Abstract
We examined the population genetic structure of the New Zealand endemic clam, Austrovenus stutchburyi, to determine (1) whether populations of this estuarine taxon are genetically subdivided and (2) if the locations of genetic boundaries were congruent with known biogeographic break points. We obtained sequences of the mitochondrial gene cytochrome c oxidase I for 372 A. stutchburyi from 29 New Zealand estuaries and conducted analyses to identify population genetic structure. We detected a pattern of genetic isolation by distance and identified six A. stutchburyi subpopulations, a greater number of subpopulations than reported for much of New Zealand’s open coast benthos. Although these data indicate that long distance dispersal may be less frequent in estuarine than in open coast taxa, partial congruence between genetic and biogeographic boundaries suggests that historical events and natural selection may also contribute to the observed population genetic structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many marine benthic invertebrates can only disperse over large distances during a pelagic larval phase (Thorson 1950; Pechenik 1999; Grantham et al. 2003). For inshore species, larvae are exported to coastal or shelf waters and develop during a period of weeks to months before returning to suitable habitat in late larval or early post-larval form (Pechenik 1999). For estuarine taxa, the process of inter-estuary dispersal is more complex. Larvae must be exported from their natal estuary, develop in coastal waters and be transported to suitable settlement habitat within another estuary when developmentally capable of settling (Bilton et al. 2002). Inter-estuary larval dispersal may be further limited by the often large distances between estuaries, the physical properties of estuarine waters and their interface with the coastal ocean (Mann 1988; Largier 1993), as well as the physiological challenges associated with a life history alternating between estuarine and coastal waters (Cognetti and Maltagliati 2000). Consequently, it has been suggested that connectivity among populations of estuarine taxa will be reduced compared to taxa occurring on the open coast and that different spatial management strategies may be needed for estuarine versus coastal taxa (Bilton et al. 2002; Watts and Johnson 2004; Pelc et al. 2009). However, determining rates of connectivity has proven challenging owing to the difficulties associated with physically tracking dispersing larvae (Levin 2006; Gawarkiewicz et al. 2007).
A number of indirect methods of estimating connectivity have been developed (Levin 2006) with population genetics one of the most widely utilized. If rates of inter-estuary gene flow are low as hypothesized, then estuarine taxa would be expected to exhibit a greater degree of genetic subdivision relative to taxa occurring on the open coast (Bilton et al. 2002; Pelc et al. 2009). However, as natural selection (Schmidt et al. 2008; Richards et al. 2010 and references therein) and patterns of historical gene flow (Avise et al. 1987; Kelly et al. 2006) can also influence population genetic structure, the interpretation of genetic patterns and estimation of gene flow has been complicated (Sotka et al. 2004).
Because larval dispersal and natural selection will also determine species’ distribution and abundance, it has been suggested that boundaries between groups of genetically distinct populations will be geographically congruent with biogeographic break points (the boundaries between taxonomically distinct communities; Avise et al. 1987). Although tested across a number of biogeographic boundaries (e.g. Avise 1992; Hare and Avise 1996; Dawson 2001, 2005; Cárdenas et al. 2009), conflicting results suggest that agreement between genetic and biogeographic boundaries may be location and taxon specific (Burton 1998; Pelc et al. 2009).
The New Zealand archipelago with its complex oceanography (Heath 1982; Laing and Chiswell 2003), dynamic geological history (Fleming 1979) and well-documented biogeography (Shears et al. 2008) provides an opportunity to examine the processes that generate population subdivision and the relationship between genetic and biogeographic boundaries. Straddling the subtropical convergence, New Zealand’s marine climate follows a steep gradient from the subtropical north to sub-Antarctic south (Laing and Chiswell 2003; Hadfield et al. 2007). The resulting temperature gradient coupled with spatial variation in swell regime, geological processes and other environmental factors (Laing and Chiswell 2003) has created regional variation in the distribution and abundance of flora and fauna. This biogeographical variation has been described in a number of classification schemes that have divided New Zealand’s coastline into distinct biogeographic regions (reviewed by Shears et al. 2008). In contrast, the influence of physical, environmental and geological processes on a population’s genetic structure is less understood.
Population genetic structure has been examined in at least 29 New Zealand coastal benthic species with the majority of studies focussed on taxa that occur on the open coast (see Ross et al. 2009 for review). Where populations have been sampled across a wide latitudinal range, there has usually been an absence of genetic structure (e.g. rock lobster: Smith and McKoy 1980; Ovenden et al. 1992, seastar: Waters and Roy 2003), or the detection of genetically divergent northern and southern populations divided through central New Zealand. Differentiation about a central boundary has now been recorded in at least ten species including open coast taxa with pelagic larvae (seastar: Waters and Roy 2004; Ayers and Waters 2005, limpets: Goldstien et al. 2006, mussel: Apte and Gardner 2002; Apte et al. 2003, chiton: Veale 2007), open coast taxa without pelagic larvae (brittle star: Sponer and Roy 2002) and estuarine taxa without pelagic larvae (Corophiid amphipods: Stevens and Hogg 2004; Knox et al. 2011, seagrass: Jones et al. 2008). While it is possible that taxa sharing this pattern of population subdivision have experienced a common set of contemporary or historical processes (Kuo and Avise 2005; Pelc et al. 2009), the identification of these processes has thus far proved elusive (see Apte and Gardner 2002; Goldstien et al. 2006).
Genetic structure in addition to the central New Zealand divergence, has only been reported for a few species, all of which exhibit characteristics of limited dispersal capacity such as non-pelagic larvae or restriction to estuarine habitats (e.g. Corophiid amphipods: Knox et al. 2011; Stevens and Hogg 2004, seagrass: Jones et al. 2008). As most of the estuarine taxa examined to date lack a pelagic larval phase, it is uncertain which characteristic (estuarine distribution vs. larval mode) is driving patterns of additional population subdivision.
The clam Austrovenus stutchburyi (Wood 1828) is the only New Zealand taxon that both disperses via pelagic larvae and is restricted to estuaries that has previously been assessed for population genetic structure (Lidgard 2001). In contrast to other estuarine taxa with limited potential for long distance dispersal, A. stutchburyi has a pelagic larval phase of 2 to 3 weeks during which dispersal among geographically discrete populations is possible. A previous analysis of allozyme polymorphism in A. stutchburyi did not detect any spatial genetic subdivision (although temporal patterns may have been detected; Lidgard 2001). This supports the notion that for previously examined taxa, the lack of pelagic larval phase rather than an estuary-restricted distribution per se might be responsible for the additional genetic structure observed (e.g. Stevens and Hogg 2004; Jones et al 2008). However, as most recent genetic studies of New Zealand taxa have used mitochondrial DNA sequences to assess population structure (Ross et al. 2009), it is uncertain whether the lack of subdivision reported for A. stutchburyi can be attributed to their pelagic larval phase or to differences in methodology (allozymes vs. mtDNA).
To determine whether populations of an estuarine species with pelagic larvae will be genetically subdivided, we conducted a population genetic analysis of A. stutchburyi using the mitochondrial cytochrome c oxidase subunit I (COI) gene. We examined the genetic structure of A. stutchburyi populations, testing for patterns of genetic isolation by distance (IBD) and performed a spatial analysis of molecular variance (SAMOVA) to identify divergent subpopulations. We also used an analysis of molecular variance (AMOVA) to determine whether genetic subdivisions are congruent with known biogeographic break points. A recent biogeographic classification of New Zealand (Shears et al. 2008; Fig. 1a) was chosen for our comparison because it incorporated results from previous biogeographic classifications and included taxa similar to those considered in our (and previous) population genetic studies. Shears et al. (2008) described a major biogeographic division through central New Zealand and another nine minor biogeographic boundaries located throughout New Zealand (Fig. 1a).
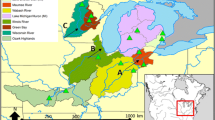
a New Zealand biogeographic provinces and regions described by Shears et al. (2008). Solid black line indicates boundary between biogeographic provinces while dashed lines indicate boundaries between biogeographic regions. Specific locations referred to in the text are named and indicated by arrows. b Sampling locations of A. stutchburyi populations and the frequencies of common cytochrome c oxidase I (COI) haplotypes at each site. Labels refer to estuary locations detailed in Table 1. Pie segment size indicates relative frequencies of COI haplotypes in each population. Haplotypes detected in fewer than eight specimens are grouped (white segments) for clarity of presentation. c Population clusters designated by SAMOVA analysis
Methods
Study Sites and Sample Collection
Between January 2007 and January 2010, we collected A. stutchburyi from 29 New Zealand estuaries (Table 1; Fig. 1b). Fourteen of the sampled estuaries were located in the North Island and 12 in the South Island. We also collected specimens from estuaries on one inshore island and the only two offshore islands where A. stutchburyi is present: Great Barrier Island (NE3) less than 20 km off the northeast coast of the North Island, the Chatham Islands (OS1) 660 km to the east of central New Zealand and the Auckland Islands (OS2) 460 km to the south of the South Island (Fig. 1b). Between 12 and 25 A. stutchburyi were collected from each estuary and stored at −80°C prior to DNA extraction.
DNA Extraction and Sequencing
We dissected a 0.25–0.50-cm2 piece of adductor mussel from each specimen and extracted genomic DNA using the Zymo Research Genomic DNA II Kit (Zymo Research Corporation, Orange, CA, USA). We then amplified a 710-bp fragment of the mitochondrial COI gene using the universal primers LCO1490 and HCO2198 (Folmer et al. 1994). PCR amplifications were conducted in 10 μl reactions containing 4.8 μl Intron i-Taq 2× PCR master mix, 5 pmol of each primer and 1 μl of unquantified template DNA. PCR reactions consisted of an initial denaturing phase of 94°C (4 min), followed by 35 cycles consisting of 94°C (60 s), 52°C (90 s) and 72°C (90 s) and a final extension period at 72°C (5 min). Unincorporated nucleotides and primers were removed by adding 2 U of exonuclease I, 0.1 U of shrimp alkaline phosphatase and 2.7 μl H2O and incubating at 37°C (30 min) then 80°C (15 min). Sequencing reactions used Big Dye terminator sequencing chemistry (Applied Biosystems) on an Applied Biosystems 3130 Genetic Analyzer. DNA strands were edited in Geneious (ver. 4.8.4) to produce an alignment of 658 bp. Sequences have been deposited in the Barcode of Life Datasystems database under project NZCOC (New Zealand Marine Bivalves) and GenBank (accession numbers JN200827-JN201198).
Population Genetic Analysis
Indices of genetic diversity were quantified using DnaSP ver. 5 (Librado and Rozas 2009). For each estuary, we calculated the number of COI haplotypes (N hap), number of segregating sites (S), haplotype diversity (H e), mean number of pairwise differences (π 1) and nucleotide diversity (π 2). The GTR model of sequence evolution was selected in jMODELTEST 0.1.1 (Posada 2008), and a GTR corrected distance matrix was generated in PAUP* 4.0 (Swofford 2000) for use in subsequent analyses. Estimates of population pairwise F ST values were then calculated in ARLEQUIN ver. 3.11 (Excoffier et al. 2005) to determine if any two populations differed significantly in their genetic composition. A Mantel test was implemented in ARLEQUIN to assess the relationship between genetic and geographic distance. Geographic distance between populations, measured in Google Earth as the shortest distance over water between two estuaries, was regressed against estimates of population pairwise F ST values to determine whether A. stutchburyi dispersal conformed to a pattern of IBD.
Population structure was further investigated using SAMOVA 1.0 (Dupanloup et al. 2002). This method is based on a simulated annealing procedure that maximizes the proportion of total genetic variance due to differences among groups of populations. SAMOVA can be used to define population clusters that are geographically homogeneous and maximally differentiated from each other without the prior assumption of subpopulation composition. Genetic variance (F ST) is partitioned into two components, F SC and F CT, indicating respectively the differentiation among populations within and among groups (note that we have adopted the ARLEQUIN and SAMOVA subscript definitions to define differentiation within and among groups of populations). SAMOVA analyses ran for 10,000 iterations from each of 100 random initial conditions for a predetermined number of subpopulations (k) ranging from 2 to 14.
We calculated Tajima’s (1989) D, Fu and Li’s (1993) F* and D* and Fu’s (1997) F S in DnaSP to test for deviation from the Wright–Fisher model of neutral evolution consistent with either non-neutral evolution or population expansion under neutral evolution. We also used the mismatch distributions of pairwise differences between all individual haplotypes (calculated in DnaSP) to further test for population stability or growth.
Comparison of Genetic and Biogeographic Boundaries
To test for congruence between genetic and biogeographic boundaries (sensu Avise et al. 1987), we compared the locations of all boundaries between A. stutchburyi subpopulations identified in SAMOVA with biogeographic break points described by Shears et al. (2008; Fig 1a). We then grouped populations according to the zonation of the biogeographic classification and performed AMOVA at both bioprovince and bioregion spatial scales (as defined by Shears et al. 2008), to assess how well this biogeographic classification represented the spatial distribution of genetic variation in A. stutchburyi.
Results
Population Genetic Analysis
Three hundred and seventy-two A. stutchburyi were sequenced for the mitochondrial COI gene. Of the 658 positions analysed, 99 were variable leading to the delineation of 125 haplotypes (Table 1). The most abundant haplotype (H2; Fig. 1b) occurred in 27 of 29 sampled populations, accounting for 30% of the total data set. Another 25 haplotypes were recorded in at least two populations (43% of data set), while the remaining 99 haplotypes were recorded only in single populations. Haplotype diversity was high throughout all populations with at least four haplotypes recorded at each location (\( \overline{H} = 0.85 \pm 0.10{\left( { \pm SD} \right)} \); Table 1). In contrast to haplotype diversity, nucleotide diversity was low with most COI sequences differing only by a small number of base changes (\( {\mathop {\ifmmode\expandafter\bar\else\expandafter\=\fi{\pi }}\nolimits_1 } = 2.8 \pm 0.93 \), \( {\mathop {\ifmmode\expandafter\bar\else\expandafter\=\fi{\pi }}\nolimits_2 } = 0.004 \pm 0.001 \); Table 1).
A plot of haplotype frequency and distribution suggested regional differences in the genetic composition of populations (Fig. 1b). In contrast to the most abundant haplotype (H2), which was detected throughout sampled populations (with the exception of sites SE6 and OS2; Fig. 1b), other haplotypes were either restricted to, or were detected more frequently, in specific regions. For example, haplotypes H1, H4 and H6 were most abundant in southern populations, H3 in lower North Island populations and OS1 to the east of the South Island and H5 in northern populations (Fig. 1b). The most dramatic shift in genetic composition appeared to be between northern and southern populations to the north of NE11 in the lower North Island.
Pairwise F ST values (Table 2) indicated that, for the most part, North and South Island populations were significantly differentiated from each other. The exceptions were NE11 which was significantly different to most North Island populations but not South Island populations and SW1 (Fig. 1b) located in the north of the South Island which was not significantly different from either lower North Island or upper South Island populations. While OS1 was significantly different from most South Island and northeast coast North Island populations, it did not differ significantly from lower North Island populations. OS2, located to the south of the South Island, was differentiated from most northern populations and the southernmost South Island populations. The two East Cape populations (NE6 and NE7; Fig. 1b) were significantly different from adjacent east coast populations.
A Mantel test revealed a highly significant positive correlation between geographic and genetic distances (F ST) among all sampled populations (P < 0.001; Fig. 2). Distance between estuaries accounted for 28% of inter-population COI variability, a result indicative of genetic IBD, implying that for A. stutchburyi, dispersal over large distances may be limited.
SAMOVA obtained its best partitioning of genetic variance when populations were assigned to six groups (Table 3). All grouping options (k = 2 to 14) produced populations clusters that were significantly differentiated. However, it was only when SAMOVA generated six or more clusters that differentiation within groups (F SC) became non-significant, indicating that populations within each cluster were similar to each other. At k = 6, populations were assigned to clusters that were spatially coherent, with the exception of cluster E in which SE6 and SW5 in the southwest were grouped with SE2 located c. 600 km to the northeast (Fig. 1c). Once k exceeded six, populations were removed one at a time from the established clusters, providing no further groupings that were informative of population structure. When k = 6 the SAMOVA assigned populations into five mainland clusters and the Auckland Islands as a sixth (Fig. 1c). Mainland populations were grouped on the northeast coast (Fig. 1c; cluster A), around East Cape (cluster B), across the lower North Island (including both SW1 in the upper South Island and OS1 to the east of the South Island; cluster C), across the upper and central South Island (included NE11 in the lower North Island; cluster D) and in the southwestern Fiordland region also incorporating SE2 (cluster E).
Tests of neutrality were all significant, indicating either natural selection or population expansion (Table 4). Further support for a recent population expansion was provided in our mismatch analysis in which the distribution of pairwise differences was a close fit under the expectations of expansion but departed from expectations under a model of population stability (Fig. 3).
Frequency distribution of pairwise differences among cytochrome oxidase subunit I (COI) haplotypes in A. stutchburyi. Solid line indicates observed frequencies while dashed and dotted lines indicate respectively the expected frequencies under models of population expansion and constant population size
Comparison of Phylogeographic and Biogeographic Boundaries
The boundaries between all SAMOVA clusters were located in close proximity to the biogeographic break points of Shears et al. (2008). The boundary between clusters C and D (Fig. 1c), which pairwise F ST values indicated to be the most significant genetic boundary (Table 2), was a close match to the major break point between biogeographic provinces (Fig. 1a). Other boundaries between SAMOVA clusters were located in proximity to more minor inter-bioregional break points. However, genetic divisions were not detected at all biogeographic break points. Shears et al. (2008) described 11 biogeographic boundaries delineating 11 bioregions within two bioprovinces. In comparison, we identified seven genetic boundaries (excluding SE2 boundaries in cluster E ; Fig. 1c), delineating five mainland A. stutchburyi subpopulations. Boundaries identified by both biogeographic and genetic methods were located around the North and East Capes, to the north of Fiordland, in the southeast of the South Island and across Cook Strait from Farewell Spit to the southeast coast of the North Island.
When A. stutchburyi populations were grouped according to the biogeographic classification (Fig. 1a), AMOVA indicated significant genetic differentiation between biogeographical provinces (F CT = 0.109, P < 0.001) and among bioregions (F CT = 0.072, P < 0.001). While these levels of differentiation were less than those detected among SAMOVA assigned population clusters (F CT = 0.114), the result suggests that this biogeographic classification provides a reasonable representation of the spatial distribution of genetic variation, especially at the larger bioprovince spatial scale. Differentiation among populations within bioregions was marginally significant (F SC = 0.018, P = 0.045 compared to F SC = −0.014NS in SAMOVA), indicating that populations within bioregions were not all genetically homogenous. When the Northeastern Portland and Portland Cook boundaries (Fig. 1a) were relocated southward (c. 150 km) to match the boundaries suggested by SAMOVA (thereby shifting NE7 and NE10 populations between bioregions; Fig. 1c), differentiation among populations within bioregions became non-significant (F SC = 0.006, P = 0.245).
Discussion
We detected a greater number of genetic subdivisions among populations of A. stutchburyi relative to most reported examples of New Zealand’s open coast benthos. SAMOVA identified six genetically distinct subpopulations, with the most significant change in the genetic composition of populations occurring through central New Zealand in the vicinity of Cook Strait. The transition across this genetic boundary was not an abrupt or complete shift as has been reported for other taxa (e.g. Goldstien et al. 2006). Instead, some COI haplotypes were found throughout New Zealand but at different frequencies in northern and southern populations, while others were restricted to either side of this genetic boundary. The boundary between clusters C and D roughly coincides with Cook Strait which separates the North and South Islands. However, as there are similarities between some upper South Island and lower North Island populations, this genetic transition does not appear to solely be a function of limited dispersal across the strait.
A similar genetic boundary has previously been reported for several species with varied developmental characteristics (planktonic larvae vs. direct developers) and habitat requirements (coastal vs. estuarine; Ross et al. 2009). Despite some inter-taxa variation in the location of this central genetic division (usually within 100 km of Cook Strait), the frequency with which the division has been detected and the taxonomic diversity of species exhibiting this population structure suggest that a similar genetic division might be found for much of New Zealand’s coastal benthos.
For most open coast taxa, this central division has been the only genetic boundary reported (e.g. Apte and Gardner 2002; Apte et al. 2003; Waters and Roy 2004; Ayers and Waters 2005; Goldstien et al. 2006). For other coastal taxa, an additional boundary has been identified about North Cape (e.g. Sypharochiton pelliserpentis; Veale 2007). In contrast, for A. stutchburyi, SAMOVA was able to identify seven genetic boundaries, of which five (North Cape, north and south of East Cape, Farewell Spit and southeast North Island) are in the vicinity of genetic divisions reported previously for other estuarine taxa (Stevens and Hogg 2004; Jones et al. 2008; Knox et al. 2011). Surprisingly, SAMOVA grouped SE2 within the E cluster despite it being located in the centre of the D cluster of populations. It is possible that this grouping is an artefact of our sample size. Alternatively, it may result from human-mediated translocation of A. stutchburyi among estuaries.
We detected a pattern of genetic IBD indicating that for A. stutchburyi, long distance dispersal among estuaries may be limited (Wright 1943). IBD has for the most part only been reported for New Zealand marine taxa lacking a planktonic larval stage (Sponer and Roy 2002; Stevens and Hogg 2004; Veale 2007), those dependent on specific host organisms (Stevens 1991), or those sampled from estuaries (Perrin et al. 2004; Stevens and Hogg 2004). The population structure we detected for A. stutchburyi is more similar to that of taxa with putatively limited dispersal than to open coast taxa with pelagic larvae (e.g. Stevens and Hogg 2004; Veale 2007; Jones et al. 2008). This suggests that distribution and habitat requirements, as well as larval characteristics, may determine patterns of gene flow among populations.
Possible Causes of Population Structure
Several mechanisms could explain the additional genetic structure (IBD) and greater number of genetic subdivisions detected in A. stutchburyi and other estuarine taxa relative to open coast taxa. For example, differences between estuarine and coastal taxa may result from limited present-day connectivity among estuarine populations (e.g. Watts and Johnson 2004). Connectivity will be restricted if larvae are retained within their natal estuary. However, while retention has been observed in species that occur in the upper reaches of an estuary (e.g. Little and Epifanio 1991; Cartaxana 1994; Paula 1998), retention is less likely for taxa such as A. stutchburyi which inhabit a broader range of estuarine habitats (Lundquist et al. 2009). A more plausible explanation is that larvae are exported from their natal estuary but fail to reach or recruit to distant populations. For some taxa, mean dispersal distances may be as little as 2–50 km (e.g. DeBoer et al. 2008; Piggott et al. 2008; Puebla et al. 2009). Where this is the case, distance between suitable habitats will be critical in determining dispersal success. Where both dispersal capacity and the distance between settlement habitats are small, gene flow over large distances can occur in small increments over many generations. However, as the distance separating suitable habitats increases, widespread gene flow becomes less likely (Alberto et al. 2010).
In some parts of New Zealand, estuaries are separated by 200–300 km of open coast (e.g. west coast of both North and South Islands and northeast coast of South Island). Three of the genetic boundaries detected in A. stutchburyi occur along such sections of coast possibly reflecting the lack of estuarine habitat in these regions. Although rocky reef is also a disjunct habitat, in New Zealand there are fewer large stretches of coastline along which no hard strata can be found. Even if patches of reef are not ideal, open coast taxa can utilize sub-optimal habitats as stepping stones between more ideal habitats (Ayre et al. 2009). Similar flexibility is unlikely for estuarine taxa, limiting their potential for dispersal where estuaries are scarce.
Conversely, the genetic composition of our offshore island populations (OS1 and OS2; Fig. 1b, c) implies that A. stutchburyi can disperse over larger distances. Genetic similarities were evident between offshore and mainland populations, particularly for the OS1 Chatham Islands population. Although c. 660 km to the southeast of the North Island, the Chatham Islands are located in the easterly flowing subtropical convergence (Heath 1982; Hadfield et al. 2007), which may facilitate the transport of larvae from mainland to Chatham Island populations. In contrast, the Auckland Islands (OS2), while geographically closer (c. 460 km), lie outside of the predominantly easterly track of sub-Antarctic water which flows past the lower South Island (Heath 1982) and appear to experience a lesser degree of connectivity.
Coastal circulation might also impede the transport of larvae among populations (e.g. Lamare 1998). It has been demonstrated that large and persistent eddies located to the north and south of East Cape (Chiswell and Booth 1999) and the semi-closed estuarine circulation typical of the fiords in southwest New Zealand (Lamare 1998) can entrain pelagic larvae. Simulations suggest that the time of entrainment could often exceed the larval duration of many benthic invertebrates (Chiswell and Roemmich 1998). As such, these hydrodynamic features may act as physical barriers to the dispersal of larvae and explain the genetic boundaries detected about East Cape and differences between Fiordland and more northerly populations.
A second possible explanation for the observed genetic subdivision is that present-day genetic boundaries are a consequence of historic dispersal barriers that no longer exist (Avise et al. 1987). Additional genetic subdivision would be expected in estuarine taxa if historic events (e.g. glaciation or topographical alteration with sea-level fluctuation) generated dispersal barriers for estuarine taxa that were easily traversed by coastal taxa on account of their greater dispersal potential. Alternatively, historic processes may have subdivided both estuarine and coastal taxa. High rates of dispersal in coastal taxa once dispersal barriers lapsed could quickly erase the genetic signatures of this subdivision, while the introgression of allopatric populations would be slower for taxa with lesser dispersal capabilities.
A third possibility is that regional environmental differences rather than patterns of gene flow are determining the genetic structure of A. stutchburyi. There is evidence for natural selection on mtDNA (Fontanillas et al. 2005; Ballard et al. 2007; Oliveira et al. 2008; Díaz-Ferguson et al. 2010) and other genetic markers (Bernardi et al. 1993; Eanes 1999) with temperature suggested as a likely selective force (Schmidt et al. 2008; Balloux et al. 2009). New Zealand encompasses a large latitudinal range and steep environmental gradients exist between northern and southern locations for variables such as air and sea surface temperature (SST). It is currently unknown whether environmental variation will generate regional differences in the genetic composition of New Zealand’s coastal and estuarine benthos. However, the detection of a similar population composition on east and west coasts, particularly in the South Island where east and west coast D cluster populations are disjunct, suggests that certain haplotypes may be favoured at specific latitudes. If survivorship or fecundity co-vary with haplotype and environmental variables such as temperature, regional variation in haplotype frequency and a major genetic divergence through central New Zealand (as detected in A. stutchburyi and other taxa) could be explained by a latitudinal gradient in SST and the relatively abrupt transition between subtropical and sub-Antarctic waters off the coast of central New Zealand (Hadfield et al. 2007). Tests of neutrality provide support for the hypothesis that A. stutchburyi are experiencing selection, possibly in conjunction with a population or range expansion. Rapid population expansion following a period of restricted abundance and distribution could further increase regional genetic differentiation, particularly if regionally restricted haplotypes evolved in response to environmental variation during periods of relative isolation. Given the ephemeral nature of estuaries when sea level fluctuates (Fleming 1979), repeated episodes of population and range expansion and contraction are a plausible scenario for estuarine taxa.
Comparison of Genetic and Biogeographic Boundaries
Congruence between biogeographic and genetic boundaries is expected and could be explained by a combination of historical and contemporary processes (Avise et al. 1987). While the degree of congruence appears to be greater for A. stutchburyi and other estuarine taxa relative to coastal species, we did not detect genetic differentiation at five of the 11 biogeographic boundaries described by Shears et al. (2008). This lack of complete congruence could result from the use of molecular markers that are inappropriate for detecting genetic variation across biogeographic break points, or where sampling resolution was inadequate to detect subtle genetic differences. Alternatively, the applicability of the hypothesis of congruence (Avise et al. 1987) may be location and taxon specific (Burton 1998).
While it is difficult to assess the suitability of molecular markers and the adequacy of sampling designs without further analyses, the idea that congruence will vary among species and biogeographic break points has already been the subject of considerable debate (e.g. Burton 1998; Dawson 2001; Pelc et al. 2009). In a review of population genetic studies across the southeast and southwest coasts of the USA, Pelc et al. (2009) found for taxa with potentially limited dispersal (estuarine taxa and direct developers), that genetic boundaries were congruent with biogeographic break points, while for open coast taxa with planktonic larvae, genetic boundaries were not. The available data suggest that a similar pattern may exist for New Zealand’s marine benthos. However, as available research has largely focussed on open coast taxa, additional studies of estuarine and direct developing species will be required to further test this hypothesis.
Conclusions and Management Implications
Our results indicate that the estuarine clam, A. stutchburyi is genetically subdivided and that genetic boundaries are partially congruent with biogeographic break points. The genetic structure we detected in A. stutchburyi was similar to that reported previously for estuarine taxa and generally greater than the structure reported for open coast taxa. Accordingly, long-distance inter-population gene flow may be more frequent in coastal compared with estuarine taxa. Historical events and environmental processes can also cause geographical variation in genetic composition and may act either individually or together with present-day dispersal to generate the genetic structure we observed. Congruence between genetic and biogeographic boundaries suggests that some of the genetic subdivisions we detected may be attributed to environmental variation or historical events. However, with a single non-recombining molecular marker (mtDNA), it will be difficult to fully determine which mechanisms are generating this subdivision (Balloux 2010).
Estuaries are one of the most highly impacted marine environments (Kennish 2002), with anthropogenic and natural disturbances often resulting in the alteration, degradation or loss of estuarine habitats and communities. Recovery will depend on the spatial and temporal scales of disturbance and the rate of recruitment from intact populations (Thrush et al. 1996, 2005). Where disturbances are estuary-wide, recovery may rely on recruitment from other estuaries. Our results indicate that dispersal among estuaries may in some cases be limited. If true, estuarine communities may be slower to recover relative to coastal taxa and more vulnerable to localised population failures. Estuarine taxa may need to be managed more conservatively and at smaller spatial scales than coastal species.
While our analyses identified six genetically distinct subpopulations, it has been suggested that analyses of mtDNA may underestimate subdivision and overestimate connectivity (Goudet et al. 1996; Buonaccorsi et al. 1999). Further analyses incorporating multiple autosomal markers will provide more reliable estimates of connectivity and subdivision (Balloux 2010). Until such data are available, environmental managers must use other tools to define population units. The partial congruence we detected with biogeographic boundaries suggests that classifications based on taxonomic diversity may provide a suitable proxy for population subdivision in estuarine taxa until genetic data become available.
References
Alberto, F., P.T. Raimondi, D.C. Reed, N.C. Coelho, R. Leblois, A. Whitmer, and E.A. Serrao. 2010. Habitat continuity and geographic distance predict population genetic differentiation in giant kelp. Ecology 91: 49–56.
Apte, S., and J.P.A. Gardner. 2002. Population genetic subdivision in the New Zealand greenshell mussel (Perna canaliculus) inferred from single-strand conformation polymorphism analysis of mitochondrial DNA. Molecular Ecology 11: 1617–1628.
Apte, S., B. Star, and J.P.A. Gardner. 2003. A comparison of genetic diversity between 432 cultured and wild populations, and a test for genetic introgression in the 433 New Zealand greenshell mussel Perna canaliculus (Gmelin 1791). Aquaculture 219: 193–220. 434.
Avise, J.C. 1992. Molecular population-structure and the biogeographic history of a regional fauna—a case-history with lessons for conservation biology. Oikos 63: 62–76.
Avise, J.C., J. Arnold, R.M. Ball, E. Bermingham, T. Lamb, J.E. Neigel, C.A. Reeb, and N.C. Saunders. 1987. Intraspecific phylogeography—the mitochondrial-DNA bridge between population-genetics and systematics. Annual Review of Ecology and Systematics 18: 489–522.
Ayers, K.L., and J.M. Waters. 2005. Marine biogeographic disjunction in central New Zealand. Marine Biology 147: 1045–1052.
Ayre, D.J., T.E. Minchinton, and C. Perrin. 2009. Does life history predict past and current connectivity for rocky intertidal invertebrates across a marine biogeographic barrier? Molecular Ecology 18: 1887–1903.
Ballard, J.W.O., R.G. Melvin, S.D. Katewa, and K. Maas. 2007. Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution 61: 1735–1747.
Balloux, F. 2010. The worm in the fruit of the mitochondrial DNA tree. Heredity 104: 419–420.
Balloux, F., L.J.L. Handley, T. Jombart, H. Liu, and A. Manica. 2009. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proceedings of the Royal Society B Biological Sciences 276: 3447–3455.
Bernardi, G., P. Sordino, and D.A. Powers. 1993. Concordant mitochondrial and nuclear DNA phylogenies for populations of the teleost fish Fundulus heteroclitus. Proceedings of the National Academy of Sciences of the United States of America 90: 9271–9274.
Bilton, D.T., J. Paula, and J.D.D. Bishop. 2002. Dispersal, genetic differentiation and speciation in estuarine organisms. Estuarine Coastal and Shelf Science 55: 937–952.
Buonaccorsi, V.P., K.S. Reece, L.W. Morgan, and J.E. Graves. 1999. Geographic distribution of molecular variance within the blue marlin (Makaira nigricans): A hierarchical analysis of allozyme, single-copy nuclear DNA, and mitochondrial DNA markers. Evolution 53: 568–579.
Burton, R.S. 1998. Intraspecific phylogeography across the Point Conception biogeographic boundary. Evolution 52: 734–745.
Cárdenas, L., J.C. Castilla, and F. Viard. 2009. A phylogeographical analysis across three biogeographical provinces of the south-eastern Pacific: The case of the marine gastropod Concholepas concholepas. Journal of Biogeography 36: 969–981.
Cartaxana, A. 1994. Distribution and migrations of the prawn Palaemon longirostris in the Mira River estuary (southwest Portugal). Estuaries 17: 685–694.
Chiswell, S.M., and J.D. Booth. 1999. Rock lobster Jasus edwardsii larval retention by the Wairarapa Eddy off New Zealand. Marine Ecology Progress Series 183: 227–240.
Chiswell, S.M., and D. Roemmich. 1998. The East Cape Current and two eddies: A mechanism for larval retention? New Zealand Journal of Marine and Freshwater Research 32: 385–397.
Cognetti, G., and F. Maltagliati. 2000. Biodiversity and adaptive mechanisms in brackish water fauna. Marine Pollution Bulletin 40: 7–14.
Dawson, M.N. 2001. Phylogeography in coastal marine animals: A solution from California? Journal of Biogeography 28: 723–736.
Dawson, M.N. 2005. Incipient speciation of Catostylus mosaicus (Scyphozoa, Rhizostomeae, Catostylidae), comparative phylogeography and biogeography in south-east Australia. Journal of Biogeography 32: 515–533.
DeBoer, T.S., M.D. Subia, M.V. Ambariyanto, K.K. Erdmann, and P.H. Barber. 2008. Phylogeography and limited genetic connectivity in the endangered boring giant clam across the coral triangle. Conservation Biology 22: 1255–1266.
Díaz-Ferguson, E., J.D. Robinson, B. Silliman, and J.P. Wares. 2010. Comparative phylogeography of North American Atlantic salt marsh communities. Estuaries and Coasts 33: 828–839.
Dupanloup, I., S. Schneider, and L. Excoffier. 2002. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology 11: 2571–2581.
Eanes, W.F. 1999. Analysis of selection on enzyme polymorphisms. Annual Review of Ecology and Systematics 30: 301–326.
Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1: 47–50.
Fleming, C.A. 1979. The geological history of New Zealand and its life. Auckland: Auckland University Press.
Folmer, O., W.R. Hoeh, M.B. Black, and R.C. Vrijenhoek. 1994. Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Molecular Marine Biology and Biotechnology 3: 294–299.
Fontanillas, P., A. Depraz, M.S. Giorgi, and N. Perrin. 2005. Nonshivering thermogenesis capacity associated to mitochondrial DNA haplotypes and gender in the greater white-toothed shrew, Crocidura russula. Molecular Ecology 14: 661–670.
Fu, Y.X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925.
Fu, Y.X., and W.H. Li. 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709.
Gawarkiewicz, G., S. Monismith, and J. Largier. 2007. Observing larval transport processes affecting population connectivity: Progress and challenges. Oceanography 20: 40–53.
Goldstien, S.J., D.R. Schiel, and N.J. Gemmell. 2006. Comparative phylogeography of coastal limpets across a marine disjunction in New Zealand. Molecular Ecology 15: 3259–3268.
Goudet, J., M. Raymond, T. Demeeus, and F. Rousset. 1996. Testing differentiation in diploid populations. Genetics 144: 1933–1940.
Grantham, B.A., G.L. Eckert, and A.L. Shanks. 2003. Dispersal potential of marine invertebrates in diverse habitats. Ecological Applications 13: S108–S116.
Hadfield, M.G., G.J. Rickard, and M.J. Uddstrom. 2007. A hydrodynamic model of Chatham Rise, New Zealand. New Zealand Journal of Marine and Freshwater Research 41: 239–264.
Hare, M.P., and J.C. Avise. 1996. Molecular genetic analysis of a stepped multilocus cline in the American oyster (Crassostrea virginica). Evolution 50: 2305–2315.
Heath, R.A. 1982. A review of the physical oceanography of the seas around New Zealand. New Zealand Journal of Marine and Freshwater Research 19: 79–124.
Jones, T.C., C.E.C. Gemmill, and C.A. Pilditch. 2008. Genetic variability of New Zealand seagrass (Zostera muelleri) assessed at multiple spatial scales. Aquatic Botany 88: 39–46.
Kelly, D.W., H.J. Macisaac, and D.D. Heath. 2006. Vicariance and dispersal effects on phylogeographic structure and speciation in a widespread estuarine invertebrate. Evolution 60: 257–267.
Kennish, M.J. 2002. Environmental threats and environmental future of estuaries. Environmental Conservation 29: 78–107.
Knox, M.A., I.D. Hogg, and C.A. Pilditch. 2011. Mitochondrial DNA (COI) variability and phylogeography of Paracorophium (Crustacea: Amphipoda) in New Zealand estuaries. Biological Journal of the Linnean Society. doi:10.1111/j.1095-8312.2011.01675.x.
Kuo, C.H., and J. Avise. 2005. Phylogeographic breaks in low-dispersal species: The emergence of concordance across gene trees. Genetica 124: 179–186.
Laing, A., and S.M. Chiswell. 2003. The ocean medium. In The living reef: The ecology of New Zealand’s rocky reefs, ed. N.L. Andrew and M.P. Francis, 24–31. Nelson: Craig Potton.
Lamare, M.D. 1998. Origin and transport of larvae of the sea urchin Evechinus chloroticus (Echinodermata: Echinoidea) in a New Zealand fiord. Marine Ecology Progress Series 174: 107–121.
Largier, J.L. 1993. Estuarine fronts: How important are they? Estuaries 16: 1–11.
Levin, L.A. 2006. Recent progress in understanding larval dispersal: New directions and digressions. Integrative and Comparative Biology 46: 282–297.
Librado, P., and J. Rozas. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphisms data. Bioinformatics 25: 1451–1452.
Lidgard C.W. 2001. Evaluating the population genetic structure of the New Zealand cockle Austrovenus stutchburyi using allozyme electrophoresis, 45 pp. MSc thesis, University of Waikato.
Little, K.T., and C.E. Epifanio. 1991. Mechanism for the re-invasion of an estuary by two species of brachyuran megalopae. Marine Ecology Progress Series 68: 235–242.
Lundquist, C.J., J.W. Oldman, and M.J. Lewis. 2009. Predicting suitability of cockle Austrovenus stutchburyi restoration sites using hydrodynamic models of larval dispersal. New Zealand Journal of Marine and Freshwater Research 43: 735–748.
Mann, R. 1988. Distribution of bivalve larvae at a frontal system in the James River, Virginia. Marine Ecology Progress Series 50: 29–44.
Oliveira, D., R. Raychoudhury, D.V. Lavrov, and J.H. Werren. 2008. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Molecular Biology and Evolution 25: 2167–2180.
Ovenden, J.R., D.J. Brasher, and R.W.G. White. 1992. Mitochondrial-DNA analyses of the red rock lobster Jasus edwardsii supports an apparent absence of population subdivision throughout Australasia. Marine Biology 112: 319–326.
Paula, J. 1998. Larval retention and dynamics of the prawns Palaemon longirostris H. Milne Edwards and Crangon crangon Linnaeus (Decapoda, Caridea) in the Mira estuary, Portugal. Invertebrate Reproduction and Development 33: 221–228.
Pechenik, J.A. 1999. On the advantages and disadvantages of larval stages in benthic marine invertebrate life cycles. Marine Ecology Progress Series 177: 269–297.
Pelc, R.A., R.R. Warner, and S.D. Gaines. 2009. Geographical patterns of genetic structure in marine species with contrasting life histories. Journal of Biogeography 36: 1881–1890.
Perrin, C., S.R. Wing, and M.S. Roy. 2004. Effects of hydrographic barriers on population genetic structure of the sea star Coscinasterias muricata (Echinodermata, Asteroidea) in the New Zealand fiords. Molecular Ecology 13: 2183–2195.
Piggott, M.P., S.C. Banks, P. Tung, and L.B. Beheregaray. 2008. Genetic evidence for different scales of connectivity in a marine mollusc. Marine Ecology Progress Series 365: 127–136.
Posada, D. 2008. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256.
Puebla, O., E. Bermingham, and F. Guichard. 2009. Estimating dispersal from genetic isolation by distance in a coral reef fish (Hypoplectrus puella). Ecology 90: 3087–3098.
Richards, C.L., J.P. Wares, and J.A. Mackie. 2010. Evaluating adaptive processes for conservation and management of estuarine and coastal resources. Estuaries and Coasts 33: 805–810.
Ross, P.M., I.D. Hogg, C.A. Pilditch, and C.J. Lundquist. 2009. Phylogeography of New Zealand's coastal benthos. New Zealand Journal of Marine and Freshwater Research 43: 1009–1027.
Schmidt, P.S., E.A. Serrao, G.A. Pearson, C. Riginos, P.D. Rawson, T.J. Hilbish, S.H. Brawley, G.C. Trussell, E. Carrington, D.S. Wethey, J.W. Grahame, F. Bonhomme, and D.M. Rand. 2008. Ecological genetics in the North Atlantic: Environmental gradients and adaptation at specific loci. Ecology 89: S91–S107.
Shears, N.T., F. Smith, R.C. Babcock, C.A.J. Duffy, and E. Villouta. 2008. Evaluation of biogeographic classification schemes for conservation planning: Application to New Zealand's coastal marine environment. Conservation Biology 22: 467–481.
Smith, P.J., and J.L. Mckoy. 1980. Genetic variation in the rock lobsters Jasus edwardsii and Jasus novaehollandiae. New Zealand Journal of Marine and Freshwater Research 14: 55–63.
Sotka, E.E., J.P. Wares, J.A. Barth, R.K. Grosberg, and S.R. Palumbi. 2004. Strong genetic clines and geographical variation in gene flow in the rocky intertidal barnacle Balanus glandula. Molecular Ecology 13: 2143–2156.
Sponer, R., and M.S. Roy. 2002. Phylogeographic analysis of the brooding brittle star Amphipholis squamata (Echinodermata) along the coast of New Zealand reveals high cryptic genetic variation and cryptic dispersal potential. Evolution 56: 1954–1967.
Stevens, P.M. 1991. A genetic-analysis of the pea crabs (Decapoda, Pinnotheridae) of New Zealand. 2. Patterns and intensity of spatial population-structure in Pinnotheres astrinicola. Marine Biology 108: 403–410.
Stevens, M.I., and I.D. Hogg. 2004. Population genetic structure of New Zealand's endemic corophiid amphipods: Evidence for allopatric speciation. Biological Journal of the Linnean Society 81: 119–133.
Swofford, D.L. 2000. PAUP*, phylogenetic analysis using parsimony (*and other methods), Version 4. Sunderland: Sinauer Associates.
Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Thorson, G. 1950. Reproductive and larval ecology of marine bottom invertebrates. Biological Reviews 25: 1–45.
Thrush, S.F., R.B. Whitlatch, R.D. Pridmore, J.E. Hewitt, V.J. Cummings, and M.R. Wilkinson. 1996. Scale-dependent recolonization: The role of sediment stability in a dynamic sandflat habitat. Ecology 77: 2472–2487.
Thrush, S.F., C.J. Lundquist, and J.E. Hewitt. 2005. Spatial and temporal scales of disturbance to the seafloor: A generalized framework for active habitat management. In Benthic habitats and the effects of fishing. American Fisheries Society Symposium, 41, ed. B.W. Barnes and J.P. Thomas, 639–649. Bethesda: American Fisheries Society.
Veale A. 2007. Phylogeography of two intertidal benthic marine invertebrates around New Zealand: The waratah anemone (Actinia tenebrosa) and the snakeskin chiton (Sypharochiton pelliserpentis), 147 pp. M.Sc. thesis, University of Auckland.
Waters, J.M., and M.S. Roy. 2003. Marine biogeography of southern Australia: Phylogeographical structure in a temperate sea-star. Journal of Biogeography 30: 1787–1796.
Waters, J.M., and M.S. Roy. 2004. Phylogeography of a high-dispersal New Zealand sea-star: Does upwelling block gene-flow? Molecular Ecology 13: 2797–2806.
Watts, R.J., and M.S. Johnson. 2004. Estuaries, lagoons and embayments: Habitats that enhance population subdivision in fishes. Marine and Freshwater Research 55: 641–651.
Wright, S. 1943. Isolation by distance. Genetics 28: 114–138.
Acknowledgments
We thank two anonymous reviewers whose thoughtful and constructive comments greatly improved the manuscript. We also thank Jonathan Banks, Alan Beu, Steve Chiswell, Nick Shears and Basil Stanton for discussions on population genetics, geology, oceanography and biogeography. Louise Chilvers, Nick Demetras, Hannah Jones, Helen Kettles, Matt Knox, Jenn Logan, John Longmore, Rebecca McLeod, Don Neale, Hazel Needham, Clarisse Niemand, Darren Parsons, Nadeesha Perera, Allison Smith, Tracey Smith, Mike Taylor, Dale Williams and Cilla Wehi provided assistance in the field or laboratory. Funding was provided by the University of Waikato Doctoral Scholarship, the Foundation for Research, Science and Technology (contracts C01X0502 and UOWX0505), a New Zealand Marine Science Society Student Research Grant, a Valder Conservation Grant and a grant from the School of Science and Engineering, University of Waikato.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ross, P.M., Hogg, I.D., Pilditch, C.A. et al. Population Genetic Structure of the New Zealand Estuarine Clam Austrovenus stutchburyi (Bivalvia: Veneridae) Reveals Population Subdivision and Partial Congruence with Biogeographic Boundaries. Estuaries and Coasts 35, 143–154 (2012). https://doi.org/10.1007/s12237-011-9429-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12237-011-9429-z