq2208451).
Author information
Authors and Affiliations
Contributions
Conception and design of study: WZ. Acquisition of data: JP. Analysis and interpretation of data: GY. Drafting the manuscript: JP. Revising the manuscript critically for important intellectual content: WZ. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors agree with the presented findings, have contributed to the work, and declare no conflict of interest.
Ethical statements
This study was passed the review of Ethics Committee of Changsha First Hospital before enrollment of patients and all participants signed informed consent (Approval Number: No. 2022-171).
Consent for publication
All participants were informed and gave written consent.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, W., Pan, J. & Ye, G. miR-135b Aggravates Fusobacterium nucleatum-Induced Cisplatin Resistance in Colorectal Cancer by Targeting KLF13. J Microbiol. 62, 63–73 (2024). https://doi.org/10.1007/s12275-023-00100-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00100-1