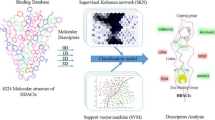
Abstract
HDAC8 inhibitors have become an attractive treatment for cancer. This study aimed to facilitate the identification of potential chemical scaffolds for the selective inhibition of histone deacetylase 8 (HDAC8) using in silico approaches. Non-linear QSAR classification and regression models of HDAC8 inhibitors were developed with support vector machine. Mean impact value-based sequential forward feature selection and grid search strategy were used for molecular descriptor selection and parameter optimization, respectively. The generated QSAR models were validated by leave-one-out cross validation and an external test set. The best QSAR classification model yielded 84 % of accuracy on the external test prediction and Matthews correlation coefficient is 0.69. The best QSAR regression model showed low root-mean-square error (0.63) and high squared correlation coefficient (0.53) for the test set. The validated QSAR models together with various drug-like properties, molecular docking and molecular dynamics simulation were sequentially used as a multi-step query in chemical database virtual screening. Finally, two hit compounds were discovered as new structural scaffolds which can be used for further in vitro and in vivo activity analyses. The strategy used in this study could be a promising computational strategy which can be utilized for other target drug design.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
HDACs super family play critical roles in the regulation of cellular metabolism, and constitute promising drug targets for treatment of a broad range of human diseases such as cardiomyopathy, osteodystrophy, neurodegenerative disorder, metabolic disorders, cardiovascular disease, aging cancer, etc. (Taylor et al. 2008). 18 HDACs enzymes have been identified and classified into four different classes based on sequence homology, function, DNA similarity, and phylogenetic analysis (Yang and Seto 2008; Lehrmann et al. 2002; Marks and Breslow 2007; Emiliani et al. 1998). Class I, Class II, and Class IV HDACs are zinc (Zn2+) dependent deacetylases, and Class III HDACs (Sirtuins) is mainly dependent on nicotinamide adenine dinucleotide (NAD+) for its deacetylation activity (Imai et al. 2000; Landry et al. 2000). The HDAC8 enzyme belongs to the class I enzymes which are found primarily in the nucleus (Valenzuela-Fernández et al. 2008). Except HDAC8, functional HDACs are found as multimeric complexes of high molecular weight and most of them are functionally inactive (Vannini et al. 2007; Bolden et al. 2006). Expression of HDAC8 notably correlates with neuroblastoma, a highly malignant childhood cancer derived from the sympathetic nervous system (Brodeur 2003; Oehme et al. 2009). Moreover, an RNA interference study showed that HDAC8 is involved in the regulation of proliferation, clonogenic growth and neuronal differentiation of neuroblastoma cells. Inv1, an abnormal fusion protein formed during acute myeloid leukemia binding HDAC8, is also associated with aberrant, constitutive genetic repression (Durst et al. 2003). These evidences prove HDAC8 as a potential target for cancer treatment. To date, a number of potential HDAC inhibitors are in clinical trials (Thangapandian et al. 2011). Therefore, HDAC8 is considered to be the best model among other mammalian HDACs from a structural biology and drug discovery perspective.
Drug discovery and development is a difficult, costly and time-consuming work. In silico virtual screening (VS) is an economical and rapid approach to retrieve potential lead in drug discovery. Currently several VS methods have been well established such as quantitative/qualitative structure–activity relationships (QSAR)-based and molecular docking-based VS (Cao et al. 2015). Obviously, different VS methods have their own advantages and disadvantages. Each of these VS methods might not perform optimally when used alone in terms of the speed and effectiveness of VS, a combination of these methods is an alternative approach. In past decades, Support vector machine (SVM) is becoming more attractive tools to develop QSAR model for VS in the drug discovery, as they reduce the complexity of experiments, screen a vast chemical library rapidly (Wan et al. 2012; Ma et al. 2010; Shi et al. 2012; Byvatov et al. 2003; Vasanthanathan et al. 2009; Han et al. 2007; Yap and Chen 2004; Wang et al. 2012; Mahé et al. 2005; Liew et al. 2009; Zhang et al. 2012; Niu 2007). During SVM model development, compounds are represented by multiple-dimensional molecular descriptors. It is unavoidable to select a subset of relevant molecular descriptors from a large amount of data, as it can bring potential benefits: facilitating data visualization and data understanding, reducing the measurement and storage requirements, reducing training and utilization times, defying the curse of dimensionality to improve prediction performance (Guyon 2003; Feature Selection Using Sequential Forward Selection and IECON 2010). In this study, a hybrid method named mean impact value-based sequential forward selection (MIV-based SFS) was used to complete the task.
This study introduced a hybrid strategy of virtual screening based on QSAR modeling and molecular docking to identify novel HDAC8 inhibitors. Two kinds of QSAR models were developed with support vector classification (SVC) and regression (SVR) based on the known HDAC8 inhibitors, which can correctly reflect the structure–activity relationship (SAR) of the existing HDAC8 inhibitors. Furthermore, the developed QSAR models were used sequentially as a two-step query for searching large databases to identify novel HDAC8 inhibitors. Molecular docking and various scrupulous drug-like properties such as Lipinski’s rule of five and ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties were employed to reduce the probability of picking false positives and nondruglike compounds, respectively. HDAC8-hit complex stability was evaluated using molecular dynamics (MD) simulation.
Materials and methods
Dataset preparation and selection of compounds
A total of 80 compounds with HDAC8 inhibitory activity values predicted under same biological assay conditions were collected from various literature resources including patents (Durst et al. 2003; Gu and Nusinzon 2006; Wu et al. 2004; Jeffrey MB, Zuomei L, Daniel D, Claire B. Methods for specifically inhibiting histone-7 and 8. US Patents 2004; Eric 2007; Walter et al. 2007; Ze-Yi et al. 2008; Joseph and Sriram 2010). This data set has included 40 inhibitors and 40 non-inhibitor compounds, which was done based on IC50 values of the compounds ranging from 0.008 to 35 µM. The IC50 values ranging from 0.008 to 0.3 µM were considered as inhibitors, the others were considered as non-inhibitors. Selection of training set compounds is pivotal for QSAR modeling which subsequently determines the quality of the generated QSAR models. The constraint random sampling (CRS) method was used to prepare training set. The training set compounds were selected based on following constraint criteria: (1) a minimum of 20 compounds were selected to avoid any chance correlation; (2) the training set should be balanced; (3) the compounds should be selected to provide clear, concise information to avoid redundancy or bias in terms of both structural features and activity range; (4) a part of the most active compounds should be included to generate reliable and rational QSAR model, and the others can be used to validate the quality of the QSAR model; (5) all data set compounds were randomly selected in combination with above criteria, which would ensure that all the compounds were selected with an equal probability. The dataset was finally divided into training and test sets containing 30 and 50 compounds, respectively. The 30 training set compounds including 15 inhibitors and 15 non-inhibitors were used in the development of classification and regression models (Fig. 1). The test set (25 inhibitors and 25 non-inhibitors) was used to validate the developed models. All the compounds in the data set were sketched in 2D structures with Accelrys Draw v4.1 (Accelrys Inc., San Diego, USA) and subsequently converted into 3D structures with Accelrys Discovery Studio v3.1 (DS) (Accelrys, San Diego, USA). Then energy minimization using CHARMM force field (Brooks et al. 1983), The Smart Minimizer option that performs 1,000 steps of Steepest Descent with a RMS gradient tolerance of 3, followed by Conjugate Gradient minimization was used with a RMS gradient of 0.1 kcal/(mol × Å).
Molecular descriptors calculation
Molecular descriptors are the final results of a logical and mathematical procedure which transforms chemical information encoded within a symbolic representation of a molecule into a useful number or the result of some standardized experiment (Yap 2011; Todeschini and Consonni 2000). To date, though thousands of descriptors can be calculated, it is only useful in medicinal chemistry perspective when they are reduced to a few set of molecular descriptors that can effectively be applied in designing novel and potent compounds. Thus the main goal of a QSAR study is not to calculate thousands of descriptors but to identify a few molecular descriptors. In this study, the collected inhibitor was represented by 20 molecular descriptors which were calculated by using ADRIANA.Code program (Molecular Networks Inc.), including global, shape and size-related descriptors (ADRIANA Code) (Table 1). Furthermore, the data set were scaled from −1 to 1 by following:
where xi is a descriptor vector of the sample data, the yi is a scaling data, which corresponds to xi, | xi | is the absolute value of the xi, n is the number of compounds.
Development of qualitative structure–activity relationship (Qualitative SAR) model
Qualitative SAR models are classification models used in drug discovery, which relate the classifier variables to a categorical value of the response variable. The Qualitative SAR models were generated using support vector classification (SVC) which was firstly proposed by V. Vapnik in 1995 (Cortes and Vapnik 2011). The whole process of SVC can be summarized as a two-step procedure: First, the sample data vectors (descriptors) are mapped to a very high-dimensional feature space by kernel function. The dimension of this space is significantly larger than dimension of the original data space. Second, the SVM classifier finds a hyperplane with the largest margin in this high-dimensional feature space with the largest margin separating classes of data. Sometimes it is not possible to find the hyperlane in high-dimensional feature space, so a tradeoff is introduced between the size of the separating margin and penalties for every vector within the margin (Byvatov et al. 2003).
Development of quantitative structure–activity relationship (Quantitative SAR) model
Quantitative SAR models are regression models used in drug discovery, which relate a set of “predictor” variables to the potency of the response variable. Support vector regression (SVR) was applied to develop Quantitative SAR models with training set compounds. Support vector machine regression is based on the structural risk minimization principle from the statistical learning theory (Niu 2007). It can be used to predict continuous values like IC50 value of ligand by introducing an alternative loss function and the results appear to be very encouraging. The SVM (SVC and SVR) calculation was used and executed in the LIBSVM 3.12 tool (Chang and Lin 2011).
Descriptor selection using MIV-based SFS method
Compound, in QSAR (Qualitative SAR and Quantitative SAR) studies, is encoded by a variety of molecular descriptors. It must be noted that usually only a subset of the calculated descriptors carries necessary information for developing a QASR model (Shahlaei 2013). Descriptor selection is aimed at finding those useful calculated descriptors for the model building. Here, a hybrid method named mean impact value-based sequential forward selection (MIV-based SFS) was used to accomplish this task.
MIV is firstly used in neural network to measure the influence of afferent neurons on efferent neurons. In this study, MIV was used as a measure reflecting the input variable of developed SVM models to prediction result (Li et al. 2012). The absolute value represents how strongly the selected molecular descriptors can affect the predictive ability of SVM model. The detailed calculation process is described below: after finishing SVM training, each of independent variable features (molecular descriptors) from training data P was increased (P1) and decreased (P2) by 10 % to get two new training data. P1 and P2 were predicted using the developed model to get two results A1 and A2. Impact value (IV) is difference between A1 and A2. Different IVs were obtained by changing the independent variables. Finally, mean of IVs (MIV) was calculated for each descriptor.
Sequential forward selection (SFS) is a data-driven model building approach which selects a most influential subset of features from the original data set for constructing a classifier that gives better performance (Guyon 2003; Haindl et al. 2006). In this approach, one variable is added to the model at a time. It involves following steps: (1) Select a classifier and the leave-one-out (LOO) test for recognition rate estimate; (2) select the first feature that has the highest LOO recognition rate among all features; (3) select the feature, among all unselected features, together with the selected features that gives the highest recognition rate; (4) repeat the previous process until you have selected enough number of features or until the recognition rate is good enough.
MIV-based SFS method selected MIV calculated using SVM as recognition rate. It begins with a model including the molecular descriptor with the greatest MIV (absolute value), and continues adding molecular descriptor to the model one at a time according to their MIV scores until the predictions of the QSAR model continue to fall.
Leave-one-out cross validation (LOO CV)
CV is a model validation technique for assessing how the results of a statistical analysis will generalize to an independent data set (Kohavi 1995). LOO CV procedure was applied to estimate the predictive capability of the developed QSAR models. In LOO CV process, a single compound from the data set was used as the test data, and the remaining compounds as the training data. This was repeated such that each sample in the data set is used once as the test data. The results were averaged as output of LOO CV.
Receiver operating characteristic (ROC) curve
A ROC curve is a metric which illustrates the performance of a binary classifier system as its discrimination threshold is varied. It is a comparison of two operating characteristics (TPR and FPR) as the criterion changes (Fawcelt 2006). For each class of a classifier, ROC applies threshold values across the interval [0, 1] to outputs. For each threshold, two values, TPR and FPR, are calculated. And the accuracy of classifier is measured by the area under the ROC curve (AUC). An area of 1 represents a perfect test; an area of 0.5 represents a worthless test. In this study, ROC curve was used to validate the accuracy of SVC model.
Grid search (GS) method
During the development of SVM modeling, a difficult issue is how to set good parameters of SVM. It is not known beforehand which parameters are best. Thus, parameter search must be done. In this stud, GS was utilized for parameter optimization. GS is straight forward but powerful method, which is exhaustive searching through a subset of the parameter space of a learning algorithm to solve problem of model selection and parameter optimization. A grid search method must be guided by some performance metric, typically measured by CV on the training set, i.e., LOO CV used in this study.
Evaluation of prediction performance
The predictive abilities from SVC and SVR were evaluated using following statistical measures. In following equations, TP is the number of true positives, TN true negatives, FP false positives, and FN false negatives, n is the number of the samples in data set, f(xi) is the predicted biological activity, and yi is the experimental biological activity. In this study, HDAC8 inhibitors were considered as ‘positive set’ and the non-inhibitors were considered ‘negative set’. The accuracy (ACC) is the degree of closeness of measurements of a quantity to that quantity’s actual (true) value. The Matthews correlation coefficient (MCC) is a measure of quality of binary classification, and it returns a value between −1 and +1. A coefficient of +1 stands for a perfect prediction, 0 represents a random prediction and −1 indicates total disagreement between prediction and observation. The true positive rate (TPR or Recall rate) is a metric of retrieved instances that are relevant. The false positive rate (FPR) measures the proportion of actual positives which are incorrectly identified. Therefore TPR and FPR are based on an understanding and measure of relevance. The root-mean-square error (RMSE) is a frequently used measure of the differences between values predicted by a model or an estimator and the values actually observed. The squared correlation coefficient (r2) is the predictive percent of behavior in the output that can be explained by the input.
-
1.
ACC can be calculated by following formula:
$$ {\text{ACC}} = \frac{{{\text{TP}} + {\text{TN}}}}{{{\text{TP}} + {\text{FP}} + {\text{TN}} + {\text{FN}}}} \times 100\% $$ -
2.
MCC can be calculated directly from the confusion matrix using the formula:
$$ {\text{MCC}} = \frac{{\left( {{\text{TP}} \times {\text{TN}}} \right) - ({\text{FP}} \times {\text{FN}})}}{{\sqrt {({\text{TP}} + {\text{FP}})({\text{TP}} + {\text{FN}})({\text{TN}} + {\text{FP}})({\text{TN}} + {\text{FN}})} }} $$ -
3.
The TPR is defined as:
$$ {\text{TPR}} = \frac{\text{TP}}{{{\text{TP}} + {\text{FN}}}} \times 100\% $$ -
4.
The FPR is defined as:
$$ {\text{FPR}} = \frac{\text{FP}}{{{\text{FP}} + {\text{TN}}}} \times 100\% $$ -
5.
The RMSE is defined as:
$$ {\text{RMSE}} = \sqrt {\frac{{\mathop \sum \nolimits_{k = 1}^{n} \left( {f(x_{i} ) - y_{i} } \right)}}{n}^{2} } $$ -
6.
The r2 is defined as:
$$ r^{2} = \frac{{\left( {{\text{n}}\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} {\text{f}}\left( {{\text{x}}_{\text{i}} } \right){\text{y}}_{\text{i}} - \mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} {\text{f}}\left( {{\text{x}}_{\text{i}} } \right)\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} {\text{y}}_{\text{i}} } \right)^{2} }}{{\left( {{\text{n}}\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} f\left( {x_{i} } \right)^{2} - \left( {\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} f\left( {x_{i} } \right)^{2} } \right)} \right)\left( {{\text{n}}\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} y_{i}^{2} - \left( {\mathop \sum \nolimits_{{{\text{k}} = 1}}^{\text{n}} {\text{y}}_{\text{i}} } \right)^{2} } \right)}} $$
Drug-like chemical database preparation and virtual screening
Virtual screening is a computational technique used in drug discovery research to search large database in order to identify novel small molecules which are most likely to bind to a drug target. The developed models were used as a two-step query to screen Maybridge database. Maybridge, a commercial chemical database containing 59,652 compounds, was employed in this study for structure-based virtual screening procedure (Maybridge). However, this database is found to have a number of non-drug-like compounds. It is worthless to screen all the compounds of these databases and then eliminate them in the later phase for their non-drug-like properties. Therefore, compounds not satisfying drug-like properties were excluded from the databases prior to SVM-based virtual screening. In order to accomplish this task, compounds in this database were subjected to various scrupulous drug-like filters such as Lipinski’s rule of five and ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties. ADMET was applied to check whether the compounds are able to cross the blood–brain barrier (BBB) and have good solubility, human intestinal absorption (HIA), and low toxicity. Here, we mainly focused on oral bioavailability, low or no hepatotoxicity, and the capacity to penetrate the BBB, which is a key decision filter for central nervous system drug discovery. The compounds that satisfied the abovementioned properties were selected for molecular docking studies. Lipinski’s rule of 5 states that clogP ≤ 5, molecular weight ≤ 500, and number of hydrogen bond acceptors ≤ 10 and donors ≤ 5. Compounds violating more than one of these rules may have problems with bioavailability. Therefore these parameters were calculated by Prepare Ligands and ADMET Descriptors protocols as available in DS v3.1 software to eliminate compounds that did not pass the above criterias. After preparation of drug-like database, the generated models were subjected to screening of this drug-like database. The retrieved hit compounds were further subjected to molecular docking process.
Structure-based molecular docking
Molecular docking is a potent method in drug discovery process, which predicts the preferred orientation of one molecule to a second when bound to each other to form a stable complex. Virtual screening followed by docking has become one of the reputed methods for drug discovery and enhancing the efficiency in lead optimization. All hit compounds retrieved from database along with two most active inhibitors (the IC50 values of Inh 1 and Inh 2 are 8 nM and 10 nM, respectively) in collected dataset were docked using GOLD (Genetic Optimization for Ligand Docking) 5.1 program from Cambridge Crystallographic Data Center, UK. GOLD uses a genetic algorithm for docking ligands into protein binding sites to explore the full range of ligand conformational flexibility with partial flexibility of protein (Verdonk et al. 2003). Protein coordinates from the crystal structure of HDAC8 (PDB ID: 2V5X) which was selected from protein databank (PDB, www.rcsb.org) with good resolution (2.0 Å) (Vannini et al. 2007). All the water molecules present in the protein structure were removed and hydrogen atoms were added. The active site was defined with a 10 Å radius around the ligand present in the crystal structure. Ten docking runs were performed per structure unless five of the 10 poses were within 1.5 Å RMSD of each other. All hit compounds were docked into HDAC8 binding site. The GOLD fitness score is calculated from the contributions of hydrogen bond and adds Van der Waals interactions between the protein and ligand, intramolecular hydrogen bonds and strains of the ligand. The interacting ability of a compound depends on the fitness score, greater the GOLD fitness score better the binding affinity. The protein–inhibitor interactions were examined by DS v3.1. Hit molecules which showed higher GOLD fitness scores and strong interaction with key residues were selected. For further validation, the binding free energies were calculated for the hit compounds together with two most active known inhibitors using the AutoDock Vina tool available in PyRx v0.8 (Trott and Olson 2010). The compounds with best binding free energies were selected.
Molecular dynamics (MD) simulation
The selected complexes from docking study were subjected to 5 ns MD simulation using GROMACS 4.5.3 package with AMBER03 force field running on a high performance Linux cluster computer (Hess et al. 2008). Topology files for the inhibitors were generated using ACPYPE (AnteChamber Python Parser interface) (Sousa da Silva 2012). The structure was solvated in a dodecahedron box with length 1 nm and the TIP3P water model was generated to perform the simulations in an aqueous environment (Berendsen et al. 1981; Jorgensen et al. 1983). The 10 Na+ counter ions were added by replacing water molecules to ensure the overall change neutrality of the simulated system. The systems were subjected to a step by step steepest descent energy minimization process until a tolerance of 1000 kJ/mol/nm, to avoid high energy interactions and steric clashes. The energy minimized system was treated for 100 ps in an equilibration run. A constant temperature and pressure of 300 K and 1 bar were achieved with the V-rescale thermostat and Parrinello–Rahman barostat (Bussi et al. 2007; Parrinello and Rahman 1981). The particle mesh Ewald (PME) method was applied to accurately determine the long-range electrostatic interactions (Essmann et al. 1995). Bonds between heavy metals and corresponding hydrogen atoms were constrained to their equilibrium bond lengths using the LINCS21 algorithm (Hess et al. 1997). The time step for the simulations was set to 2 fs and the coordinate data were written to the file every 10 ps. All the analyses of the MD simulations were carried out by GROMACS and DS v3.1 software.
Results and discussion
Strategy for screening novel HDAC8 inhibitors
Virtual screening is a useful computational technique for drug design as it is a cost-effective and time saving process. Virtual screening methods can be divided into two broad categories: structure-based and ligand-based methods. To date, the three-dimensional (3D) structure of HDAC8 as a target receptor and its binding sites are available. Molecular docking is a highly effective structure-based technique for screening HDAC8 inhibitors. A set of active inhibitors are available, and it is possible to compute their shared information. Keeping this in view, an innovative hybrid strategy integrating structure-based and ligand-based approaches to identify novel HDAC8 inhibitors is presented in this study.
This strategy (Fig. 2) begins with preparation of drug-like database which is further bound to the HDAC8 protein thus predicting the binding conformations and molecular interactions. In next step, SVC and SVR models were applied sequentially to drug-like compounds, so that hit compounds with high activity were passed for further molecular docking. On the basis of the binding mode analysis, hit compounds with good binding characteristics and showing strong interaction with crucial amino acids at active site of HDAC8 were selected as final hits. 5 ns MD simulation was used to check the stability of HDAC8-inhibitor complex.
MIV calculation and descriptors selection based on MIV-based SFS
First of all, MIV of each molecular descriptor was calculated using SVR with RBF kernel functions (Table 2). The global descriptor TPSA and XlogP have the greatest MIVs. The variance explained values were 13.33 % for TPSA and 10.96 % for XlogP, which suggested that they are more relevant than the others for developing QSAR models. Overall MIVs are very small values. Thus, any single descriptor is not good enough to develop QSAR models with high performance and a combination of descriptors are needed for developing SVM models.
Here, MIV-base SFS method was selected due to its good performance in selecting of relevant descriptors. During the development of models, this method added descriptors to the developed model one at a time. This method began with the first descriptor with the greatest MIV (TPSA) which was firstly added to the model. Then the descriptor with the second greatest MIV (XlogP) was added to the model together with the first one. New descriptor, which gave the greatest MIV among all unselected descriptors, was progressively added to the QSAR model. New descriptors were added until the LOO CV results of model decreased successively three times (Fig. 3).
Construction and validation of SVC model (Qualitative SAR model)
Non-linear classification model was developed by SVC with RBF kernel function based on 30 training set compounds. In SVC, two factors, input descriptors and parameters of SVC (C and g), can affect prediction of SVC model. The optimized combination of these two factors would improve the performance of the model significantly. The optimal combination of parameters was found by GS method. To improve generalization quality of SVC model, a process of LOO CV of the whole training set was performed. The number of descriptors was firstly selected through average accuracy of LOO CV (Fig. 3a). During SVC modeling, the model developed with six descriptors (TPSA, XlogP, Span, Rgyr, InertiaY and HDon) showed best LOO CV result (average accuracy was 76.67 %). The next three descriptors decreased the prediction, which demonstrated that the first six descriptors are of great influence in classifying the HDAC8 inhibitors. The trend of decreasing prediction percentage with increased number of descriptors has shown the negative influence of other descriptors, which satisfied the stopping criteria of MIV-based SFS. Thus, this process was stopped by nine descriptors. Meanwhile this model was chosen as SVC model for virtual screening. The values of the optimal parameters C and g were 78.79 and 4.59, respectively.
After LOO CV, the generated SVC model was firstly validated by training set (Table 3). The model gave accuracy of 93.33 %, MCC of 0.87, TPR of 100 %, and FPR of 13.33 %. Of all 30 compounds, 28 were correctly predicted and only two non-inhibitor (compound 18 and compound 22) were wrongly predicted (Table 4). The developed SVC model is not just to classify the training set correctly but also to verify whether the model is capable of classifying external compounds which are outside of the training set accurately. Thus, to verify the generalization quality of the developed model, test set containing 25 inhibitors and 25 non-inhibitors was further predicted as an independent validation. The accuracy, MCC, TPR, and FPR for test set were 84, 0.69, 92, and 24 %, respectively (Table 3, Table 5). The SVC model was further validated by ROC curve (Fig. 4). The values of AUC were 0.99 for training set and 0.83 for test set. These results clearly demonstrated that the established SVC model achieved high robust and good utility, implying the SVC model can be used as a screening tool for retrieving HDAC8 inhibitors.
Construction and validation of SVR model (Quantitative SAR model)
Non-linear regression model was built using SVR with RBF kernel function based on the training set. The pIC50 (−log IC50) values were considered as dependent variable and the molecular descriptors were considered as independent variables. During SVR modeling, the generated models were first validated by LOO CV, which can improve generalization quality of models. The average value of RMSE was used to measure differences between pIC50 values predicted by SVR model and actual pIC50 values. The GS method attempted to minimize the RMSE value by identifying good parameters (C, g, and loss epsilon insensitive function ε). And descriptors were selected using MIV-based SFS. The average value of RMSE of the model built with eight descriptors containing TPSA, XlogP, Span, Rgyr, InertiaY, HDon, NAtoms, and InertiaZ was the minimal (0.62) and the process of selecting descriptors was stopped when three new descriptors (9th, 10th, and 11th) increased the RMSE value (Fig. 3b). Thus, a robust Quantitative SAR model was obtained selecting eight descriptors and those good parameters gave the lowest average value of RMSE. The values of optimal parameters C, g, and ε were 1024, 0.09, and 0.21, respectively.
In order to assess the predictive ability of the SVR model just built, training and test sets were used and the activity of each compound in training and test sets was estimated by SVR model. All compounds in training and test sets were classified relatively into two groups based on their pIC50 values: Group 1 (Inhibitor) pIC50 > = 0.52; Group 2 (non-inhibitor) pIC50 < 0.52. The SVR model can correctly predict 23 out of 30 training set compounds, achieving 76.67 % accuracy (Table 4), and also showed low RMSE and high r2 of 0.42 and 0.74, respectively. In addition, the values of RMSE and r2 for the test set were 0.63 and 0.53, respectively. Although the statistical parameters for the test set were not so excellent as that for the training set, the SRV model can predict with up to 76 % accuracy on the test set (38 out of 50 test set compounds) (Table 5). Except few compounds, all remaining inhibitors were predicted correspondingly and non-inhibitors were estimated as group 2 (non-inhibitors). The SVR model was able to estimate the activities of compounds in their own activity ranges. This result suggested that the SVR model not only fit for training set compounds but also the external test set compounds. Thus, the developed SVR model can be used as an estimator for HDAC8 inhibitor screening.
Database virtual screening
Virtual database screening using QSAR models deals with the quick search of large libraries of small-molecule discover drug target. This approach serves an advantage over any de novo design methods by providing a set of compounds directly for the biological testing. Both the validated QSAR models (SVC and SVR models) were used as a two-step filter in database screening. Maybridge database containing 59,652 compounds has been utilized in database screening. Prior to ligand-based virtual screening, this database was transformed to drug-like database by Prepare Ligands and ADMET Descriptors protocols of DS v3.1. Prepare Ligands protocol eradicated the duplicate structures, fixed bad valencies, and calculated 3D coordinates of all the compounds. ADMET Descriptors protocol calculated various properties such as aqueous solubility, blood brain barrier penetration, CYP2D6 binding, hepatotoxicity, intestinal absorption, and plasma protein binding. Calculating ADMET descriptors early in the development of a drug is important to avoid elimination of compounds with unfavorable ADMET characteristics later in the drug development process. Finally, 4,741 drug-like compounds were selected and applied subsequently in ligand-based virtual screening. The drug-like compounds fitting with two QSAR models were identified as hit compounds for further molecular docking study. SVC model has identified 1084 hit compounds. The hit compounds resulted from this step were subsequently subject to SVR model for predicting biological activity (pIC50 value). 30 out of 4,741 drug-like compounds, which were predicted with a high probability of activity through SVM prediction, were selected for molecular docking study.
Next, these 30 compounds along with 2 most active inhibitors in data set were docked into the active site of preprocessed protein structure of HDAC8 using GOLD software and the binding free energy was calculated by AutoDock Vina (The detail process was in supplementary materials). 2 out of 30 compounds were identified as hit compounds on the basis of strong binding interactions at active site of target protein, good GOLD fitness docking score, and favorable binding free energy (S1, S2 and S3).
The two hit compounds resulted in the molecular docking were further subjected to molecular dynamics simulation study to examine the stability of HDAC8-hit complex (The detail process was in supplementary materials). The observations revealed that these two hit compounds were as stable as two most active inhibitors in our data set (S4). It indicates that these two hit compounds with new structural scaffolds have high probability of activity and can be reasonably used for further in vitro and in vivo biological activity analyses.
In this day and age, the discovery of novel chemical entities is becoming increasingly difficult, costly and time-consuming, medicinal chemists have always struggled with the difficult problem of identifying the compounds with a high probability of activity from thousands or millions of possible molecules. Virtual screening allows chemists to reduce a huge virtual library to a more manageable size. In this study, we discussed and reported the SVM-based QSAR modeling approach in combination with molecular docking and molecular dynamics simulation, which would facilitate discovery of new structural scaffolds of HDAC8. This study showed that SVM is a very powerful QSAR modeling technique to ligand-based virtual screening. Besides that, the two hit compounds were identified in this study as new structural scaffolds for HDAC8 inhibitors, and can be reasonably selected for testing biological activity by in vitro and in vivo analyses. Even if these two new structural scaffolds are not HDAC8 inhibitors until validate their actual biological activity, they are still interesting and useful for the further HDAC8-based drug design and the chemists who devoted themselves to discovery of HDAC8 inhibitors.
Conclusion
In this study, a hybrid protocol of virtual screening method based on two QSAR models and molecular docking was utilized to discover potential HDAC8 inhibitors. As the first step, a druglike database based on Maybridge was prepared. From results of QSAR-based virtual screening, 2 final hits were selected according to their binding characteristics and interactions with crucial amino acids. Subsequently, 5 ns MD simulation was used to check their complex stability.
This study further suggested that combination of SVC and SVR has the capacity to rapidly discover potential HDAC8 inhibitors and MIV-based SFS method is a useful descriptor selection routine for developing SVM model. On the whole, two final hits can be used as potential HDAC8 inhibitors for further in vivo studies. The developed models could be a fast and effective tool to assist discovery of novel HDAC8 inhibitors. The strategy used in this study could be a promising computational approach and may be generally applicable to other target drug designs.
References
ADRIANA.Code (2004) Molecular Networks Inc. Available from: www.molecular-networks.com
Berendsen HJC, Postma JPM, Van Gunsteren WF, Hermans J (1981) Interaction models for water in relation to protein hydration. Intermol forces 11:331–342
Bolden JE, Peart MJ, Johnstone RW (2006) Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5:769–784
Brodeur GM (2003) Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 3:203–216
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101
Byvatov E, Fechner U, Sadowski J, Schneider G (2003) Comparison of support vector and artificial neural network system for drug/nondrug classification. J Chem Inf Comput Sci 43:1882–1889
Cao GP, Arooj M, Thangapandian S, Park C, Arulalapperumal V, Kim Y, Kwon YJ, Kim HH, Suh JK, Lee KW (2015) A lazy learning-based QSAR classification study for screening potential histone deacetylase 8 (HDAC8) inhibitors. SAR QSAR Environ Res 26:397–420
Chang CC, Lin CJ (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2:1–27
Cortes C, Vapnik V (2011) Support-vector networks. Mach Learn 20:273–297
Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW (2003) The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol Cell Biol 23:607–619
Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci 95:2795–2800
Dizhong C, Weiping D, Kand S, Hong YS, Eric, TS, Niefang Y, Yong Z (2007) Benzimidazole derivatives: preparation and pharma-ceutical applications. US Patents 2007/0043043 A1, 22 Feb 2007
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Fawcelt T (2006) An Introduction to ROC Analysis. Pattern Recogn Lett 27:861–874
Gu W, Nusinzon I (2006) Smith RDJr, Horvath CM, Silverman, RB. Carbonyl-sulfurcontaining analogs of suberoylanilide hydroxam-ic acid: potent inhibition of histone deacetylases. Bioorg Med Chem 14:3320–3329
Guyon I (2003) An introduction to variable and feature selection. J Mach Learn Res 3:1157–1182
Haindl M, Somol P, Ververidis D, Kotropoulos C (2006) Feature selection based on mutual correlation. Progress in pattern recognition, image analysis and applications. Springer, Berlin, pp 569–577
Han LY, Zheng CJ, Xie B, Jia J, Ma XH, Zhu F, Lin HH, Chen X, Chen YZ (2007) Support vector machines approach for predicting druggable proteins: recent progress in its exploration and investigation of its usefulness. Drug Discovery Today 12:7–8
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Hess B, Kutzner C, van der Spoel D (2008) GROMACS 4: algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput 4:435–447
Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800
Jeffrey MB, Zuomei L, Daniel D, Claire B (2004) Methods for specifically inhibiting histone-7 and 8. US Patents 2004/0072770 A1, 15 April 2004
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926
Joseph, JB, Sriram B (2010) Uses of selective inhibitors of HDAC8 for treatment of T-cell proliferative disorders. US Patents 7,820,711, 26 Oct 2010
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the fourteenth international joint conference on artificial intelligence 1995; 1995 August 20–25; Quebec Canada. California: Morgan Kaufmann
Landry J, Slama J, Sternglanz R (2000) Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem Bioph Res Co 278:685–690
Lehrmann H, Pritchard LL, Harel-Bellan A (2002) Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv Cancer Res 86:41–65
Li HZ, Tao W, Gao T, Li H, Lu YH, Su ZM (2012) Improving the accuracy of DFT calculation for homolysis bond dissociation energies of Y—NO Bond via back propagation neural network based on mean impact value. Chem J Chinese U 33:346–352
Liew CY, Ma XH, Liu X, Yap CW (2009) SVM model for virtual screening of Lck inhibitors. J Chem Inf Model 49:877–885
Ma XH, Wang R, Tan CY, Jiang YY, Lu T (2010) Virtual screening of selective multitarget kinase inhibitors by combinatorial support vector machines. Mol Pharm 7:1545–1560
Mahé P, Ueda N, Akutsu T, Perret JL, Vert JP (2005) Graph kernels for molecular structure-activity relationship analysis with support vector machines. J Chem Inf Model 45:939–951
Marcano-Cedeño A (2010) Feature selection using sequential forward selection and classification applying artificial metaplasticity neural network. IECON 2010—36th annual conference on IEEE industrial electronics society, 2845–2850
Marks PA, Breslow R (2007) Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol 25:84–90
Niu B, Lu WC, Yang SS, Cai YD, Li GZ (2007) Support vector machine for SAR/QSAR of phenethy-amines. Acta Pharmacol Sin 28:1075–1086
Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, von Deimling A, Fischer M, Witt O (2009) Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res 15:91–99
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182
Shahlaei M (2013) Descriptor selection methods in quantitative structure-activity relationship studies: a review study. Chem Rev 113:8093–8103
Shi Z, Ma XH, Qin C, Jia J, Jiang YY (2012) Combinatorial support vector machines approach for virtual screening of selective multi-target serotonin reuptake inhibitors from large compound libraries. J Mol Graph Model 32:49–66
Sousa da Silva AW (2012) Vranken WF. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes 5:1–8
Taylor D, Maxwell M, Luthi-Carter R, Kazantsev A (2008) Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci 65:4000–4018
Thangapandian S, John S, Lee Y, Kim S, Lee KW (2011) Dynamic structure-based pharmacophore model development: a new and effective addition in the histone deacetylase 8 (HDAC8) inhibitor discovery. Int J Mol Sci 12:9440–9462
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley-VCH, Weinheim, p 9
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Valenzuela-Fernández A, Cabrero JR, Serrador JM, Sánchez-Madrid F (2008) HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions error. Trends Cell Biol 18:291–297
Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfí A, De Francesco R, Steinkühler C, Di Marco S (2007) Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO Rep 8:879–884
Vasanthanathan P, Taboureau O, Oostenbrink C, Vermeulen NP, Olsen L, Jørgensen FS (2009) Classification of cytochrome P450 1A2 inhibitors and noninhibitors by machine learning techniques. Drug Metab Dispos 37:658–664
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Improved protein-ligand docking using GOLD. Proteins 52:609–623
Walter S, Haishan W, Zheng Y (2007) Biaryl linked hydroxamates: preparation and pharmaceutical applications. US Patents 2007/0167499 A1, 2007 Jul 19
Wan HL, Wang ZR, Li LL, Cheng C, Ji P, Liu JJ, Zhang H, Zou J, Yang SY (2012) Discovery of novel Bruton’s tyrosine kinase inhibitors using a hybrid protocol of virtual screening approaches based on SVM model, pharmacophore and molecular docking. Chem Biol Drug Des 80:366–373
Wang M, Wang K, Yan A, Yu C (2012) Classification of HCV NS5B polymerase inhibitors using support vector machine. Int J Mol Sci 13:4033–4047
Wu TY, Hassig C, Wu Y, Ding S, Schultz PG (2004) Design, synthesis, and activity of HDAC inhibitors with a N-formyl hydroxyl-amine head group. Bioorg Med Chem Lett 14:449–453
Maybridge; Maybridge Chemical Co., Cornwall, UK. Available from: www.maybridge.com
Yang XY, Seto E (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9:206–218
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32:1466–1477
Yap CW, Chen YZ (2004) Prediction of cytochrome P450 3A4, 2D6, and 2C9 inhibitors and substrates by using support vector machines. J Chem Inf Model 45:982–992
Ze-Yi L, Haishan W, Yan Z (2008) Aclyurea connected and sul-fonamide connected hydroxamates. US Patents 2008/0070954 A1.48, 20 March 2008
Zhang J, Han B, Wei X, Tan C, Chen Y, Jiang Y (2012) A two-step target binding and selectivity support vector machines approach for virtual screening of dopamine receptor subtype-selective ligands. PLoS ONE 7:e39076
Acknowledgments
This research was supported by a grant from Marine Biotechnology Program (PJT200671) Funded by Ministry of Oceans and Fisheries, Korea. And this work was also supported by the Next-Generation BioGreen 21 Program (PJ01106202) from Rural Development Administration (RDA) of Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that this article content has no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, G.P., Thangapandian, S., Son, M. et al. QSAR modeling to design selective histone deacetylase 8 (HDAC8) inhibitors. Arch. Pharm. Res. 39, 1356–1369 (2016). https://doi.org/10.1007/s12272-015-0705-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-015-0705-5