Abstract
Cells that migrate away from a central tumour into brain tissue are responsible for inefficient glioblastoma treatment. This migratory behaviour depends partially on lysosomal cysteine cathepsins. Reportedly, the expression of cathepsins B, L and S gradually increases in the progression from benign astrocytoma to the malignant glioblastoma, although their specific roles in glioma progression have not been revealed. The aim of this study was to clarify their specific contribution to glioblastoma cell invasion. The differences between the matrix invading cells and non-invading core cells from spheroids derived from glioblastoma cell culture and from glioblastoma patients’ biopsies, and embedded in type I collagen, have been studied at the mRNA, protein and cathepsin activity levels. Analyses of the two types of cells showed that the three cathepsins were up-regulated post-translationally, their specific activities increasing in the invading cells. The cystatin levels were also differentially altered, resulting in higher ratio of cathepsins B and L to stefin B in the invading cells. However, using specific synthetic inhibitors and silencing strategies revealed that only cathepsin B activity was involved in the invasion of glioblastoma cells, confirming previous notion of cathepsin B as tumour invasiveness biomarker. Our data support the concept of specific roles of cysteine cathepsins in cancer progression. Finally the study points out on the complexity of protease regulation and the need to include functional proteomics in the systems biology approaches to understand the processes associated with glioma invasion and progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiformae (GBM) is the most common and most malignant form of brain tumour in adult patients, with a mean life expectancy of about 18 months after first diagnosis [1]. GBMs are characterized by rapid cell proliferation, a high level of single-cell invasiveness into the surrounding brain and increased vascularity. This leads to renewed tumour growth and poor prognosis [2].
The invasive nature of GBM leads to local destruction and dissolution of the adjacent normal tissue, associated with increased focal degradation of underlying extracellular matrix creating tracks for cells to migrate on [3, 4]. The invasion process involves the expression of numerous genes that have been found altered in the invasive vs. non-invasive carcinoma [5] and in other cancers [6]. In GBM, the increased levels of cell motility gene expression and down-regulation of proaptoptiotic and proliferation gene transcription in the invasive cell populations was first found by Mariani et al. [7]. Recently, two signatures of “migrating” and “stationary” glioma cells and a gene signature, capable of classifying glioma cultures based on their migration rate were revealed [8]. The increasingly migratory phenotype is induced by the extracellular matrix during the ECM invasion in vitro [9] and in vivo by the brain microenvironment [10].
Increased expression and activation of proteolytic enzymes at the leading edges of tumours and in invadopodia have been reported, involving podosome formation in the migrating cells [11, 12]. It is well documented that proteolytic enzymes are up-regulated at protein and activity levels in almost all types of cancer, although their various roles may be pro- or anti-tumorigenic [13]. Cysteine cathepsins (Cats) B, L and S, aspartic CatD, plasminogen activator-plasmin system and various clans of matrix metalloproteases have been shown to play a role in GBM invasion, and some were demonstrated to have a prognostic impact for patient survival [3, 14, 15]. Cysteine cathepsins comprise the largest family of lysosomal enzymes with 11 proteases, structurally grouped in CatB-like and CatL-like enzymes (http://www.merops.ec.uk). Increased levels of CatB mRNA in glioblastoma were first observed by Rempel et al [16]. Later, we confirmed that CatB expression in glioma progression is gradually increased, [17] whereas in clinical studies we reported that CatB in the tumour and endothelial cells had an impact on patient survival [18, 19]. Collectively, these studies confirmed that CatB is particularly important for glioma invasion and progression. CatL also correlated with glioma progression [20] but no prognostic impact was found, neither was it expressed in the endothelial cells [19]. CatS has been studied in GBMs mostly by Flannery et al. [21, 22] who recently demonstrated that highly expressed CatS protein in WHO grade IV brain tumours was an independent predictor of survival in multivariate analysis. However, the roles of both cathepsins L and S in gliomas are still not clear.
The ultimate regulation of the cysteine cathepsins is by binding to their inhibitors, called cystatins. This superfamily comprises several families, of which the cystatin family comprises the extracellular cystatins, and intracellular stefins, all these playing a role in cancer progression [23]. In brain tumours, stefin A mRNA was detected in benign, but not in malignant tissues, indicating possible downregulation of this inhibitor [17]. Both stefin A and B proteins were only faintly immunolabelled in glioma tumour sections [19]. In contrast, cystatin C is quite an abundant protein in cerebrospinal fluid [24], but no clear association of cystatin C with glioma progression has been found [25]. Interestingly, multivariate analysis including cystatin C demonstrated a highly significant prognostic impact of low cystatin C expression on shorter disease-free survival of glioma patients [26] and low cystatin C expression was connected to high glioma invasiveness in vitro [27]. The final effectiveness of the cysteine cathepsins is therefore defined by their enzymatic activity, which is not only regulated by the levels of the enzymes and their endogenous inhibitory proteins, but by alterations in their localization.
In this study, we first aimed to define the abundance of CatB, CatL and CatS at all expression levels in invasive versus non-invasive cells of GBM spheroids in the in vitro invasion assay. Secondly, we aimed to establish the balance between the cathepsins and cystatins, possibly affecting the cathepsins’ activities. Finally, we interfered with this natural balance by selectively inhibiting cysteine cathepsins by synthetic inhibitors, in order to define their contribution to U87-MG cell invasion as well as to the invasion of primary GBM cells migrating away from the tumour-derived spheroids of GBM patients.
Materials and Methods
Cell Culture and Spheroid Formation
Human glioblastoma cell line U87-MG was obtained from ATCC (Manassas VA, USA) and SNB-19 human glioblastoma cell line was obtained from DSMZ (Braunschweig, Germany). Both cell lines were cultured in DMEM medium (Sigma-Aldrich, Steinheim, Germany) supplemented with 10% foetal bovine serum (FBS), 1% non-essential amino acids, 1% penicillin/streptomycin, 2.5 mM L-glutamine (all PAA Laboratories, Linz, Austria).
Cell culture-derived spheroids were prepared using the hanging drop technique. 2000 cells (U87-MG or SNB-19) were grown in 20 µL culture medium drops for 2 days (3 days for SNB-19) on the lid of a water-filled culture dish. The multi-cellular aggregates that formed were then transferred to agar-coated wells on a 96-well plate for 1 day, when spheroids of 400–500 μm in diameter formed. All cells and spheroids were cultured under standard conditions at 37°C in humidified atmosphere with 5% CO2.
Patients and Tumour Samples
The patients were operated on at the Department of Neurosurgery, University Clinical Centre of Ljubljana. Five were diagnosed as WHO grade IV glioblastoma (GBM) and one as WHO grade II pleomorphic xanthoastrocytoma (PXA) by established histopathology protocols. Tumour sample data are shown in Table 1. This study was approved by the National Medical Ethics Committee of the Republic of Slovenia.
Within 60 minutes after surgical removal the tumour samples were transported on ice to the cell-culture laboratory. There, they were immediately cut into <0.5 mm pieces. Tumour-derived spheroids were obtained by selecting very small pieces and placing them in agar-coated wells of a 96-well plate filled with primary culture medium (DMEM medium supplemented with 10% FBS, 1% penicillin/streptomycin, 2.5 mM L-glutamine, 10 mM HEPES). Spheroids formed after 4–19 days of incubation and those of 300–600 μm in diameter were selected for further experiments.
The tumour-derived spheroids were cultured under standard conditions at 37°C in a humidified atmosphere with 5% CO2.
3D-Matrix Invasion Assay and Separation of Invading and Non-Invading Cell Populations
Individual spheroids were embedded in 50 µL drops of the selected matrix — 1.0 mg/mL rat tail collagen type I or 1.0 mg/mL Matrigel (both BD Bioscience, Bedford MA, USA). After incubating for 30–45 minutes at 37°C the matrix was covered with cell culture medium. In the experiments for comparing expression of cathepsins and inhibitors at mRNA and activity levels in spheroids invading collagen or Matrigel, higher concentrations of matrices (3.0 mg/mL) were used. The spheroid diameter and cell invasion distance were measured under a light microscope using an ocular micrometer. Invasion distance was defined as the distance from the edge of the spheroid to the population of the cells most distant from the spheroid. The invasion was monitored for up to 7 days.
Invading and non-invading cells were separated by washing the invasion assays twice in 1 × PBS. Using a fine needle the central spheroids (i.e. non-invading cells) were cut out of the matrix under the light microscope and aspirated into separate tubes, thus separating them from the cells invading the matrix. The separated cell populations were then processed separately for RNA and protein isolation as described below.
RNA Isolation and Reverse Transcription, Quantitative Real-Time-PCR
RNA was isolated with TRIzol reagent (GIBCO Products, Invitrogen, Grand Island NY, USA) added directly to the separated cell populations, without removal of the matrix, as described by the manufacturer, and stored at -80°C until further use. RNA was quantified with Quant-iT RiboGreen RNA Kit (Molecular Probes, Invitrogen, Eugene OR, USA) and 1.0 μg of each RNA sample was reverse transcribed to cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City CA, USA). All the procedures were performed according to manufacturers’ protocols.
Quantitative real-time-PCR assays were performed on ABI Prism 7900 HT Sequence Detection System using TaqMan Universal PCR Master Mix. Human GAPDH was used as internal control (all Applied Biosystems). Sequences of forward primers, reverse primers and probes (in this order) used for detecting gene-specific mRNA were as followed: for CatB: 5’-CTC TATg AAT CCC ATg Tag ggT gC-3’, 5’-CCT gTT TgT Agg TCg ggC Tg-3’ and 5’-CCC TgT gAg CAC CAC gTC AAC gg-3’; for CatL: 5’-TCA ggA ATA Cag ggA Agg gAA A-3’, 5’-TCC Tgg gCT TAC ggT TTT gA-3’ and 5’-CAC Tgg TCA TgT CTC CAA Agg CgT TCA T-3’; for StefA: 5’-ggA ggC TTA TCT gAg gCC AAA-3’, 5’-CAA gCT gTg gTT TAA CCT TAT CAA CA-3’ and 5’-CCg CCA CTC CAg AAA TCC Agg AgA-3’; for StefB: 5’-gCC gAg ACC CAg CAC ATC-3, 5’-ggC CTT AAA CAC Agg gAA CTT CT-3 and 5’-ACC Agg TgA ggT CCC AgC TTg AAg AgA-3; for CysC: 5’-gAC AAC TgC CCC TTC CAT gA-3, 5’-gCA CAg CgT AAA TCT ggA AAg A-3 and 5’-CAg CCA CAT CTg AAA Agg AAA gCA TTC Tg-3. All the probes were 5’-FAM 3’-TAMRA modified. For CatS, Hs00175403_m1 TaqMan Gene Expression Assay (Applied Biosystems) was used.
The mRNA data were calculated as fold differences in expression levels (F) between distinguished cell populations or samples, as in Demuth et al. [8]; F = 2(CPIh - CPIg) - (CPIIh - CPIIg), where CPI is the first, and CPII the second cell population/sample, h the Ct value of the internal control (i.e. GAPDH) and g the Ct value of the gene of interest. F ≥ 1.50 means up-regulation / high expression and F ≤ 0.75 down-regulation / lower expression of the selected gene in the first cell population / sample. F values between 1.50 and 0.75 are regarded as non-significant differences.
Protein Extraction
Samples were washed with 1 × PBS, treated with 25 CDU/mL of collagenase I (Sigma-Aldrich) for 45–60 min at 37°C to degrade the collagen then washed in 1 × PBS again. The collagenase step was omitted with the samples used for activity assays, as it damaged the cathepsins’ activities. Cells were then homogenized by 2 min sonication in 50 mM Tris buffer, pH 6.9, supplemented with 0.05% (V/V) Brij 35, 0.5 mM dithiothreitol, 5 mM EDTA, 0.5 mM paramethylsulphonyl fluoride and 10 mM pepstatin A (all Sigma-Aldrich). The homogenates were centrifuged at 12.000 × g for 30 min at 4°C. The supernatants were aliquoted and stored at -80°C until used.
ELISA Tests
Total protein was quantitfied using the Bio-Rad protein assay (Bio-Rad Labs, Munich, Germany) according to the manufacturer’s protocol.
Human CatB, CatL, CatS, StefA, StefB and CysC sandwich ELISA kits were used as suggested by the manufacturer (provided by Janko Kos, Faculty of Pharmacy, University of Ljubljana, Ljubljana, Slovenia). The protein samples were diluted to 20 mg protein/mL for CatB, StefB, CysC ELISA tests and to 80 mgprotein/mL for CatL, CatS and StefA ELISA tests. Rabbit anti-human CatB polyclonal antibodies (pAb), sheep anti-human CatL pAb, murine anti-human CatS monoclonal antibodies (mAb), mouse anti-human StefA mAb, mouse anti-human StefB mAb and rabbit anti-human CysC mAb were used as capture antibodies. The detection antibodies: sheep anti-human CatB pAb, sheep anti-human CatL pAb, murine anti-human CatS mAb, mouse anti-human StefA mAb, mouse anti-human StefB mAb and mouse anti-human Cys mAb were all horseradish peroxidase conjugated. Purified native human CatB, CatL, CatS, StefA, StefB and CysC were used as the standards. Minimum detection limits of the tests were: 0.03 nmol/L for CatB, 0.06 nmol/L for CatL, 0.03 nmol/L for CatS, 0.09 nmol/L for StefA, 0.09 nmol/L for StefB and 0.02 nmol/L for CysC. The tests detect both precursor and active forms of the cathepsins as well as enzyme-inhibitor complexes.
Enzyme Activity Assays
Cat B and CatL activities were measured as described in Bervar et al. [28] and Zajc et al. [29] respectively, with slight modifications. Duplicates of water diluted protein samples were supplemented with activation buffer (0.4 M phosphate buffer pH 6.0, 2.5 mM fresh DTE for CatB; 0.34 M acetate buffer, pH 4.2, 2.0 mM fresh DTE for CatL; all Sigma-Aldrich) and incubated 30 min, 37°C to allow full activation of the cathepsins. To measure specific cathepsin activity water was added to one of the duplicates and specific inhibitor (60 μM Ca-074, Peptide Institute, Osaka, Japan, for CatB; 2 μM Clik 148, provided by N. Katunuma, Tokushima Bunri University, Tokyo, Japan for CatL) to the other. Activity buffer (0.4 M phosphate buffer pH 6.0, 2.5 mM fresh DTE for CatB; 0.34 M acetate buffer pH 5.5, fresh 2.5 mM DTE for CatL; all Sigma-Aldrich) was then added and the reaction started by adding specific substrate (100 µM Z-RR-AMC for CatB, 100 µM Z-FR-AMC for CatL; both Bachem, Bubendorf, Switzerland). After 90 min at 37°C the reaction was stopped with 1 mM iodoacetic acid and the released 7-AMC measured on a spectrofluorimeter (Tecan, Groedig, Austria). Specific cathepsin activity was calculated as the difference in 7-AMC release in presence and absence of the specific cathepsin inhibitor.
CatS activity was measured as described by Flannery et al. [21] Water diluted protein samples were supplemented with inactivation buffer (100 mM phosphate buffer, pH 7.5) for 60 min at 37°C to fully inactivate CatB and CatL, which might otherwise affect the measurements. The pH value was then returned to 6.0 using 500 mM MES buffer. Reaction buffer (200 mM MES, 200 mM EDTA, pH 6.0, fresh 1 mM DTT, all Sigma-Aldrich) was added and the reaction started by adding specific substrate (100 µM Z-VVR-AMC, Peptide Institute). After 90 min at 37°C the reaction was stopped with 100 mM acetate buffer, pH 4.3 and the released 7-AMC measured on a spectrofluorimeter.
Each activity assay was performed in triplicate, and controls with omitted sample (blanks) were performed. Specific activities were expressed in enzyme units (EU) per μg of total DNA (determined by PicoGreen dsDNA Quantitation Kit, Molecular Probes according to manufacturer’s protocols), with one EU being the amount of the enzyme releasing 1 nM of 7-AMC per minute.
Inhibition of Spheroid Invasion Using Chemical Inhibitors
3-D collagen matrix invasion assays were set up as described earlier in Methods. The specific inhibitors were added in non-toxic concentrations to the collagen matrix and to the cell culture media. The media were changed daily and invasion followed for 3 days. The cell-membrane soluble inhibitors were used in the following final concentrations: for CatB, 1.0 μM Ca074-Me (Peptide Institute, Osaka, Japan), for CatL, 0.5 μM Clik 148 (provided by N. Katunuma, Tokushima Bunri University, Tokyo, Japan) and for CatS, 1.0 μM Z-FL-COCHO (Calbiochem, San Diego CA, USA).
Toxicity Assay
The non-toxic concentrations of synthetic inhibitors were established by MTT assay in monolayer culture and confirmed by clonogenicity testing in spheroids.
For MTT assay, U87-MG cells were harvested at 80% confluence and plated to a 96-well plate. The specific inhibitors were added in test concentrations to the culture media. Cells were incubated for 21 h at 37°C, 5%CO2, then MTT (1-(4,5 dimethyltiazol-2-yl)-2,5 diphenyl tetrazolium bromide, Sigma-Aldrich) was added to 0.5 mg/mL final concentration. After an additional 3 h at 37°C, the formazan crystals (corresponding to cell viability) were pelleted by centrifugation and dissolved in dimethyl sulfoxide. Absorbance was measured at 570 nm (reference filter 690 nm) on a spectrofluorimeter (Tecan). The tested inhibitor concentration was considered non-toxic if cell viability was ≥ 95% of the control value.
For clonogenicity testing 3-D collagen matrix invasion assays were set up as described above. Specific inhibitors were added in test concentrations to the collagen matrix and to the cell culture media. The media were changed daily and invasion followed for 3 days. The spheroids in collagen were then washed 3-times in 1 × PBS and the collagen degraded using 25 CDU/mL of collagenase I (Sigma-Aldrich). Spheroids were degraded to single cell suspensions using 0.25% trypsin / 2 mM EDTA (Sigma-Aldrich). 2000 cells were seeded on 60 mm tissue culture treated Petri dishes and incubated at 37°C, 5% CO2 for 14 days. Cultures were then washed with 1 × PBS, fixed and stained with 2.0 g crystal violet / 400 mL 95% ethanol. The number of colonies formed (>50 cells) was counted. Plating efficiency was calculated for control and treated spheroids and survival % compared to the control established. The tested inhibitor concentration was considered non-toxic if survival was ≥ 95% that of the control.
CatB Silencing
U87-MG cells in monolayer were transfected with the CatB si-RNA with sense: r(ggA UCA CUg Cgg AAU CgA A)dTdT, antisense: r(UUC gAU UCC gCA gUg AUC C)dTdg, and target sequence: CAg gAT CAC TgC ggA ATC gAA; using the RNAiFect trasfection reagent (all Qiagen, Hilden, Germany), according to the manufacturer's instructions. The non-si RNA with sense: UUC UCC gAA CgU gUC ACg UdT dT; antisense: ACg UgA CAC gUU Cgg AgA AdT dT and target sequence: AAT TCT CCg AAC gTg TCA CgT (Qiagen) was used as an empty vector control. After 1 day in culture the transfected cells were trypsinized and 20 μL hanging drops with 2000 cells prepared. After 2 days incubation the cell aggregates were transferred to agar-coated wells for 1 additional day to obtain the spheroids.
Immunohistochemistry
After 3 days of invasion the U87-MG spheroids in 1.0 mg/mL type I collagen were washed in PBS, fixed for 3 h in formalin and dehydrated in a series of 50%, 80%, 96% and absolute ethanol, 30 min each. They were washed in xylene twice for 30 min and finally embedded in paraffin. Paraffin blocks were cut on a microtome to 5 μm sections, mounted on cover glass and dried overnight. Series of every 10th section were haematoxylin & eosin stained and checked under the light microscope to select high quality sections for immuno-fluorescence staining.
To prepare for immuno fluorescence staining, the sections were de-waxed in xylene and rehydrated in an inverted ethanol series to tap water. Sections were boiled 3 × 6 min in 10 mM sodium citrate buffer (pH 6.0) for antigen retrieval. Non-specific staining was blocked with 10%FBS 1%BSA for 2 h. Primary antibodies used were monoclonal mouse anti-human CatB IgG (clone 3E1, 65.1 μg/mL), monoclonal mouse anti-human CatL IgG (clone N135, 153.0 μg/mL), monoclonal mouse anti-human CatS IgG (clone 1E3, 100.0 μg/mL) and monoclonal mouse anti-human StefB IgG (clone A6/2, 110.0 μg/mL), all provided by Janko Kos, Faculty of Pharmacy, University of Ljubljana, Ljubljana, Slovenia. Sections were incubated with primary antibodies overnight at 4°C. The secondary layer of goat anti-mouse IgG Alexa Fluor 488 (1:200, Molecular Probes, Eugene OR, USA) was applied for 1 h at room temperature in the dark. The sections were then mounted with ProLong Gold Antifade Reagent (Molecular Probes) and stored at -20°C in the dark until use. Negative controls with omitted primary antibodies and blank (no antibody) sections were also performed. The green fluorescence staining was observed under the Zeiss LSM 510 confocal microscope, using the 63× oil objective. To visualize the cells, DIC pictures of the same areas were superimposed.
Statistical Analysis
All the statistical analysis was performed with Excel 2002 (Microsoft Corp., Redmond WA, USA). The statistical significances of the differences observed were calculated as standard t-test with assumed two-tailed distribution and unequal variance. The correlations between average fold differences in cathepsins’ activity and average invasion distances were calculated with Prism 5.01 (GraphPad Software Inc., San Diego CA, USA) as Spearman’s non-parametric correlations.
Results
Spheroid Invasion into Collagen and Matrigel
Two types of spheroids were used in this study: those prepared from glioma tumour samples (henceforth tumour-derived spheroids) and those prepared from established GBM cell lines, U87-MG and SNB19. The spheroids were embedded in collagen type I matrix and in Matrigel (both at 1.0 mg/mL concentration). Invasion distances were measured after 1, 3 and 7 days of invasion into protein matrices, as described in Materials and Methods. Cells from U87-MG spheroids were the most invasive, invading more than two-fold faster than the cells from any of the tumour-derived spheroids (Fig. 1a). The difference in invasion efficiency in collagen ranged around 3.5 fold between the most and the least invasive glioma samples. Pleomorphic xanthoastrocytoma (WHO grade II, PXA 061108, see Table 1) also showed low invasive potential. Both U87-MG and SNB19 derived spheroids invaded 30–50% faster in type I collagen than in the Matrigel at all time points measured (Fig. 1b, c). Collagen was therefore used in further in vitro invasion experiments.
Spheroid invasion in a 3D-invasion model. a Comparison of spheroids from different origins – spheroids from U87-MG cell line and tumour-derived spheroids (Ο U87-MG, GBM 050915, Δ GBM 080110, ♦ GBM 080107, ■ GBM 070103, ▲ GBM 061123, ◊ PXA 061108). Maximum invasion distances after 1, 3 and 7 days in 1.0 mg/mL collagen type I matrix. b Spheroids from U87-MG and (c) spheroids from SNB-19 cell lines in different matrices- 1.0 mg/mL collagen type I and 1.0 mg/mL Matrigel. Invasion distances in Matrigel (interrupted line) were significantly lower (* p < 0.05, ** p < 0.001) than in collagen (full line) at all time points
The Effect of Extracellular Matrices on Expression of Cathepsins and Cystatins
mRNA Levels
The expression levels of cathepsins and cystatins in U87-MG and SNB19 spheroids embedded in higher concentrations (3.0 mg/mL) of collagen I and Matrigel were compared to assess the possible effect of matrix on their expression at mRNA levels. These data are represented as fold differences (F) between the spheroids in the two matrices, with F ≥ 1.50 meaning up-regulation / higher expression and F ≤ 0.75 down-regulation / lower expression in collagen matrix (shown in Table 2). Significantly higher CatB expression was observed in both U87-MG and SNB19 derived spheroids (F = 1.52 and 1.74, respectively), embedded for 3 days in type I collagen relative to that in Matrigel. CatS expression in U87-MG spheroids in collagen was also higher (F = 1.53), whereas StefA expression was lower in SNB19 spheroids in collagen than in Matrigel (F = 0.36). No significant differences were observed for CatL, StefB and CysC mRNA expression in different matrices.
Cathepsin Activity Levels
Activity levels of CatB in U87-MG spheroids invading different matrices for 3 days, were significantly (p < 0.05) higher by 4.3 fold in Matrigel and 8.4 fold in collagen than in freely floating spheroids (Fig. 3a). No significant differences were found for either CatL or CatS activity. Comparing the spheroids in both types of matrix, CatB and CatS activities were significantly lower (49% and 73%, respectively; p < 0.05) in Matrigel than in collagen invading spheroids.
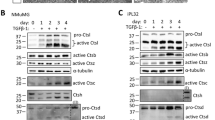
Protein levels of cathepsins and cystatins. Protein levels were determined in the cell extracts by ELISA, as described in Material and Methods and presented in the non-invading (central spheroid) and invading cells (outer rim of cells invading the matrix) from U87-MG spheroids after 3 days in type I collagen. Trends to increased CatL levels (grey columns) and decreased StefB levels (black columns) can be observed in the invading cells. No change was observed for CatB (white columns). CatS, StefA and CysC protein levels were below the detection limit of the ELISA
Activities of cathepsins. a Comparison of freely floating and embedded U87-MG spheroids. 3.0 mg/mL Matrigel matrix (grey columns) or 3.0 mg/mL type I collagen (black columns) were used. CatB activity was significantly higher (* p < 0.05) in the Matrigel and collagen embedded spheroids than in freely floating spheroids (white columns). Comparison of CatB and CatS activities in the spheroids embedded in different matrices shows, that these were significantly higher (+ p < 0.05) in the collagen than in the Matrigel. There were no significant differences in CatL activities. b–d Comparison of invading cells (black columns) and non-invading cells (white columns) in collagen embedded U87-MG spheroids. CatB b and c CatL activities were significantly higher in the invading cells than in the non-invading cells after 1, 3 and 7 days in collagen (* p < 0.05, ** p < 0.001). d While the trend to the increased CatS activity remained high at all time points, it was significantly increased only at day 1. e Differences between U87-MG and tumour-derived spheroids. Average fold differences (F) in cathepsin activities (in invading cells vs. non-invading cells) in U87-MG and tumour-derived spheroids after 7 days in collagen are shown. For comparison, the corresponding invasion distances after 7 days in collagen are indicated (short lines). Fold differences in CatB (black dots), but not CatL (white dots) or CatS (grey dots) activity correlated significantly with the invasion distances (r = 0.83, p < 0.05)
When comparing mRNA and activity levels, there was a good correlation between RNA and activity up-regulation in GBM cells in the presence of collagen matrix for CatB and CatS, whereas CatL mRNA and activity level were not affected by this matrix.
Expression of Cathepsins and Cystatins in Invading Cells vs. Non-Invading Cells
mRNA Levels
Invading cells were separated from the bulk of the spheroid, i.e. the cells not invading the matrix after 1, 3 and 7 days of invasion into the collagen matrix, as described in Materials and Methods. Total mRNA and total proteins were extracted and the levels of cathepsins and cystatins were measured.
For U87-MG spheroids and tumour-derived spheroids invading the collagen matrix, fold differences (F) in mRNA expression between the invading and non-invading cells were calculated. In U87-MG spheroids (Table 3) CatB and StefA were up-regulated in the invading cells at the first day of invasion (F = 1.50 and 1.56, respectively), while no significant differences were observed for other proteins. The expression of StefA was further up-regulated at day 3 (F = 2.53) but not after 7 days. CatS was also significantly up-regulated (F = 16.0) at day 3 and further (F = 11.2) at day 7.
When comparing tumour-derived spheroids from the one WHO II sample and five GBM samples (Table 4), no consistent pattern was observed in a single gene expression. For example, CatB was down regulated in the invading cells of two tumour samples, unchanged in two samples and up-regulated in two samples, once even to an extremely high level (F = 808). Similarly, CatL was up-regulated twice, not changed in the invading cells of three samples and down-regulated in one sample. Also, no consistent patterns in the cathepsin and cystatin levels were found in the invading cells among the different tumour-derived spheroids.
Protein Levels
Only CatB, CatL and StefB were present at the protein level in detectable quantities in U87-MG spheroids in collagen after 3 days of invasion (Fig. 2). CatS, StefA and CysC were below the detection limits of the ELISA tests used. Due to high variability in the measurements, no significant differences could be measured in any of the proteins although trends to high CatL increase (~70%) and StefB decrease (~50%) were observed in the invading cells compared to non-invading cells. However, molar ratios of CatB:StefB and of CatL:StefB changed greatly in favour of the cathepsins in the invading cells, from 2.7 to 4.2 for CatB and from 0.4 to 1.5 for CatL.
Low amounts of protein material obtained from the invading cells in the tumour-derived spheroids did not allow the comparisons at the protein level in those samples.
Activities
Comparing the invading and non-invading cells from U87-MG spheroids embedded in collagen matrix, CatB and CatL activities were significantly higher (4–7 fold and 9–14 fold, respectively; p < 0.05) in the former than in the latter at all time-points measured (Figs. 3b, c). For CatS the differences were also high (2–7 fold), however significant only after the first day (Fig. 3d). In general, the specific activities of CatB and CatS were comparable, whereas CatL activities were on average 3–10 times lower.
With the tumour-derived GBM spheroids average fold differences in activity between non-invading and invading cells for U87-MG and tumour-derived spheroids in collagen matrix were expressed relative to the corresponding average invasion distances in the same samples (Fig. 3e). Strong correlation with increased CatB activity in the invading cells was found (r = 0.83, p < 0.05). No such correlations were found for CatL or CatS activities. In general, all the cathepsin activities in the invading cells were higher than in the non-invading cells, however, due to high inter-sample variability, most of these differences were insignificant (data not shown).
Inhibition of Invasion of U87-MG Cell Lines and in Tumour-Derived GBM Cells
The synthetic inhibitors, CA074-Me for CatB, Clik 148 for CatL and Z-FFL-COCHO, were added to the U87-MG spheroid culture at low, non-toxic concentrations at which the inhibitors are selective for CatB, CatL and CatS, respectively. Their toxicity, i.e. the effect on cell viability was tested using MTT and clonogenity assays (see Materials and Methods). Only the CatB inhibitor significantly inhibited (~30%, p < 0.001) invasion into collagen (Fig. 4a), while CatL and CatS inhibitors had no effect on invasion.
Inhibition of U87-MG spheroid invasion in collagen. a Non- toxic concentrations of specific CatB (1.0 μM Ca074-Me, ●), CatL (0.5 μM Clik 148, ♦) and CatS (1.0 μM Z-FL-COCHO, ▲) synthetic inhibitors were used. Only CatB inhibition significantly decreased invasion distances (** p < 0.001) compared to the non-treated control (`). b The invasion of U87-MGsiCatB clone with silenced CatB (black columns) was significantly lower than in wild-type U87-MG cells (white columns) and empty vector controle U87-MGsiv (grey columns), (* p < 0.05)
To confirm this result we transiently down-regulated CatB expression by silencing it in U87-MG cells. This was shown to down regulate CatB mRNA up to 50% (data not shown). After 1 and 2 days of invasion the invasion of U87-MGsiCatB spheroids with silenced CatB into collagen matrix was significantly lower than that of empty-vector control U87-MGsiv and wild type U87-MG spheroids (Fig. 4b).
Localization of Cathepsins and Cystatins in the Non-Invading and Invading Cells
Paraffin sections of U87-MG spheroids were prepared after 3 days of invasion in collagen and their morphology examined by haematoxylin & eosin staining. Sections where spheroids were cut through the core and where invading single cells were observed were selected for immuno-fluorescence staining of cathepsins and stefins. Lower cell density was observed in the spheroid core than in the highly density outer rim (Fig. 5a). Only cell surface-bound cathepsins were stained. The staining of CatB (Fig. 5b) was more intense than that of CatL and CatS (Fig. 5c, d respectively). No staining of StefB was observed (data not shown). The negative controls were also performed (Figs. 5e, f).
Paraffin sections of U87-MG spheroids in collagen. a Haematoxylin & eosin staining. In spheroid core lower cell density compared to the outer rim was observed. Single cells were invading from the spheroid. b–f Cell surface immunofluorescence staining of cathepsins. Paraffin sections of spheroids were incubated with primary mouse anti-human CatB, CatL and CatS antibodies overnight at 4°C. Goat anti-mouse Alexa Fluor 488 antibodies were used as secondary layer (see Materials & Methods). Localization of cathepsins is indicated by green fluorescence and the DIC image of the same areas is superimposed to visualize the spheroids. b Cell surface bound CatB staining was the most intense. c CatL and d CatS staining were much weaker. Negative controls: e secondary antibody only and f no antibodies- blank
All three cathepsins were distributed more or less evenly on the surface of non-invading core cells as opposed to matrix invading cells (Fig. 6a, c, e). In the invading cells, CatB expression was focused mainly at the progressive front of the cells (Fig. 6b). Similarly, CatL was focused mainly but not entirely at the progressive front of the invading cells (Fig. 6d). No such focus could be observed for CatS (Fig. 6f).
Cell surface immunofluorescence staining of cathepsins in non-invading and invading U87-MG cells in collagen. Paraffin sections of spheroids were incubated with primary mouse anti-human CatB, CatL and CatS antibodies overnight at 4°C. Goat anti-mouse Alexa Fluor 488 antibodies were used as secondary layer (see Materials & Methods). a CatB was expressed all around the surface of the non-invading cells. b Focus of CatB expression mostly at the progressing front of the invading cells could be observed. c Similarly, CatL was expressed all around the non-invading cells, while d partial focus to the progresing front of the invading cells could be observed. e No focus could be observed in either non-invading or f invading cells for CatS. The direction of invasion is indicated by yellow arrows (i.e. away from the spheroid)
Discussion
The genetic heterogeneity and the invasive behaviour of GBM cells prevent all therapeutic approaches from being successful [1]. Thus there is a need to find new targets to design specific anti-invasive drugs that could be used in adjuvant post- or pre-operative therapeutic regimens.
In this study, we have focused on the role of the three most investigated cysteine cathepsins in glioma invasion, CatB, CatL and CatS. We used an in vitro 3D spheroids migration model, which mimics better the in vivo glioma growth and invasion than the standard 2D models as it enables formation of two distinct environments and therefore induces differences in the originally homogenous population of cells. In the spheroid core the cells are in contact only to each other while in the outer cellular rim the cell-matrix interactions also occur, enabling these cells to invade the matrix [30]. Collagen I was selected as the standard matrix protein, as it is frequently used in various glioma invasion models, although it clearly differs from the glioma microenvironment in vivo [9, 31, 32]. Recently, Demuth et al. [8] used the 2D radial migration assay in collagen I, separating stationary (core) and migratory (rim) cells, indicating transcriptional plasticity of established cultured GBM cell lines, including the subpopulation with “migratory transcriptome signature”. Surprisingly, no protease genes were found within the group of migratory genes. Similarly, in our 3D experimental model the non-invading core spheroid and the matrix invading cellular rim were separated, but the mRNA levels of cysteine cathepsins and their inhibitors were not significantly different between the two. However, alteration and induction of all four classes of proteases at mRNA, protein and activity levels has been extensively documented in a variety of cancers [6, 13] including glioma [3, 14, 15]. One of the multiple roles of proteases in tumour progression is to facilitate invasion by a cascade of enzyme reactions, including in particular CatB, the urokinase-(uPA)-plasminogen system and transmembrane MT1-MMP and other matrix metalloproteases. These were reported to be activated at the cell surface and pericellularly to promote focal dissolution of ECM during invadopodia formation [11]. Moreover, in glioma, simultaneous RNA silencing of uPAR and CatB in SNB19 cells not only inhibited invasion but also induced caspase-8 mediated apoptosis [33]. The surprising absence of proteases genes in the migratory transcriptome in the above GBM model [8], as also confirmed by our data, can be either due to the lack of tumour microenvironment effects on the protease transcriptome or, more likely, to the fact that proteases in the actively invading cells are activated only post-translationally during migration through the ECM.
In our model we found only the activity of CatB, but not CatL or CatS, to be relevant to glioblastoma invasion. This result supports the numerous findings, using transgenic mouse cancer models in combination with deletion of a specific cathepsin gene [34, 35] that these three enzymes are not redundant and that their activities are indeed associated with specific processes, depending strongly on the stage and type of the tumour and its microenvironment. CatB expression has been found to be up-regulated in malignant glioma cells [16–18] and in other cellular compartments of the GBM microenvironment [19]. Later clinical studies confirmed that CatB levels correlated with the evidence for clinical invasion [18], presumably related to prognostic impact on the survival of GBM patients [19]. Moreover, it was reported that down-regulation of this enzyme reduces GBM invasion, tumour growth and angiogenesis in experimental animals [36]. Interestingly, CatB was also highly expressed in endothelial cells of newly formed tumour vasculature, indicating its role in the invasion of these cells during of their out-bursting in angiogenesis. This is supported experimentally by inhibition of capillary formation in vitro using selective CatB inhibitors [37]. The present study demonstrates significant impairment of GBM invasion rate either by inhibiting CatB activity and / or by silencing its expression in U87-MGsiCatB clone. Interestingly, in the actively invading cells, only CatB activity was increased but no up-regulation of CatB at the transcriptional and translational levels was observed. We may speculate that this activation resulted from the specific effects of the matrix collagen I. This matrix was also found to induce CatB in melanoma [38] and possibly in cells metastasizing to the bone [39]. Based on our imaging of the collagen invading cells, we hypothesize that, most probably, plasma associated focal redistribution of CatB is responsible for the increased cell invasiveness, with the added contribution of the intra lysosomal CatB to the invasiveness demonstrated in living glioma cells [40] and in carcinoma cells [28].
In addition, down-regulation of cytosolic endogenous inhibitors, such as stefin B, may be responsible for increased CatB activity. Indeed the ratio between the two proteins changed in the invading cells in favour of CatB activity. The levels of stefin B have been found to be relevant in other types of cancer [15] and recently this inhibitor was found to be a significant prognostic factor for meningioma progression (Lah et al., submitted). In our experimental design, focusing on the matrix invading cells, there were no differences in any of the cystatins, although stefin A was too low and cystatin C was secreted and neither could be detected at the protein level. Taken together, these results confirm that activation of CatB at the post-translational level is a way of facilitating invasion of GBM cells in vitro.
Cat L was first correlated with glioma progression by Sivarapathi et al. [20] confirmed later by our studies [17]. Abundant expression of CatL in GBM cells was shown in vivo, but no prognostic value of CatL was found, supporting the lack of association with GBM invasion [19]. This is in line with the fact, that in our hands, CatL inhibition did not affect GBM cell invasion. Other processes, such as tumour proliferation [41], tumour cell senescence [42] and apoptosis [43] in glioma may be mediated by up-regulated CatL. For example, we have recently confirmed its role as an anti-apoptotic enzyme, protecting U87-MG cells against STS and TNF alpha induced apoptosis [29]. Its up-regulation may possibly induce higher resistance of the cells during invasion to apoptosis [29, 44], which would be in line with the suggestion by Mariani et al. [7] of an inverse relationship between the two processes. CatS protein expression was reported to be highly increased in grade IV glioma and high CatS protein levels in GBM were associated with significantly shorter survival, CatS being an independent predictor even in multivariate analysis [21, 22]. Interestingly, CatS was also expressed in macrophages and microglia, known to possibly promote GBM invasion, but not in normal astrocytes, oligodendrocytes, neurons and endothelial cells [45]. However its role in the invasion has so far not been confirmed, either in this study using selective CatS inhibitors or by others.
In conclusion, this study reveals the important role of CatB, but not CatL and CatS, in facilitating glioma cell invasion. Secondly, CatB activity is further induced post-translationally in the glioma cells during the invasion process. This may be due to altered levels of endogenous cystatins, and/or to the effects of collagen I. Finally the study points out the complexity of protease regulation and the need to include functional proteomics in the systems biology approaches to understand the processes associated with glioma invasion and progression.
Abbreviations
- CatB:
-
cathepsin B
- CatL:
-
cathepsin L
- CatS:
-
cathepsin S
- CysC:
-
cystatin C
- ECM:
-
extracellular matrix
- GBM:
-
Glioblastoma (multiformae)
- PXA:
-
Pleomorphic xanthoastrocytoma
- StefA:
-
stefin A
- StefB:
-
stefin B
References
Ohgaki H, Kleihues P (2005) Epidemiology and etiology of gliomas. Acta Neuropathol 109(1):93–108
Sathornsumetee S, Rich NJ (2006) New treatment strategies for malignant gliomas. Expert Rev Anticancer Ther 6(7):1087–1104
Rao JS (2003) Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer 3(7):489–501
Demuth T, Berens M (2004) Molecular mechanisms of glioma cell migration and invasion. J Neurooncol 70(2):217–228
Wang W, Goswami S, Sahai E et al (2005) Tumour cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biology 15(3):138–145
Sahai E (2005) Mechanisms of cancer cell invasion. Curr Opin Genet Dev 15(1):87–96
Mariani L, Beaudry C, McDonough WS et al (2001) Glioma cell motility is associated with reduced transcription of proapoptotic and proliferation genes: a cDNA microarray analysis. J Neurooncol 53(2):161–176
Demuth T, Rennert JL, Hoelzinger DB et al (2008) Glioma cells on the run- the migratory transcriptome of 10 human glioma cell lines. BMC Genomics 9:54
Berens ME, Rief MD, Loo MA, Giese A (1994) The role of extracellular matrix in human astrocytoma migration and proliferation studied in a microliter scale assay. Clin Exp Metastasis 12(6):405–415
Hoelzinger DB, Nakada M, Demuth T et al (2008) Autotaxin: a secereted autocrine/paracrine factor that promotes glioma invasion. J Neurooncol 86(3):297–309
Wolf K, Friedl P (2005) Functional imaging of pericellular proteolysis in cancer cell invasion. Biochemie 87(3–4):315–320
Tu C, Ortega–Cava CF, Chen G et al (2008) Lysosomal cathepsins B participates in the podosome-mediated extracellular matrix degradation and invasion via secreted lysosomes in the v-Src fibroblasts. Cancer Res 86(22):9147–9156
Lopez–Otin C, Matrisian LM (2007) Emerging roles of proteases in tumour suppression. Nature Rev Cancer 7(10):800–808
Levičar N, Nuttall RL, Lah TT (2003) Proteases in brain tumour progression. Acta Neurochir (Wien) 145(9):825–838
Lah TT, Duran Alonso MB, Van Noorden CJ (2006) Antiprotease therapy in cancer: hot or not? Expert Opin Biol Ther 6(3):257–279
Rempel SA, Rosenblum ML, Mikkelsen T et al (1994) CathepsinB expression and localization in glioma progression and invasion. Cancer Res 54(23):6027–6031
Lah TT, Strojnik T, Levičar N et al (2000) Clinical and experimental studies of cysteine cathepsins and their inhibitors in human brain tumors. Int J Biol Markers 15(1):90–93
Strojnik T, Kos J, Židanik B, Lah TT (1999) Cathepsin B immunohistochemical staining in tumour and endothelial cells is a new prognostic factor for survival in patients with brain tumours. Clin Cancer Res 5(3):559–567
Strojnik T, Kavalar R, Trinkaus M, Lah TT (2005) Cathepsin L in glioma progression: comparison with cathepsin B. Cancer Detect Prev 29(5):448–455
Sivarapathi M, Yamamoto M, Nicolson GL et al (1996) Expression and immunohistochemical localization of cathepsin L during progression of human gliomas. Clin Exp Metastasis 14(1):27–34
Flannery T, Gibson D, Mirakhur M et al (2003) The clinical significance of cathepsin S expression in human astrocytomas. Am J Pathol 163(1):175–182
Flannery T, McQuaid S, McGoohan C et al (2006) Cathepsin S expression: An independent prognostic factor in glioblastoma tumours-A pilot study. Int J Cancer 119(4):854–860
Kos J, Lah TT (2006) Cystatins in cancer. In: Zerovnik E, Kopitar-Jerala N (eds) Human Stefins and Cystatins. Nova Science Publishers Inc, New York
Abrahamson M (1994) Cystatins. Methods Enzymol 244:685–700
Lignelid H, Collins VP, Jacobsson B (1997) Cystatin C and transthyretin expression in normal and neoplastic tissues of the human brain and pituitary. Acta Neuropathol 93(5):494–500
Nakabayashi H, Hara M, Shimuzu K (2005) Clinicopathologic significance of cystatin C expression in gliomas. Hum Pathol 36(9):1008–1015
Konduri SD, Yanamandra N, Siddique K et al (2002) Modulation of cystatin C expression impairs the invasive and tumorigenic potential of human glioblastoma cells. Oncogene 21(57):8705–8712
Bervar A, Zajc I, Sever B et al (2003) Invasiveness of transformed human breast epithelial cell lines is related to cathepsin B and inhibited by cysteine proteinase inhibitors. Biol Chem 384(3):447–455
Zajc I, Hreljac I, Lah T (2006) Cathepsin L affects apoptosis of glioblastoma cells: a potential implication in the design of cancer therapeutics. Anticancer Res 26(5A):3357-64
Hegedüs B, Marga F, Jakab K et al (2006) The Interplay of Cell-Cell and Cell-Matrix Interactions in the Invasive Properties of Brain Tumors. Biophys J 91(7):2708–2716
Corcoran A, De Ridder LI, Del Duca D et al (2003) Evolution of the brain tumour spheroid model: transcending current model limitations. Acta Neurochir (Wien) 145(9):819–824
Rubenstein BM, Kaufman LJ (2008) The role of extracellular matrix in glioma invasion:a cellular potts model approach. Biophys J 95(12):5661–5680
Gondi CS, Kandhukuri N, Kondraganti S et al (2006) RNA interference–mediated simultaneous down-regulation of urokinase-type plasminogen activator receptor and cathepsin B induces caspase-mediated apoptosis in SNB 19 human glioma cells. Mol Cancer Ther 5(12):3197–3208
Gocheva V, Zeng W, Ke D et al (2006) Distinct role for cysteine cathepsin genes in multistage tumourigenesis. Genes Dev 20(5):543–556
Reinheckel T, Gocheva V, Peters C, Joyce JA (2008) Roles of cysteine proteases in tumour progression: Analysis of cysteine cathepsins knockout mice in cancer models. In: Edwards D, Hoyer-Hansen G, Blasi F, Sloane BF (eds) The Cancer Degradome. Springer Science + Business Media, New York
Lakka C, Gondi CS, Yanamandra N et al (2004) Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumour cell invasion, tumour growth and angiogenesis. Oncogene 23(27):4681–4689
Premzl A, Zavasnik-Bergant V, Turk V, Kos J (2003) Intracellular and extracellular cathepsin B facilitate invasion of MCF-10A neoT cells through reconstituted extracellular matrix in vitro. Exp Cell Res 283(2):206–214
Klose A, Wilbrand-Hennes A, Zigrino P et al (2006) Contact of high-invasive, but not low-invasive, melanoma cells to native collagen I induces the release of mature cathepsin B. Int J Cancer 118(11):2735–2743
Podgorski I, Linebough BE, Sameni M et al (2005) Bone microenvironment modulates expression and activity of cathepsin B in prostate cancer. Neoplasia 7(3):207–223
Sameni M, Dosescu J, Sloane BF (2001) Imaging proteolysis by living human glioma cells. Biol Chem 382(5):785–788
Zhu DM, Uckun FM (2000) Z-Phe-Gly-NHO-Bz, an inhibitor of cysteine cathepsins, induces apoptosis in human cancer cells. Clin Cancer Res 6(5):2064–2069
Felbor U, Kesseler B, Mothes W et al (2002) Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci USA 99(12):7883–7888
Levičar N, Dewey RA, Daley E et al (2003) Selective supression of cathepsin L by antisense cDNA impairs human brain tumor cell invasion in vitro and promotes apoptosis. Cancer Gene Ther 10(2):141–151
Castino R, Pace D, Démoz M et al (2002) Lysosomal proteases as potential targets for the induction of apoptotic cell death in human neuroblastomas. Int J Cancer 97(6):775–779
Bellail AC, Hunter SB, Brat DJ et al (2004) Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol 36(6):1046–1069
Acknowledgements
We are grateful to Seyed Y. Ardebili, M.D., for providing us with human tumour samples, to Dr. Janko Kos for providing the ELISA kits and primary antibodies, to Dr. Nobuhiko Katunuma for providing Clik 148, to Dr. Irena Zajc and Dr. Simon Caserman for their scientific and technical contributions to this work, and to Dr. Roger Pain for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gole, B., Durán Alonso, M.B., Dolenc, V. et al. Post-Translational Regulation of Cathepsin B, but not of Other Cysteine Cathepsins, Contributes to Increased Glioblastoma Cell Invasiveness In Vitro . Pathol. Oncol. Res. 15, 711–723 (2009). https://doi.org/10.1007/s12253-009-9175-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-009-9175-8