Abstract
The inhibition of histone deacetylase (HDAC) has become a well-recognized target for cancer therapy. Until now only five HDAC inhibitors i.e., SAHA, romidepsin (FK-228), belinostat, chidamide, and panobinostat have been approved by FDA. The first four of them are being employed for the treatment of cutaneous T-cell lymphoma and last one is being used for the treatment of multiple myeloma. In the present study, structure and ligand-based computational approaches were selected to design novel histone deacetylase inhibitors. A ligand-based pharmacophore model was developed employing phase module that exhibited five similar features and the generated pharmacophore models were validated by enrichment studies using the decoy set. Atom-based 3D-QSAR model was developed and validated using internal, partial least square (PLS) and external validation methods. The best 3D-QSAR model exhibited high value of regression coefficient for training set (R2) = 0.926 and test (R2) = 0.699, cross-validated coefficient (rcv2) = 0.967 and R2 pred = 0.6578 with low root mean standard deviation (RMSE) = 0.4963. Additionally, the selected pharmacophore model (AAADR.12514) was employed as a 3D query for virtual screening against the ZINC database. The hit compounds were subsequently subjected to ligand–receptor interaction studies. Further, HDAC receptor-ligand complex were subjected to MM-GBSA and molecular dynamic simulation to evaluate the binding energy, strain energy, stability, and electrostatics of complex. Moreover, ADMET studies were also performed on resulted molecules. The outcome of these studies could be utilized for identification of novel lead for the development of HDAC inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is the deadliest ailment and the second leading cause of death worldwide, which involves complex phenomenon like unregulated cell growth and proliferation, loss of apoptosis, and metastasis due to aberrant regulation of gene expression (Siegel et al. 2017; Suzuki and Miyata 2005). There are two enzymes i.e., Histone acetyl-transferases (HATs) and histone deacetylase (HDACs) playing an important role in the dynamic equilibrium between acetylation and deacetylation of histone proteins leading to appropriate regulation of gene transcription and gene expression of eukaryotes at DNA level (Singh et al. 2016). An unbalance of them can result in the abnormalities in proliferation and differentiation of normal cells and then lead to the initiation of tumor (Rajak et al. 2014; Singh et al. 2017b). The enhanced HDAC activity may play a crucial function in the pathogenesis of cancer (Minucci and Pelicci 2006; Singh et al. 2017a). The HDACs have significant role in cancer cell biology and resides in nucleus, as well as in cytoplasm (Binaschi et al. 2010). It is well known that the process of histone deacetylation is mainly associated with transcriptional repression, while histone hyperacetylation is involved in gene expression (Taunton et al. 1996). Therefore, inhibition of HDAC activity could promote histone hyperacetylation, resulting in transcriptional activation of suppressed genes, which is correlated with apoptosis, differentiation, or cell cycle arrest in tumor cells (Mahlknecht and Hoelzer 2000; Zhu et al. 2010).
HDACs are the collection of zinc-dependent metalloenzymes (Wu et al. 2016), divided into four structural classes on the basis of their homology to yeast proteins. Subtypes HDAC1, 2, 3, and 8 are included in class I and exhibits homology to the yeast protein rpd3. Class II can be divided into subtypes HDAC4, 5, 7, and 9 and Class IIb can be subdivided into HDAC6 and 10. Subtype HDAC11 is included in Class IV. The enzymatic activity of HDACs is based on Zn2+-dependent mechanism and NAD+-dependent mechanisms. Class I, Class II, and Class IV HDACs follows Zn2+dependent mechanism while the class III HDAC enzymes (or sirtuins) operates by a NAD+-dependent mechanism. The class III comprises of seven subtypes in humans (SIRT1 to SIRT 7) and they demonstrate homology to the yeast protein Sir2 (Gray and Ekstrom 2001; Minucci and Pelicci 2006). A broad range of chemical structures including hydroxamic acids, electrophilic ketones, benzamides, mercaptoamides, and thiols, as well as small fatty acids have showed inhibitory potential for Zn2+ dependent HDACs (class I and II) (Biel et al. 2005; Kozikowski et al. 2007; Marks et al. 2004; Schafer et al. 2008) Among them hydroxamic acid derivatives are effective in nanomolar concentration (TSA IC50: 12 nM and SAHA IC50:110–370 nM) (Bieliauskas et al. 2007; Chen et al. 2009; Gopalan et al. 2013) (Fig. 1). However, TSA and other HDAC inhibitors have a clear disadvantage that their production is costly and profound concerns remain about their toxicity, non-specificity, and side effects (Yoshida et al. 1990). Thus, there has been an increasing attention in developing novel HDAC inhibitors with lesser toxicity, better pharmacokinetic properties, and improved anticancer activity. 3D QSAR, pharmacophore modeling, and virtual screening may be a better solution for the fulfillment of aforementioned goal (Patel et al. 2016).
In the present work, 3D-QSAR models were developed using Pharmacophore alignment and Scoring module (PHASE) on hydroxamic acid derivatives as HDAC inhibitors for antineoplastic activity. The efficiency and reliability of 3D-QSAR models were checked by PLS analysis and external validation methods. The obtained QSAR contour maps were used as visual guide for the designing of novel and active molecules. The combination of virtual screening-based docking was used in rational drug design to get better docking results and an information about the inhibitory mechanisms. Virtual screening of ZINC database with the help of developed pharmacophore models was performed to explore novel and potential HDAC inhibitors. Furthermore, MM-GBSA and molecular dynamic (MD) simulation were performed for the assessment of accuracy of docking. Absorption, distribution, metabolism, excretion, and toxicity (ADMET) studies were also performed to conform the drug-like property of top scored molecules. The strategy used in this work could be a promising computational strategy which could be utilized for the design of novel HDAC inhibitors.
Methodology
The molecular modeling studies (3D QSAR, pharmacophore modeling, virtual screening, docking, MM-GBSA and ADMET) were executed using Schrodinger interface (Maestro version 9.3) LLC, New York software and MD simulation was performed using Accelrys Discovery Studio 4.1 software.
Dataset
The available dataset of 51 compounds having hydroxamic acid based scaffolds with wide structural diversity was selected for developing 3D-QSAR models with same biological assay (fluorescence assay) method (Jin et al. 2015; Su et al. 2009; Wang et al. 2017). All the molecular structures and activity data employed for 3D-QSAR study are described in Table 1. The inhibitory activity (IC50 value in moles/liter) for each compound was transformed into negative logarithm of IC50 (pIC50). The pIC50 values were employed as dependent variable for the QSAR analysis.
Pharmacophore modeling
The ligand-based pharmacophore models were developed using ‘PHASE’ v3.4 (Schrödinger 2012). It is a widely used technique for common pharmacophore recognition and 3D QSAR model development. In phase, the development of pharmacophore models were initiated with clean up of all 51 ligands. In the second step, conformers of all ligands were produced with the help of conformer generation macromodel search method with maximum number of conformers 1000 per structure and structures were minimized using OPLS_2005 force field with 100 minimization steps (Watts et al. 2010). In the next step, sites were generated for all the ligand molecules that gave us different Pharmacophore hypotheses based upon the activity threshold of active and inactive molecules. Each of these hypotheses contained a maximum of six features namely, hydrogen bond donor (D), hydrogen bond acceptor (A), aromatic ring (R), hydrophobic group (H), positively charged group (P), and negatively charged group (N), which were characterized by a group of chemical structure. Generated hypotheses were ranked on the basis of survival, survival inactive, post-hoc scores, vector, volume, and site scores (Table 2) (Sallam et al. 2013). The best pharmacophore models were recognized by aligning the pharmacophore with active ligands, with maximum adjusted survival score and used for additional 3D-QSAR studies (Nair et al. 2012).
Atom-based 3D-QSAR modeling
QSAR modeling was performed using the selected hypothesis by dividing the dataset into a training set (41) and a test set (10) in such a manner that 80% of the compounds fall under the training set and the residual 20% compounds were taken as the test set ranging from maximally active to moderate and then least active (based on their pIC50) compounds. In the PHASE module, two options are available for alignment of 3D structure of molecules: (i) the pharmacophore-based alignment and (ii) the atom-based alignment (Teli and Rajanikant 2012). In the present study, an atom-based QSAR model was employed, which is more beneficial in describing the SAR (structure activity relationship). The atom-based QSAR models were generated for selected hypothesis by maintaining 1 A° grid spacing and 5 as maximum number of PLS factors with good statistics and predictive ability. In score hypotheses step, the hypotheses were carefully chosen with the help of the activity data for all ligands and visualization of the QSAR results helped us to explore the optimization of the core structures (Sallam et al. 2013).
Pharmacophore-based virtual screening and docking
The virtual screening is a fast and precise database searching method applied for the identification of novel and potential lead molecules, appropriate for additional development (Lengauer et al. 2004). It has been accomplished on the basis of best pharmacophore obtained through comparison of various pharmacophoric features. The chemical database having novel chemical structures with desired pharmacophoric features was obtained from commercially available database (ZINC database-zincpharmer.csb.pitt.edu.) and constraints such as root mean square deviation (RMSD) kept <1, with rotatable bonds <10 and the range of molecular weight <500 dalton were used to achieve the best similarity search of 3D database. In the screening of database, molecules must match at least three sites for the hypotheses with three or four pharmacophoric sites and match a minimum of at least four sites for hypotheses of five or more pharmacophoric sites. The virtual screening workflow (maestro v9.3) was employed for this study. Initially, ligands were filtered using Qikprop (prefilterd by Lipinski rule of five), ligands with reactive functional groups were removed, then the preparation of ligands was done. The chemical database ligands with the best scores were then subjected to molecular docking studies, such as High throughput virtual screening (HTVS), SP (standard precision), and XP (extra precision) to estimate ligand-protein binding interactions.
Molecular docking was assessed with free-binding energy MM-GBSA (Molecular Mechanics-Generalized Born Surface Area). MM/GBSA is a post-scoring technique that re-arrange the docked protein-ligand complexes on the basis of their relative binding free energy.
MD simulation
MD simulation of receptor-ligand complex (cocrystal ligand SAHA and compound 26 with PDB: 4LXZ) was performed using the simulation tool of Accelrys Discovery Studio 4.1 software, to check the stability, conformational changes, and interaction of structure during the simulation process. For molecular simulation study, the protein (4LXZ) was selected on the basis of their relevance to cancer, structural resemblance of co-crystallized ligand (SAHA) with the selected dataset compounds and better resolution (1.85 Å) (Wang et al. 2017). MD simulation study consists of several steps i.e., preparation of protein structure, solvation of protein structure, standard dynamics cascade analysis, and MD trajectory analysis. The prepared protein was assigned using the CHARMm force field parameters. After solvation of protein structure 26418 water molecules, 70 sodium and 82 chloride ions were added, which retained the system neutral and kept the salt concentration of the system 0.145 with 0.1 kcal/mol RMS gradient. Standard dynamics cascade protocol includes minimization, minimization 2, heating, equilibration, and production. Energy minimization were performed using 1000 steps of steepest descent and 2000 steps of adopted basis NR algorithm without assigning any constraint. Subsequently, for equilibration and production steps, the SHAKE constraint with Leapfrog Verlet dynamic algorithm was employed. Finally, the production and equilibration MD simulations were carried out for 200 and 20 ps, respectively, with explicit periodic boundary conditions, in an NPT ensemble (T = 300 K, thermostat relaxation time = 2.0 fs; P = 1 atm), target temperature 300 K and temperature coupling decay time 5 ps. SHAKE algorithm was employed to secure all available covalent bonds having hydrogens and Particle-Mesh-Ewald Technique (York et al. 1993) was used for electrostatic studies.
Results and discussion
Pharmacophore model
The comparison of survival score indicates that pharmacophore model (AAADR.12514) has best survival score in comparison of all generated ligand-based pharmacophore models and showed the best alignment over most active compound along with the distance (Å) (Fig. 2a).The ligand-based pharmacophoric model (AAADR.12514) exhibited five key features including three hydrogen bond acceptors, one hydrogen bond donar and one aromatic rings with highest survival score of 3.937. For validation of the developed pharmacophore model (AAADR.12514), a small database including 117 compounds was prepared through zincpharmer.csb.pitt.edu., consisting of 17 active molecules and 100 decoys for HDAC inhibitors. Decoy sets were considered on the basis of their similarity to the active ligands with respect to five physical descriptors (molecular weight, total hydrogen bond donors, total hydrogen bond acceptors, number of rotational bonds and the octanol-water partition coefficient) without being chemically identical to any of the active ligands. Several statistical parameters such as accuracy, precision, specificity, sensitivity, Receiver operating characteristic curve (ROC), Area under curve (AUC), Robust initial enhancement (RIE), Boltzmann-enhanced discrimination of receiver operating characteristic (BEDROC, α = 20), Enrichment factor (EF 5%) and GH (goodness of hit) (Truchon and Bayly 2007) were calculated for selected AAADR.12514 hypothesis. These are some essential parameters, which influences the fitness of the pharmacophore hypothesis. The accuracy, sensitivity, specificity, ROC, AUC, RIE, BEDROC (α = 20) and EF (5%) of the best pharmacophore model (AAADR.12514) for the decoy set were found to be 0.99, 0.94, 0.95, 0.96, 0.89, 4.78, 0.73 and 4.6, respectively, which were within the standard statistical limits and assisted the envisaging capability of the top score pharmacophoric model (Fig. 2b) indicating the ROC performance of top score pharmacophore model (AAADR.12514).
Phase Pharmacophore model generation and validation. a alignment of best pharmacophore (AAADR.12514) onto the most active compound 26 with fit value of 3.0 Å (aromatic ring (orange circle), three hydrogen acceptor (red sphere), and one hydrogen donor (light blue sphere) with distance in Å). b receiver operating curve (ROC) plot of best pharmacophore model (AAADR.12514)
3D-QSAR modeling
The 3D-QSAR models were prepared to set up a statistical relationship between the 3D spatial arrangement of the pharmacophoric features and the cytotoxic activity of HDAC inhibitors. A total of 51 compounds under study were divided into training set (41compounds) and a test set (10 compounds). The compounds were divided in test and training set as per their structural features and their biological activity threshold. Five featured Pharmacophoric hypothesis (AAADR.12514) with high value of survival score was chosen for atom-based 3D-QSAR model generation (PLS model) using Schrödinger v9.3 with five PLS factors. The predicted activities of training and test set molecules along with fitness using AAADR.12514 pharmacophore hypothesis are presented in Table 3 and the residual values were calculated by subtraction of predicted activity from observed activity and sum of residual was also calculated and found as 1.256 for 3D-QSAR model AAADR.12514. Scatter plots of actual vs. predicted activities showed that pIC50 values were successfully predicted for both training (Fig. 3a) and test (Fig. 3b) set compounds and it could be employed to identify the existence of outliers from a QSAR model.
3D-QSAR contour maps analysis
The QSAR contour maps received from top pharmacophore model (AAADR.12514) showed how 3D-QSAR studies could recognize the features vital for the interaction between test molecules and receptor. These maps provided information regarding positions that are essential for a particular physicochemical property to augment the cytotoxic activity of a ligand. These insights could be understood by comparing the contour maps of the most and least active compounds (Figs. 4a–f). The QSAR model displayed 3D characteristics in the form of boxes or cubes. The blue cubes showed positive coefficients that are favorable, while yellow cubes showed negative coefficients that are unfavorable features for activity. Finally, it can be concluded that these maps hints about desirable or undesirable functional groups at particular positions in a ligand. Figures 4a, b shows the hydrogen bond donor contour map of most active and inactive molecule, respectively. Figure 4a indicates the blue favorable boxes around the NH group nearer to the benzene ring (donor feature, D9) and blue boxes seen around the hydroxyl group of cap region, thus illustrating that additional donor groups at these positions (blue cubes) could increase biological activity. Although, in the contour map of most inactive molecule, yellow unfavorable boxes were observed nearer to the benzene ring (donor feature D9), indicating the biological inactiveness of the molecule (Fig. 4b).
Comparison of the contour maps of the most active compound 26 and least active compound 51: The hydrogen bond donor effects in a most active compound 26 and b the least active compound 51; The hydrophobic effects in c most active compound 26 and d the least active compound 51; The electron withdrawing effects in e most active compound 26 and f the least active compound 51. Blue cubes indicates favorable regions while yellow cubes indicates unfavorable region for the activity
Figures 4c, d shows the hydrophobic contour map of most active and inactive molecule, respectively. Fig. 4c indicates the blue favorable boxes around the para-position of benzene ring (hydrophobic feature, R14) of the molecule, indicating the biological activeness of the molecule. While, in the least active compound (Fig. 4d), the presence of yellow unfavorable boxes around the para-position of benzene ring (hydrophobic feature, R14) and near to benzene ring of linker, illustrates the weak biological activity of molecule.
Figures 4e, f shows the electron withdrawing contour map of most active and inactive molecule, respectively. Figure 4e shows the blue favorable boxes around the both carbonyl groups nearer to the benzene ring and carbonyl group of pyrrole ring (A4, A5, and A6), suggesting substitution at this position with electron withdrawing group increases the activity. While, in the least active compound (Fig. 4f), the yellow unfavorable boxes were found around A4, A5, and A6, illustrating the weak biological activity of molecule.
Statistical parameters of 3D-QSAR models
Internal validation and PLS analyses
A good 3D-QSAR model should exhibit reliable predictability that can be validated by both validation methods i.e., internal and external validation. The predictive power of the generated 3D-QSAR model (AAADR.12514) was analyzed using a test set of 10 compounds and the statistical significance of the model was achieved using a PLS factor of 5. The robustness of model in predicting the active molecules were considered depending upon different internal and PLS parameters including regression coefficient for the training set (Q2), regression coefficient for test set (R2), the standard deviation (SD), the root mean square error (RMSE), the variance ratio (F), significance level of variance ratio (P), the Pearson correlation coefficient (Pearson-r), chi square (χ2), and the stability of model. The regression coefficient for test set (R2) and training set (Q2) were 0.926 and 0.699, respectively, indicating that model had good internal predictive ability. Additionally, the high stability of model 0.7412, low SD 0.2154, low RMSE 0.4963, high Pearson-r 0.8811, chi square (χ2) 0.0761, and high F-value 87.5 with smaller P-value 8.997e–019, supported the significance of the selected model (Table 4). All the internal statistical parameters and PLS parameters were well within the range but, external statistical validation was also essential for greater reliability of selected model.
External validation
A reliable 3D-QSAR model should also exhibit good correlation between predicted and observed activities, which can be indicated by external validation parameters i.e., correlation coefficient R2 (or r2). The best predictive ability of selected 3D-QSAR model (AAADR.12514) was supported by a value of R2 = 0.930 (or r2 = 0.718).
The predictive capability of the 3D-QSAR model (AAADR.12514) was also favored by a value of R2 pred (R2 pred = 0.6578). The predictive capability of the selected model was also proved by the values of r2 − r02 / r2 = −0.0282, r2 − ŕ02 / r2 = −0.3328, K = 0.4810, K̍’ = 0.6990, r02 = 0.9570 and r0′2 = 0.7180 that were attained by calculating the correlation coefficient of regression lines of the scatter plot. Additionally, the external predictability and acceptability of selected model was also confirmed by rm2 and Rp2. The values of rm2 and Rp2 were 0.7772 and 0.5018, respectively (Table 5), which were within the standard statistical limits. Therefore, from all of the external validation parameters, the reliability of QSAR model was confirmed for the prediction of a new molecule.
Virtual screening and docking
Best pharmacophore hypothesis was further employed against the Zinc database for 3D search query. The hit compounds were obtained by overlapping their chemical functional features with corresponding features of the best pharmacophoric model (AAADR.12514). As a result of searching the features against Zinc database, a total of 5238 compounds were obtained and these screened compounds again tested for drug likeliness by utilizing Qikprop and Lipinski’s rule of five. After screening, total 1775 compounds were obtained and further subjected to HTVS (high throughput virtual screening). In virtual screening workflow, 30% of best compounds were selected for SP (standard precision) docking and after SP docking, 30% of best compounds were selected for XP (extra precision) docking. The structure of best hits after virtual screening i.e., ZINC10600215, ZINC12802221, ZINC17192586, ZINC17189715, and ZINC12548600 are shown in Fig. 5. The XP, SP, and HTVS dock score, glide energy, potential energy-OPLS-2005, and RMSD of all best five hits i.e., ZINC10600215, ZINC12802221, ZINC17192586, ZINC17189715, and ZINC12548600 in the active site of 4LXZ are shown in Table 6.
On the basis of docking, screened hit molecule ZINC10600215 showed best scores and H bond interactions as compared to crystal ligand (SAHA) (Figs. 6a, b). The 2d interaction pattern of top scored molecule ZINC10600215 in the active site of protein (4LXZ) were compared with the crystal ligand (SAHA) and are shown in Figs. 6c, d. For the validation of docking, co-crystallized ligand was redocked and its RMSD was calculated to validate the docking process (Fig. 7).
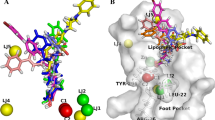
a Docked pose of compound ZINC10600215 in the active site of 4LXZ showing the hydrogen bond interaction (pink dotted lines) with TYR 308 and HIS 140; b Docked pose of crystal ligand SAHA in the active site of 4LXZ showing the hydrogen bond interaction with TYR 308, HIS 145, HIS 146, and ASP 104; c 2D interaction diagram of compound ZINC10600215; and d 2d interaction diagram of crystal ligand SAHA
The Fig. 6d shows the interaction of crystal ligand (SAHA) with (C = O…..H-Tyr308 O-H…..N-His145, N-H…..N-His146 and N-H…..O-Asp104), in addition the hydrophobic site of catalytic pocket (Phe210 and Phe155) showed hydrophobic interaction with aliphatic and aromatic parts of the compound. After visual inspection of interaction pattern of top scored molecule ZINC10600215 (Fig. 6c), it was found that it interacted similarly with the residue in the catalytic pocket (C = O…..H-Tyr308, N-H…..N-His146 and N-H…..O Asp104). The aliphatic and aromatic parts of compound also showed similar hydrophobic interaction with hydrophobic site of the receptor (Phe210 and Phe155).
MM/GBSA-based rescoring
For rescoring of already docked molecules, MM/GBSA (Molecular Mechanics-Generalized Born Surface Area) study was performed using Prime 3.1 module. Best five molecules with dG_Bind (free-binding energy), dG Bind_Solv (free solvation energy) are shown in (Table 7). Top scored molecules ZINC10600215 impart highly negative free-binding energy (−56.9548) than other compounds, it means, ZINC10600215 exhibit high binding affinity to the receptor as compared to other molecules. Overall result shows that the energy profile of top scored molecules ZINC10600215, exhibited interesting correlation with the crystal ligand (SAHA).
MD simulation
Protein-ligand complex (docked pose of top ranked molecule ZINC10600215 and SAHA with PDB ID: 4LXZ) was simulated for 5000 ps (5 ns). Temperature vs. time and total energy vs. time plot obtained after production step are shown in (Fig. 8). Electrostatics of protein-ligand complex was studied by employing spherical cutoff method and it exhibited kinetic energy change from 4332.59 to 4381.51 kcal/mol and potential energy change from −3487.92 to −3585.70 kcal/mol (Fig. 9) and final RMS gradient after simulation was 18.662 (kcal/(mol × Å)).
Trajectory snapshots were collected at every 2 ps during the simulation. The stability of complex, conformational changes and strength of interaction were examined by calculating RMSD, root mean square fluctuation (RMSF), heat map, and hydrogen bond interactions map attained throughout MD trajectories are shown in Fig. 10. The protein backbone residues exhibited average RMSD and RMSF values of 1.73 Å and 0.57 Å, respectively. After MD simulation of protein-ligand complex (top scored molecule ZINC10600215 and SAHA with PDB ID: 4LXZ) exhibited average RMSD value 1.48 Å and 1.46 Å, respectively ((Fig. 10d, e).
a Backbone RMSD of 4LXZ during molecular dynamic simulation; b RMSF values of 4LXZ which represents local changes along the protein chain during molecular dynamic simulation; c RMSD plot of top scored molecule ZINC10600215, 4LXZ backbone and reference compound (SAHA) during MD simulation; d RMSF plot of top scored molecule ZINC10600215, 4LXZ backbone, and reference compound (SAHA), which represents local changes during molecular dynamic simulation; e Map represents h-bond and hydrophobic interactions of ZINC10600215with different residue of enzyme 4LXZ
Top scored molecule ZINC10600215-4LXZ interactions were observed in docking analyses and found stable with Tyr308, His146, and Asp104 during last 0.1 ns simulation. Apart from H-bonds, hydrophobic interaction with Phe210 was reliable during MD simulation (Fig. 10f). The stable ligand–receptor interactions in the course of MD simulation confirmed the results of earlier docking studies.
ADME and toxicity evaluation
ADMET studies were performed for top 5 hits using Qikprop module of Schrodinger v9.3. The results of analysis included various pharmacokinetic parameters required for ADMET and are shown in Table 8. Some important pharmacokinetic parameters like Aq. Solubility, octanol/water partition coefficient, percent human oral absorption, blood-brain-barrier (BBB) penetration, apparent Caco-2 cell permeability, and blockage of HERG K + channels were estimated for top 5 scored molecules. All top five molecules exhibited good octanol/water partition coefficient (QPlogPo/w) values, which were the indication of better absorption and distribution capability of drug. The predicted value of partition coefficient for all compounds ranged between 0.973 and 2.583, which were in acceptable range. Apparent Caco-2 cell permeability (QPPCaco) is an important parameter for understanding the cell permeability. The QPPCaco of top five compounds ranged from 46.454 to 114.046, which was in an acceptable range. All of the screened top molecules exhibited good pharmacokinetic drug-like properties and were within the standard limit for human use.
Conclusion
The present work describes the design of novel scaffold of HDAC inhibitors by combining different in silico methods. The ligand-based pharmacophore was developed using Maestro v9.3. The developed pharmacophore model was validated by enrichment studies using the decoy set and the model exhibited five pharmacophoric features i.e., three hydrogen bond acceptor group, one hydrogen bond donor group, and one aromatic ring. The 3D QSAR model developed using the pharmacophore-based alignment showed high values of regression coefficient for training set (R2) = 0.926 and test (Q2) = 0.699, cross-validated coefficient (rcv2) = 0.967 and R2 pred = 0.6578 with low RMSD = 0.4963. Additionally, the best pharmacophore model (AAADR.12514) was further utilized in virtual screening against the available Zinc database for 3D search query and five top scored molecules were obtained. Finally, the relative binding free energy and binding affinity of hit molecules were confirmed using MM-GBSA technique and MD simulation. The pharmacokinetic properties of all hit molecules were compared with the crystal ligand SAHA. After MD simulation, docked pose of top scored molecules ZINC10600215 exhibited similar binding interactions. It can be concluded that the combination of the 3D QSAR, pharmacophore modeling, virtual screening, MM-GBSA, MD simulation, and ADMET approaches are capable to recognize new hit molecules from chemical databases, which might be a promising lead in the discovery of novel HDAC inhibitors.
References
Biel M, Wascholowski V, Giannis A (2005) Epigenetics—an epicenter of gene regulation: histones and histone-modifying enzymes. Angew Chem Int Ed Engl 44:3186–216
Bieliauskas AV, Weerasinghe SV, Pflum MK (2007) Structural requirements of HDAC inhibitors: SAHA analogs functionalized adjacent to the hydroxamic acid. Bioorg Med Chem Lett 17:2216–9
Binaschi M, Boldetti A, Gianni M, Maggi CA, Gensini M, Bigioni M, Parlani M, Giolitti A, Fratelli M, Valli C, Terao M, Garattini E (2010) Antiproliferative and differentiating activities of a novel series of histone deacetylase inhibitors. ACS Med Chem Lett 1:411–5
Chen Y, Li H, Tang W, Zhu C, Jiang Y, Zou J, Yu Q, You Q (2009) 3D-QSAR studies of HDACs inhibitors using pharmacophore-based alignment. Eur J Med Chem 44:2868–76
Gopalan B, Ponpandian T, Kachhadia V, Bharathimohan K, Vignesh R, Sivasudar V, Narayanan S, Mandar B, Praveen R, Saranya N, Rajagopal S, Rajagopal S (2013) Discovery of adamantane based highly potent HDAC inhibitors. Bioorg Med Chem Lett 23:2532–7
Gray SG, Ekstrom TJ (2001) The human histone deacetylase family. Exp Cell Res 262:75–83
Jin K, Li S, Li X, Zhang J, Xu W, Li X (2015) Design, synthesis and preliminary biological evaluation of indoline-2,3-dione derivatives as novel HDAC inhibitors. Bioorg Med Chem 23:4728–36
Kozikowski AP, Chen Y, Gaysin A, Chen B, D’Annibale MA, Suto CM, Langley BC (2007) Functional differences in epigenetic modulators-superiority of mercaptoacetamide-based histone deacetylase inhibitors relative to hydroxamates in cortical neuron neuroprotection studies. J Med Chem 50:3054–61
Lengauer T, Lemmen C, Rarey M, Zimmermann M (2004) Novel technologies for virtual screening. Drug Discov Today 9:27–34
Mahlknecht U, Hoelzer D (2000) Histone acetylation modifiers in the pathogenesis of malignant disease. Mol Med 6:623–44
Marks PA, Richon VM, Miller T, Kelly WK (2004) Histone deacetylase inhibitors. Adv Cancer Res 91:137–68
Minucci S, Pelicci PG (2006) Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6:38–51
Nair SB, Teli MK, Pradeep H, Rajanikant GK (2012) Computational identification of novel histone deacetylase inhibitors by docking based QSAR. Comput Biol Med 42:697–705
Patel P, Singh A, Patel VK, Jain DK, Veerasamy R, Rajak H (2016) Pharmacophore based 3D-QSAR, virtual screening and docking studies on novel series of HDAC inhibitors with thiophen linker as anticancer agents. Comb Chem High Throughput Screen 19:735–51
Rajak H, Singh A, Raghuwanshi K, Kumar R, Dewangan PK, Veerasamy R, Sharma PC, Dixit A, Mishra P (2014) A structural insight into hydroxamic acid based histone deacetylase inhibitors for the presence of anticancer activity. Curr Med Chem 21:2642–64
Sallam AA, Houssen WE, Gissendanner CR, Orabi KY, Foudah AI, El Sayed KA (2013) Bioguided discovery and pharmacophore modeling of the mycotoxic indole diterpene alkaloids penitrems as breast cancer proliferation, migration, and invasion inhibitors. MedChemComm 4:1360–9
Schafer S, Saunders L, Eliseeva E, Velena A, Jung M, Schwienhorst A, Strasser A, Dickmanns A, Ficner R, Schlimme S, Sippl W, Verdin E, Jung M (2008) Phenylalanine-containing hydroxamic acids as selective inhibitors of class IIb histone deacetylases (HDACs). Bioorg Med Chem 16:2011–33
Schrödinger (2012) Phase, version 3.4, schrodinger, LLC, New York
Siegel RL, Miller KD, Jemal A (2017) Cancer Statistics, 2017. CA Cancer J Clin 67:7–30
Singh A, Patel P, Jageshwar, Patel VK, Jain DK, Kamal M and Rajak H (2017a) The safety, efficacy and therapeutic potential of histone deacetylase inhibitors with special reference to panobinostat in gastrointestinal tumors: a review of preclinical and clinical studies. Curr Cancer Drug Targets https://doi.org/10.2174/1568009617666170630124643.
Singh A, Patel P, Patel VK, Jain DK, Veerasamy R, Sharma PC, Rajak H (2017b) Histone deacetylase inhibitors for the treatment of colorectal cancer: recent progress and future prospects. Curr Cancer Drug Targets 17:456–66
Singh A, Patel VK, Jain DK, Patel P, Rajak H (2016) Panobinostat as pan-deacetylase inhibitor for the treatment of pancreatic cancer: recent progress and future prospects. Oncol Ther 4:73–89
Su H, Yu L, Nebbioso A, Carafa V, Chen Y, Altucci L, You Q (2009) Novel N-hydroxybenzamide-based HDAC inhibitors with branched CAP group. Bioorg Med Chem Lett 19:6284–8
Suzuki T, Miyata N (2005) Non-hydroxamate histone deacetylase inhibitors. Curr Med Chem 12:2867–80
Taunton J, Hassig CA, Schreiber SL (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408–11
Teli MK, Rajanikant GK (2012) Pharmacophore generation and atom-based 3D-QSAR of novel quinoline-3-carbonitrile derivatives as Tpl2 kinase inhibitors. J Enzym Inhib Med Chem 27:558–70
Truchon JF, Bayly CI (2007) Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J Chem Inf Model 47:488–508
Wang X, Li X, Li J, Hou J, Qu Y, Yu C, He F, Xu W, Wu J (2017) Design, synthesis, and preliminary bioactivity evaluation of N(1) -hydroxyterephthalamide derivatives with indole cap as novel histone deacetylase inhibitors. Chem Biol Drug Des 89:38–46
Watts KS, Dalal P, Murphy RB, Sherman W, Friesner RA, Shelley JC (2010) ConfGen: a conformational search method for efficient generation of bioactive conformers. J Chem Inf Model 50:534–46
Wu Q, Sharma S, Cui H, LeBlanc SE, Zhang H, Muthuswami R, Nickerson JA, Imbalzano AN (2016) Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget 7:27158–75
York DM, Darden TA, Pedersen LG, Anderson MW (1993) Molecular dynamics simulation of HIV-1 protease in a crystalline environment and in solution. Biochemistry 32:1443–1453
Yoshida M, Kijima M, Akita M, Beppu T (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265:17174–9
Zhu Y, Li HF, Lu S, Zheng YX, Wu Z, Tang WF, Zhou X, Lu T (2010) Investigation on the isoform selectivity of histone deacetylase inhibitors using chemical feature based pharmacophore and docking approaches. Eur J Med Chem 45:1777–91
Acknowledgements
PP is thankful to University Grants Commission (UGC), New Delhi for providing Junior Research Fellowship (JRF). HR is highly grateful to Department of Science and Technology-Science and Engineering Research Board (DST-SERB), New Delhi, India for providing financial assistance in the form of ‘Start up Grant for Young Scientist’ (Grant No. SERB/LS-385/2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Patel, P., Rajak, H. Development of hydroxamic acid derivatives as anticancer agent with the application of 3D-QSAR, docking and molecular dynamics simulations studies. Med Chem Res 27, 2100–2115 (2018). https://doi.org/10.1007/s00044-018-2219-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-018-2219-4