Abstract
Identification of quantitative trait loci (QTL) responsible for stress tolerance could help to develop new tolerant potato cultivars through markers-assisted selection. The objective of the research was to identify and map loci that may play major role in the control of osmotic stress tolerance in potato under in vitro vegetative growth conditions. Mannitol-induced osmotic stress response of cv. White Lady and breeding line S440 as well as their 85F1 progeny for root number and root length under in vitro conditions was investigated. Interval mapping based on 200 informative markers produced a molecular marker map comprising 13 linkage groups for White Lady and 14 for S440. In total, 14 QTLs with LOD > 2 were identified from which 6 were confirmed as major QTLs. Of those six, three were associated with root length and three with root number. For root length, one major QTL was identified on chromosome XII (LOD = 4.8), which explained 52.3 % of the phenotypic variance. Two other major QTLs from an unidentified chromosome explained 64.9 and 51 % of the phenotypic variance with LODs of 5.34 and 4.8, respectively. For root number, one QTL with 19.2 % of the phenotypic variance (LOD of 2.9) was tentatively identified on chromosome IX, while another was putatively identified on chromosome XII (LOD of 2.4), together explaining 26.8 % of the phenotypic variance.
Resumen
La identificación de loci de rasgos cuantitativos (QTL) responsables de la tolerancia al agobio podría ayudar a desarrollar nuevas variedades de papa tolerantes mediante selección asistida por marcadores. El objetivo de la investigación fue identificar y crear un mapa de loci que pudiera jugar un papel importante en el control de la tolerancia al agobio osmótico en papa bajo condiciones de crecimiento vegetativo in vitro. Se investigó la respuesta del agobio osmótico inducido por manitol de la variedad White Lady y de la línea de mejoramiento S440, así como de su progenie 85 de F1, para número y longitud de raíces, bajo condiciones in vitro. Mapeando a intervalos con base en 200 marcadores informativos se produjo un mapa de marcadores moleculares compuesto por 13 grupos ligados para White Lady y 14 para S440. En total, se identificaron 14 QTL con LOD > 2, de los cuales 6 fueron confirmados como QTL’s mayores. De esos seis, se asociaron tres con longitud de raíz y tres con número de raíces. Para longitud de raíz, se identificó un QTL mayor en el cromosoma XII (LOD = 4.8), lo que explicó 52.3 % de la varianza fenotípica. Otros dos QTL mayores de un cromosoma no identificado explicaron 64.9 y 51 % de la varianza fenotípica con LODs de 5.34 y 4.8, respectivamente. Para el número de raíces, un QTL con 19.2 % de la varianza fenotípica (LOD de 2.9) se identificó tentativamente en el cromosoma IX, mientras que otro se identificó presumiblemente en el cromosoma XII (LOD de 2.4), juntos explicando 26.8 % de la varianza fenotípica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Compared with other species, potato is very sensitive to water-stress because of its shallower root system (Iwama and Yamaguchi 2006), and has been classified as moderately salt-tolerant to moderately salt-sensitive (Maas 1985). Improvement in root traits (root number and root length) is considered to be important for developing osmotic stress tolerant genotypes (Rossouw and Waghmarae 1995; Iwama and Yamaguchi 2006; Lahlou and Ledent 2005). Larger and deeper roots have been shown to contribute to osmotic tolerance in many crops, including potato (Schafleitner et al. 2007; Lahlou and Ledent 2005). The accumulation of polyols (mannitol, sorbitol, inositol and their derivatives) is considered to be related to drought and salinity stress tolerance in many plant species (Peuke et al. 2002; Sakthivelu et al. 2008; Ehsanpour and Razavizadeh 2005; Mohamed et al. 2000; Watanabe et al. 2000; Dobranszki et al. 2003).
The evaluation of osmotic stress tolerance in potato genotypes (Solanum spp.) in conventional field trials is rather time consuming and labour intensive. The results are often confounded by many field and environmental variations (Ingram et al. 1994; Erusha et al. 2002; Iwama and Yamaguchi 2006; Georgieva et al. 2004). Simulation of osmotic stress under in vitro tissue culture conditions can minimize environmental variation due to the use of defined nutrient media, controlled conditions and homogeneity of stress application. In 1993, Leport et al. stated that in vitro selection for stress tolerance will have a significant place in the strategy for establishing plant systems with optimal stress reaction and output in the future. Applying osmotic stress during the regeneration phase was found to be the most efficient for the selection for drought tolerance (Hsissou and Bouharmont 1994). Gopal and Iwama (2007) investigated in vitro screening of potato against osmotic stress mediated through sorbitol and mannitol. Their results demonstrated that osmotic factors in culture media adversely affected plantlet growth, and genotypes differed in their responses. They concluded that in vitro screening of potato under specific and limited osmotic stress conditions may provide a system capable of effectively differentiating genotypes for their expected root mass production under field conditions.
Abiotic stress tolerance in plants is known to be a quantitatively inherited trait. It is generally under the control of a small number of quantitative trait loci (QTLs). The identified advantageous marker alleles could help to introduce new varieties having higher tolerance to osmotic stress by marker assisted selection of genotypes with higher phenotypic value (Bálint et al. 2008). Recently, progress has been made in developing the theory of linkage analysis and quantitative trait locus mapping in autotetraploid species for a full-sib family derived from crossing of two parents (Luo et al. 2001; Hackett et al. 2001). Bradshaw et al. (2008) applied interval mapping of quantitative trait loci for 16 yield, agronomic and quality traits in potato, in total identifying 39 QTLs.
Genetic linkage maps are essential to identify a QTL and are valuable in basic genetic studies or in applied breeding programs, especially for the identification and selection of genotypes with specific combinations of favourable traits. To develop a linkage map by classical genetic markers such as morphological traits, several segregating populations are needed, as only a limited number of loci segregates in each population. With the initiation of molecular markers, unlimited amounts of segregating loci have become available from a single cross. For outbreeding species like potato, heterozygous parents are used to obtain segregating populations, and mapping can be carried out in the F1 progeny of a single cross.
In the present study our aim was to identify and map major QTLs affecting the osmotic stress tolerance in tetraploid potato during vegetative growth under in vitro conditions. Osmotic stress tolerance was characterized by root length and root number, namely, the higher these values the higher the osmotic stress tolerance of that genotype was considered. According to our knowledge, this is the first report to applying SCoT markers (Collard and Mackill 2009) and Intron targeting markers, for the construction of a linkage map in tetraploid potato.
Materials and Methods
Plant Material
The mapping population used in this study was obtained from a single cross between Hungarian potato cultivar White Lady (WL) and the breeding line S440 (male parent). Based on their reaction under field conditions, these parental genotypes differ in their reaction to natural heat and drought stress. WL is more tolerant than S440. Eighty five F1 genotypes were randomly selected from the segregating population and used to investigate osmotic stress, QTL analysis and mapping.
Osmotic Stress Assay
To discriminate stress sensitive and tolerant parents, the in vitro osmotic stress conditions were optimized. At first 40, three leaf stage shoot tips of WL and S440 grown on basic MS media were cultured individually in glass tubes containing 10 ml basic MS salts medium supplemented with different concentration of mannitol (0 (without mannitol) as control, 0.15 mol/dm3, 0.2 mol/dm3 and 0.3 mol/dm3, ten plants/concentration) at 16/8 hours illumination and 20 °C. The study was carried out as a factorial experiment based on a completely randomized design, 10 replications and two factors. After 10 days, data were recorded for root number and root length. In the second experiment osmotic stress tolerance of 85 F1 genotypes was tested under the same conditions but only at the selected 0.3 M concentration of mannitol.
Genomic DNA Isolation
Genomic DNA was extracted from 80 mg of leaf and stem tissue of in vitro plants using the procedure of Walbot and Warren (1988).
Molecular Marker Assays
A total of 57 primer pairs including 13 SCoT, 10 Intron targeting (IT), 11 ISSR, 2 SSR, 1 SCAR and 20 RAPD were used to discover polymorphic fragments using an Eppendorf Mastercycler ep 384 (Eppendorf, Germany) and a Robocycler (Stratagene, USA).
IT Analysis
PCR was conducted with a final reaction volume of 12 μL. PCR reaction mixture contained 40 ng DNA as template, 1.2 μl from each 10 PicoM 12-mer primer, 1.2 μl 2 mM dNTP (Fermentas, Lithuania), 1.5 μl 10× PCR buffer (1 mM Tris-HCl, pH 8.8 at 25 °C, 1.5 mM MgCl2, 50 mM KCl and 0.1 % Triton X-100) and 0.29 U of DyNAzyme II (Finnzymes, Finland) polymerase. The reaction was performed based on the following profile: 3 min at 94 °C, followed by 35 cycles at 94 °C for 1 min, 51 °C for 1 min and 72 °C for 1 min. The final extension step was at 72 °C for 10 min. Amplified PCR products were mixed with 5 μl BPB dye (99.5 % de-ionsied formamide, 10 mM EDTA pH 8, 0.05 % bromphenol-blue, xylene-cyanol dye solution, 1 μl steril H2O) and separated on 1.5 % agarose gels (Promega, USA) in 0.5× TBA ( Tris-HCl, Boric acid, EDTA). After electrophoresis, amplified bands were visualized by ethidium-bromide staining, and documented with a GenGenius Bio Imaging System (Syngene, UK).
SCAR Analysis
PCR reaction mixture, analysis and documentation of amplified products were conducted as described for IT primers. Amplification conditions were 1 min initial denaturation step at 94 °C, followed by 35 cycles of 30 s at 94 °C, 1 min at 54 °C, and 1 min at 72 °C. The reactions were completed by a final extension step of 10 min at 72 °C.
SSR Analysis
The simple sequence repeat (SSR) markers (Milbourne et al. 1998) localized on potato chromosome XI and XII were tested. The PCR reaction was performed using 25 μL reaction mixtures in a Robocycler (Stratagene, USA) with 96-well microtiter plates containing the following components: 50 ng DNA, 0.5 μL 10 mM dNTP (Fermentas, Lithuania), 2.5 μL 10× Taq buffer (Fermentas), 2 μL 25 mM MgCl2 (Fermentas), 1 μl from each 10 PicoM primer, 0.5 u Taq DNA polymerase (Fermentas). PCR was carried out by initially denaturing template DNA at 94 °C for 4 min, followed by 35 cycles at 94 °C for 30 s, 51 °C for 30 s, and 72 °C for 1 min. The final extension step was at 72 °C for 5 min. PCR products were separated on a 1.5 % agarose gel (Promega, USA) in 0.5× TBE buffer and were stained with ethidium-bromide.
SCoT, ISSR and RAPD Analysis
Amplification conditions, PCR reaction mixture, analysis and documentation of amplified products were conducted according to Gorji et al. (2011).
Construction of Linkage Map
Preliminary cluster analysis was done on each parent for the markers identified as simplex, using the simple matching coefficient. These identified markers were located on the same chromosome. All simplex, duplex and multi-allelic markers were then analyzed by group average cluster analysis to partition them into LGs (Luo et al. 2001). Markers were analysed on the two parents separately. For each LG, recombination frequencies and LOD scores between every pair of markers were calculated for all possible phases using the expectation-maximization (EM) algorithm, as described by Luo et al. (2001). A simulated annealing algorithm (Hackett et al. 2003) was used to identify the order with the minimum value of the weighted least squares criterion (Stam 1993) and to calculate map distances between the markers. A permutation test (Churchill and Doerge 1994) was used to establish a 99 % threshold for declaring a simplex to double-simplex linkage. TetraploidMap software was used to analyse data (Hackett and Luo 2003).
Statistical Analysis
Amplified products were scored as present (1) or absent (0) to form a binary matrix. Ambiguous bands were discarded. QTL analysis, detection of homologous chromosomes and a permutation test were performed using TetraploidMap software (Hackett and Luo 2003). Statistical analysis was carried out using the SPS V11.5 and SAS system V8.
Results
Investigation of Parents
Results of the factorial analysis showed that under the examined conditions the mean root number was significantly different between WL (6.98/plant) and S440 (5.93/plant) (p> <alpha>= 0.05). WL had the same root number irrespective of applied mannitol concentration, while the root number of S440 gradually decreased with increasing osmotic stress. However, this decrease was significant for 0.3 mol/dm3 mannitol only. The root number for S440 was greater than for WL under the 0.0 mol/dm3 mannitol concentration (control condition), but was not significant. However, root number was lower at each level of osmotic stress, indicating its higher susceptibility. The results also revealed that the difference between WL and S440 was only significant under severe osmotic stress (Table 1).
For root length, the two parents did not differ from each other significantly at any of the tested mannitol concentrations. However, the increasing mannitol concentration significantly decreased the root length for both parents. The highest decrease was recorded at 0.3 mol/dm3 mannitol. The result also showed that at each level of osmotic stress the root length of S440 was higher than that of WL but this difference was not significant (Table 1).
F1 Genotypes
The variance analysis indicated that the effect of osmotic stress on root number was highly significant at genotype level (p < 0.0001). WL, genotype 457, 103 and 460 produced significantly more roots than the others. Altogether, 15 genotypes showed greater values than the parent S440 but it was only significant for genotype 457, 103 and 460.
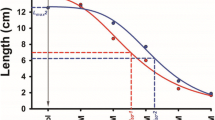
For root length, significant differences were detected between genotypes (p < 0.0001). Thirteen and nineteen genotypes had longer roots than S440 and WL, respectively. However, it was only significant for genotype 448 (Fig. 1). In general, genotypes with a higher number of roots usually also had longer roots (Fig. 1). Analysis of regression for curve estimation between root number and root length showed that observed data are fitted to a quadratic trend (Fig. 2). A Pearson correlation coefficient revealed that there is a positive and significant correlation between root number and root length (r = 0.66).
Curve estimation between root number and root length using White Lady, S440 and 85 F1 genotypes. The regression lines for the linear and quadratic trends are described by the equations: \( {\text{RN}} = 0.923 + 1.587 \times \,{\text{and}}\,{\text{RN}} = - 0.181 + 3.4 \times - 0.61{ \times^2} \), where RN is root number and x is root length (r 2 = 0.60)
Construction of Genetic Linkage Map
White Lady
Eighty nine markers were used to construct a partial linkage map of WL. Of these, 65 segregated in a 1:1 ratio (P < 0.001) and 24 segregated in a 5:1 ratio with P < 0.01. A single linkage clustering and an average linkage clustering were used to recognize the number of linkage groups. The result of single linkage clustering identified 12 groups at a similarity of 0.93. The average linkage clustering formed 12 LGs at a similarity of 0.31. Therefore, a linkage map was built on the distribution of 104 (15 ITs, 2 SSRs, 8 SCoTs, 5 ISSRs, 1 SCAR and 58 RAPDs) markers into 12 co-segregation groups with 2–20 markers. Any marker that showed a poor fit in the ordering was moved to another group, to a new group or was excluded altogether, leading to the construction of 13 LGs. IT and SSR markers were used to identify different LGs corresponding to the chromosomes. Following this analysis, three LGs were identified corresponding to three chromosomes:
Chromosome VII
This group has an IT marker (f79) which was amplified with a primer pair that was designed based on the EST sequence having full homology to chromosome VII of potato. F79 was found to be a simplex marker and linked in repulsion with the duplex marker f52 and simplex marker f99. The LOD and recombination frequency (RF) of f79 with f99 were 0.59 and 0.21, and those were 1.21 and 0.26 with f52, respectively. The map is shown in Fig. 3(i).
Overall Linkage Groups and QTLs (highlighted). The two parents are shown separately but the four homologous chromosomes are merged into a single map. Anchore markers are underlined, WL white lady, a LGIIa (Chromosome IX), b LGIIb (Chromosome IX, c LGVII (Chromosome XII), d LGX, e LGII (Chromosome XI), f LGV, g LGIX (Chromosome XII), h LGX, i LGXII (chromosome VII)
Chromosome XI
An anchor-IT marker, f78, and four other IT markers and a RAPD marker were found in this group. The primers for identification of f78 were designed based on the Ry1In gene sequence, which shows high homology to chromosome XI of potato. F78 was found to be a simplex marker and linked in coupling with the duplex RAPD marker f42, and in repulsion with 2 simplex markers (f84 and f89) and 2 duplex markers (f83 and f87). F83, f84, f87 and f89 were also IT markers but the chromosomal position of those markers could not be determined by the homology test. The range of LOD score and recombination frequency between f78 and other markers were from 0.57 to 0.93 and from 0.002 to 0.28, respectively. The map is shown in Fig. 3(e).
Chromosome XII
This LG includes 10 markers of which 5 are IT, 1 SSR, 1 SCAR, and 3 RAPD. The Rysto gene was previously also located in this group. F23 (Cat260) is an anchor marker for chromosome XII (Cernak et al. 2008) and maps in the same location with f24 (IT) and f25 (SCAR) markers. F23 is a simplex marker and linked in coupling with 7 simplex markers (f28, f37, f80, f26, f24, f25 and f27) and a duplex marker f77 (RAPD). F23 also linked in repulsion with the duplex marker f58 (RAPD). The LOD score of f23 with other markers in this group was usually high. The maximum LOD was in association with f24 and f25 (22.58) and the minimum was associated with f58 (0.41). The range of recombination frequency between f23 and other markers was from 0 (f24 and f25) to 0.31 (f58). F26 (STM0003) is also an anchor marker for chromosome XII (Milbourne et al. 1998) and maps at 6 cM apart from f23 and was 5 cM distant from Rysto on the other side. F26 is also a simplex marker and linked in coupling with 7 simplex markers (f28, f37, f80, f23, f24, f25 and f27) and a duplex marker f77. F26 also linked in repulsion with the duplex marker f58. The minimum and maximum LOD between f26 and other markers in this group ranged from 0.98 (f77) to 14.14 (f23, f24 and f25), and the recombination frequency also ranged from 0.0029 (f77) to 0.29 (f28) (data not shown). The map is shown in Fig. 3(g).
S440
A total of 111 markers were used to construct the genetic linkage map of S440. Among them 95 were found to be simplex, while 16 were duplex markers. The result of the single linkage clustering revealed 12 co-segregation groups at a similarity of 0.96. The average linkage clustering formed 12 linkage groups at a similarity of 0.33. Therefore, we first created a linkage map with 12 homology group comprising 3–23 markers. Two groups were subsequently divided, following more detailed assessment of recombination frequencies inside the groups. From 14 LGs, we recognized three linkage groups corresponding to two chromosomes (IX and XII) based on the IT markers. The map is shown in Fig. 3.
Chromosomes Identified by ITs
Chromosome IX
A simplex (m100) and a duplex (m101) IT marker with high homology to chromosome IX mapped in different LGs. We assume that these two LGs represent chromosome IX, and were marked with A and B until supplementary marker data are obtained (Fig. 3a and b). The m101 is a duplex marker and is linked in coupling with two duplex (m48 and m129) and two simplex markers (m54 and m58). The m57 simplex marker was also linked in repulsion with m101. The recombination frequency of m101 with other markers in this LG was low and ranged from 0.0006 to 0.132. The LOD was scored from 1.32 to 6.92. All markers linked with m101 were RAPDs except m129, which was an ISSR marker. The m100 is a simplex marker and linked in repulsion with seven other simplex markers (m24, m82, m49, m61, m27, m16 and m45), of which m16 and m24 are ISSR and the remaining are RAPD markers. The recombination frequency of m100 with other markers ranged from 0.03 to 0.24. The LOD between m100 with other markers was generally low and scored from 0.45 to 1.53.
Chromosomes XII
The simplex IT marker m97 identifies and aligns the S440 map of chromosome XII (Fig. 3c). One duplex and 6 simplex markers are linked in repulsion with m97. Four simplex markers are also linked in coupling with m97. The recombination frequency of m97 with other markers in this group ranged from 0.21 to 0.44. Only the LOD score of two markers (m33 and m71) were more than 3 in connection to m97. We estimate that the designation of these as chromosome XII is tentative (because of its RF) until further mapping data are obtained.
QTL mapping
Root Length
Nine QTLs with LOD > 2 were identified for root length. From those, six were for WL and three were for S440. The results of a permutation test confirmed 3 of those as major QTLs (Fig. 4). All these QTLs have two copies of dominant alleles for increasing resistance to osmotic stress. From these QTLs, 2 are in homology groups V and IX of WL and one is in homology group X of S440. The homology group IX was identified as chromosome XII using anchor markers, as outlined above. The QTLs on linkage group V of WL and X of S440 were closely linked in coupling to simplex ISSR marker f13 (2 cM) and SCoT marker m12 (4 cM), and individually explain 64.9 and 54.8 % of phenotypic variance with maximum LOD 5.34 and 5.1, respectively. The QTL on chromosome XII of WL was closely linked in coupling to RAPD marker f58 (2 cM) and individually explains 52.3 % of the phenotypic variance with maximum LOD 4.8. It is also closely linked to a QTL (RN1) of root number (<0.3 cM) that was not recognized as a major QTL based on a permutation test but individually explains 29.74 % of phenotypic variance with LOD 2.23. The locations of the QTLs are shown in Fig. 3. The positions, LOD scores and percentage phenotypic variance explained for the full model of six QTL genotypes is shown in Table 2.
Results of a permutation test for major QTLs. a, b and d show the LOD profile for root length. c, e and f show the LOD profile for root number. 90 % and 95 % thresholds are shown as horizontal lines in the upper panel. FM profile of full model, SM profile of simple model. LG linkage group, WL White Lady
Root Number
Five QTLs with LOD > 2 were detected for root number. From these QTLs, 3 were identified as being in White Lady and 2 in S440. Three of the QTLs were recognized as major QTLs based on the permutation test results (Fig. 4). Two of these major QTLs were in S440 and one in WL. The QTL in WL is in homology group X and closely linked in coupling to duplex RAPD markers f54 (3 cM) and f20 (4 cM) and confer resistance to osmotic stress. It has two copies of dominant alleles on a chromosome in homology group X and maps in 46 cM of a QTL of root length (RL4), which explains 54.5 % of the phenotypic variance with a maximum LOD of 3.92. The maximum LOD and variance explained for the QTL related to root number in that linkage group were 2.87 and 19.19 %, respectively (Table 2.). There are two copies of dominant alleles on chromosomes in homology groups VII and X of S440. The first one narrowly linked in coupling to simplex RAPD markers m33 (2 cM) and m51 (2 cM). It individually explains 26.8 % of the phenotypic variance with maximum LOD 2.4. It is also mapped closely (2 cM) to a QTL (RL8) for root length, which explains 38.4 % of the phenotypic variance with LOD 2.65. The QTL in homology group X individually explains 43.1 % of phenotypic variance (LOD 3.14) and closely (3 cM) linked in coupling to the simplex ISSR marker m123. It is linked to a major QTL of root length (RL9) that explains 54.76 % of variance with maximum LOD 5.1. Linkage between QTLs of root number and root length infer that they could in fact be the same allele as the two traits are associated. However, there are inadequate bridging markers to decide whether the QTLs in homology group X are the same locus in the two parents.
Assessment of Markers Closely Mapped to QTLs
Comparison of molecular data for markers which closely map to major QTLs and the result of in vitro assessment using 15 tolerant and 15 susceptible genotypes revealed that marker m12 was able to verify 55 % of in vitro results. The rate for f13, f20, f58, m51 and f54 were 55, 50, 45, 42 and 40 %, respectively.
Discussion
Osmotic stresses are among the major abiotic stress factors affecting significantly the success of plant production. Evaluation of osmotic stress tolerance/susceptibility is an essential step in the process of plant improvement. In the present study, we evaluated the osmotic stress response of two potato genotypes and their 85 F1 progenies (cv.WL, female parent and S440, male parent) under in vitro condition. The results showed that there is a difference between the parents in term of root number and root length when different concentrations of mannitol were used. Related to root number, S440 proved to be more susceptible to osmotic stress than WL. This phenomenon is in agreement with field behaviour of parents where WL has less tuber defects and lower yield decrease to drought and heat stresses. The concentration of 0.3 mol/dm3 mannitol was found to be appropriate for the discrimination of sensitive and tolerant genotypes to osmotic stress based on factorial analysis. For root number, the difference between sensitive and tolerant potato cultivars under in vitro and osmotic stress conditions was previously reported by Dobranszki et al. (2003). Our result revealed that root length is more susceptible to osmotic stress than root number. In our study there was no significant difference between WL and S440 for root length at different levels of mannitol. As the root number of S440 was comparable to WL under normal conditions, and was significantly less at the maximum concentration of mannitol, we assume that a higher concentration of mannitol (above 0.3 mol/dm3) may also discriminate WL and S440 based on root length. Root mass should be increased in tolerant potatoes under osmotic stress. Larger and deeper roots have been shown to contribute to drought tolerance in potato and many other crops (Schafleitner et al. 2007; Lahlou and Ledent 2005). Maruyama et al. (2008) declared that the root-bending ratio of lettuce (Lactuca sativa L.), which is an indicator of growth sustainability under stress (Howden and Cobbett 1992; Wu et al. 1996), was not affected at mannitol concentrations up to 5.0 % (~280 mM) but declined with increasing mannitol concentrations ranging from 5.0 % to 12.0 %.
The result of the F1 genotypes showed that a significant difference (<0.0001) exists between genotypes for root number and root length at 0.3 M mannitol concentration. Transgressive segregation for root length was found in the present population and only one genotype (448) produced significantly longer roots than the parents. Lilley et al. (1996) previously reported a transgressive segregation in rice under osmotic stress condition.
Genetic Linkage Map and QTLs
A genetic linkage map represents the relative order of genetic markers along chromosomes. The first genetic linkage map in a non-inbred species was developed for potato, using the segregation data from only one of the parents (Bonierbale et al. 1988). Later, Gebhardt et al. (1989) used the approach with allelic bridges to align the parental maps. Other genetic linkage maps in potato were constructed by Jacobs et al. 1995; Van Eck et al. 1995; Milbourne et al. 1998; Brugmans et al. 2006; Bradshaw et al. 2008, and have enabled the localization of resistance genes, quality traits and QTLs. A complete linkage map of potato would comprise 12 sets of four homologous chromosomes (homology or LGs), and the 12 LGs would be numbered from I to XII (Dong et al. 2000; Celebi-Toprak et al. 2005; Bradshaw et al. 2008). In the present study, 13 LG groups were established for WL and 14 LG groups for S440 after omitting double-simplex markers. Double-simplex markers are very uninformative about recombination and should be omitted from the linkage maps of the parents (Bradshaw et al. 2008). The 13 WL groups (total map length 951 cM) were aligned with those from S440 (total map length 1,096 cM). There were insufficient bridging markers between WL and S440 to be accurately aligning the parental maps. For that reason QTL models were also fitted to the two parents separately. The same result was previously reported by Bradshaw et al. (2008) for tetraploid potato. Furthermore the map we developed was not an improvement on the previously published one and, consequently, we could not compare our map with it.
The linkage map for chromosomes XII was the most appropriate because it was identified by two well characterized chromosome specific IT and SSR marker. Bradshaw et al. (2008) identified chromosomes IV and V as the most acceptable with two well characterized chromosome specific SSR markers. However, there were characterized chromosome specific markers for chromosomes VII and XI of WL and IX and XII of S440. As a rule of thumb that at least three putatively homologous markers per LG should be used for chromosome identification (Rouppe van der Voort et al. 1997), chromosomal identification of those LGs is tentative, with varying degrees of ambiguity until they can be associated with well characterized SSR, IT or other chromosome specific markers.
For mapping and QTL analysis in potato and for sufficiently following the products of meiosis in the offspring, it is important to have adequate simplex markers on each of the 48 chromosomes of both parents. Duplex markers are necessary for identifying homologous chromosomes. As recombination frequencies from simplex/duplex markers in coupling and repulsion are estimated with equal accuracy, in theory, a single duplex marker per LG would be sufficient (Bradshaw et al. 2008). In the present study, we used 65 simplex and 24 duplex markers for mapping of WL and 95 simplex and 16 duplex markers for mapping of S440. Hackett et al. (1998) used 14 simplex and 4 duplex markers per set of four homologous chromosomes in their simulation study and concluded that a population size of at least 150, and preferably 250, should be used to identify homologous chromosomes. Hence we just constructed an overall map for each LG. Our results also showed that there was no even distribution of duplex markers for parents. This result is in agreement with the results of Bradshaw et al. (2008) in potato. They suggested that increasing the number of primer combinations should provide more than enough duplex markers present in one parent and absent in the other. It remains to be seen if in future SNPs will provide the required density of markers more cheaply and easily but the same problems have to be faced over linkage phase.
The use of genetic linkage maps can help to identify the loci contributing to adaptive changes in populations. Maps with a density of markers around 20–30 cM are sufficient for detecting the presence of QTLs, and increasing marker density allows more precise positioning of the QTLs (Darvasi and Soller 1994). Moreover, the introduction of DNA-based molecular markers allows the identification of genetic factors (QTLs) underpinning the variation of quantitative traits (Tanksley 1993; Quarrie 1996; Ribaut et al. 1997; Tuberosa et al. 1998; Sari-Gorla et al. 1999). Although QTLs for root characteristics have been extensively analyzed in other plants, such as rice (Yadav et al. 1997; Price et al. 2000), limited information is available for potato. This study indicates the possibility of using in vitro tests to identify QTLs for traits of the root system in tetraploid potato. A total of 14 QTLs with LOD > 2 were identified. As probably only a small portion of the genome was actually sampled we suppose that more QTLs relevant to osmotic stress should probably exist. Bradshaw et al. (2008) confirmed that some QTLs with a large effect may have been missing through inadequate marker coverage on some chromosomes but it is more likely that many QTLs of minor effect remain undetected.
Out of 14 QTLs, only 6 of them were confirmed as a major QTL based on the permutation test. Of those, 3 QTLs were for root length and 3 for root number. Three QTLs for root length explained 51, 52.3 and 64.9 % of phenotypic variance, individually. This range of phenotypic variance was previously reported by Bradshaw et al. (2008) for maturity (one QTL), after cooking blackening (six QTLs) and tuber shape (four QTLs) of potato. Three major QTLs for root number explained 19.2, 26.8 and 43.1 % of variation, independently. This range of phenotypic variance previously accounted for four QTLs of sprouting and fry colour of potato (Bradshaw et al. 2008). For each major QTL, we identified close markers that mapped at 2 to 4 cM from them. These markers were able to verify 40 % to 55 % of in vitro results. The development of near-isogenic lines at this QTL region would provide a valuable opportunity to validate and further characterize its effects on other quantity traits such as yield under osmotic stress conditions. Based on the phenotypic variance of 8 remaining QTLs, these were found not be major QTLs based on the permutation test being relatively high and ranged from 18.2 % to 54.5 %. QTLs with minor to intermediate effects, rather than ones of large effect, have been reported by other researchers for some of the traits. Bonierbale et al. (1993) identified 26 loci from seven of the potato LGs where restriction fragment length polymorphisms (RFLPs) were associated with measured yield characteristics in diverse tetraploid germplasm. Freyre et al. (1994) identified six QTLs (one on each of six LGs) for dormancy in a population derived from diploid (S. tuberosum x S. chacoense) x S. phureja. In the same population a total of ten putative QTLs for specific gravity (DM) were located on six LGs (Freyre and Douches 1994). Van den Berg et al. (1996a, b) detected QTLs affecting tuberization and dormancy in reciprocal backcrosses between S. tuberosum and S. berthaultii. Eleven distinct loci on seven LGs were associated with variation in tuberisation. Most of the loci had small effects but a QTL explaining 27 % of the variance, which was found on LG V (tuberisation in long days could be interpreted as early maturity). QTLs were detected on nine LGs that affected tuber dormancy at 13 °C in the dark, either alone or through epistatic interactions. Schaefer-Pregl et al. (1998) identified eight putative QTLs for tuber yield on eight LGs using RFLP markers on two different crosses among dihaploid breeding lines. Menendez et al. (2002) mapped QTLs for cold sweetening in diploid potato. QTLs for sugar content were located on all potato chromosomes with some explaining >10 % of the variability for reducing sugars located on six LGs.
In conclusion, our study demonstrates that root number and root length are appropriate traits to study of osmotic stress under in vitro conditions and are suitable to identify QTLs responsible for osmotic stress tolerance. Identified markers that were closely mapped with major QTLs could be used for the selection of osmotic stress tolerant genotypes. Nevertheless, further experiments are needed to confirm their usability to discriminate resistant/susceptible genotypes with known root mass production under field conditions.
References
Bálint, A.F., F. Szira, A. Börner, and G. Galiba. 2008. Segregation- and association based mapping of loci influencing osmotic tolerance in barley. Acta Biologica Szegediensis 52(1): 101–102.
Bonierbale, M.W., R.L. Plaisted, and S.D. Tanksley. 1988. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120: 1095–1103.
Bonierbale, M.W., R.L. Plaisted, and S.D. Tanksley. 1993. A test of the maximum heterozygosity hypothesis using molecular markers in tetraploid potatoes. Theoretical and Applied Genetics 86: 481–491.
Bradshaw, J.E., C.A. Hackett, B. Pande, R. Waugh, and G.J. Bryan. 2008. QTL mapping of yield, agronomic and quality traits in tetraploid potato (Solanum tuberosum subsp. tuberosum). Theoretical and Applied Genetics 116: 193–211.
Brugmans, B., G.B. Ronald, A. Hutten, O. Nico, G.F. Rookmaker-Richard, J. Visser, and H. van Eck. 2006. Exploitation of a marker dense linkage map of potato for positional cloning of a wart disease resistance gene. Theoretical and Applied Genetics 112: 269–277.
Celebi-Toprak, F., J.A. Watanabe, and K.N. Watanabe. 2005. Molecular markers in identification of genotypic variation. In Genetic improvement of Solanaceous crops: potato, vol 1, ed. M.K. Razdan and A.K. Mattoo, 115–141. Enfield: Science Publishers.
Cernak, I., K. Decsi, S. Nagy, I. Wolf, Z. Polgar, G. Gulyas, Y. Hirata, and J. Taller. 2008. Development of a locus-specific marker and localization of the Ry sto gene based on linkage to a catalase gene on chromosome XII in the tetraploid potato genome. Breeding Science 58: 309–314.
Churchill, G.A., and R.W. Doerge. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971.
Collard, B.C.Y., and D.J. Mackill. 2009. Start Codon Targeted (SCoT) polymorphism: A simple novel DNA marker technique for generating gene- targeted markers in plants. Plant Molecular Biology 27: 86–93.
Darvasi, A., and M. Soller. 1994. Optimum spacing of markers for determining linkage between marker loci and quantitative trait loci. Theoretical and Applied Genetics 89: 351–357.
Dobranszki, J., K.M. Tabori, and A.T. Hudak. 2003. Growth and developmental responses of potato to osmotic stress under in vitro conditions. Acta Biologica Hungarica 54(3–4): 365–372.
Dong, F., S. Song, S.K. Naess, J.P. Helgeson, C. Gebhardt, and J. Jiang. 2000. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theoretical and Applied Genetics 101: 1001–1007.
Ehsanpour, A.A., and R. Razavizadeh. 2005. Effect of UV-C on drought tolerance of alfalfa (Medicago sativa) callus. American Journal of Biochemistry and Biotechnology 1(2): 107–110.
Erusha, K.S., R.C. Shearman, T.P. Roirdan, and L.A. Wit. 2002. Kentucky bluegrass cultivar root and top growth responses when grown in hydroponics. Crop Science 42: 848–852.
Freyre, R., and D.S. Douches. 1994. Development of a model for marker assisted selection of specific gravity in diploid potato across environments. Crop Science 34: 1361–1368.
Freyre, R., S. Warnke, B. Sosinski, and D.S. Douches. 1994. Quantitative trait locus analysis of tuber dormancy in diploid potato (Solanum spp.). Theoretical and Applied Genetics 89: 474–480.
Gebhardt, C., E. Ritter, T. Debener, U. Schachtschabel, B. Walkemeier, and F. Salamini. 1989. RFLP analysis and linkage mapping in Solanum toberosum. Theoretical and Applied Genetics 78: 65–75.
Georgieva, M., D. Djilianov, T. Konstantinova, and D. Parvanova. 2004. Screening of Bulgarian raspberry cultivars and elites for osmotic tolerance in vitro. Biotechnology and Biotechnological Equipment 95–98.
Gopal, J., and K. Iwama. 2007. In vitro screening of potato against water-stress mediated through sorbitol and polyethylene glycol. Plant Cell Reports 26: 693–700.
Gorji, M.A., P. Poczai, Z. Polgar, and J. Taller. 2011. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. American Journal of Potato Research 88(3): 226–237.
Hackett, C.A., and Z.W. Luo. 2003. TetraploidMap: Construction of a linkage map in autotetraploid species. Journal of Heredity 94: 358–359.
Hackett, C.A., J.E. Bradshaw, R.C. Meyer, J.W. McNicol, D. Milbourne, and R. Waugh. 1998. Linkage analysis in tetraploid species: A simulation study. Genetics Research 71: 143–154.
Hackett, C.A., J.E. Bradshaw, and J.W. McNicol. 2001. Interval mapping of QTLs in autotetraploid species. Genetics 159: 1819–1832.
Hackett, C.A., B. Pande, and G.J. Bryan. 2003. Constructing linkage maps in autotetraploid species using simulated annealing. Theoretical and Applied Genetics 106: 1107–1115.
Howden, R., and C.S. Cobbett. 1992. Cadmium-sensitive mutants of Arabidopsis thaliana. Plant Physiology 99: 100–107.
Hsissou, D., and J. Bouharmont. 1994. In vitro selection and characterization of drought-tolerant plants of durum wheat (Triticum durum Desf). Agronomy 2: 65–70.
Ingram, K.T., F.D. Bueno, O.S. Namuco, E.B. Yambao, and C.A. Beyrouty. 1994. Rice root traits for drought resistance and their genetic variation. In Rice roots: nutrient and water use, ed. G.J.D. Kirk, 67–77. Manila: IRRI.
Iwama, K., and J. Yamaguchi. 2006. Abiotic stresses. In Handbook of potato production, improvement and postharvest management, ed. J. Gopal and S.M. Khurana, 231–278. New York: Food Product Press.
Jacobs, J.M.E., H. van Eck, P. Arens, B. Verkerk-Bakker, B.T. Hekkert, H.J.M. Bastlaanssen, A. El-Kharbotly, A. Pereira, E. Jacobsen, and W.J. Stiekema. 1995. A genetic map of potato (Solanum tuberosum) integrating molecular markers, including transposons, and classical markers. Theoretical and Applied Genetics 91: 289–300.
Lahlou, O., and J.F. Ledent. 2005. Root mass and depth, stolons and roots formed on stolons in four cultivars of potato under water stress. European Journal of Agronomy 22: 159–173.
Leport, L., M. Petrivalsky, and M. Chappart. 1993. Effectors for the osmoinduced proline response in higher plants. Plant Physiology and Biochemistry 31: 911–922.
Lilley, J.M., M.M. Ludlow, S.R. McCouch, and J.C. O’Toole. 1996. Locating QTL for osmotic adjustment and dehydration tolerance in rice. Journal of Experimental Botany 302: 1427–1436.
Luo, Z.W., C.A. Hackett, J.E. Bradshaw, J.W. McNicol, and D.M. Milbourne. 2001. Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics 157: 1369–1385.
Maas, E.V. 1985. Crop tolerance to saline sprinkling water. Plant and Soil 89: 273–284.
Maruyama, H., R. Koyama, T. Oi, M. Yagi, M. Takeda, M. Kanechi, N. Inagaki, and Y. Uno. 2008. In vitro evaluation of osmotic stress tolerance using a novel root recovery assay. Plant Cell Tissue and Organ Culture 95: 101–106.
Menendez, C.M., E. Ritter, R. Schaefer-Pregl, B. Walkemeier, A. Kalde, F. Salamini, and C. Gebhardt. 2002. Cold sweetening in diploid potato: Mapping quantitative trait loci and candidate genes. Genetics 162: 1423–1434.
Milbourne, D., R. Meyer, A. Collins, L. Ramsay, C. Gebhardt, and R. Waugh. 1998. Isolation, characterization and mapping of simple sequence repeat loci in potato. Molecular and General Genetics 259: 233–245.
Mohamed, M.A.H., P.J.C. Harris, and J. Henderson. 2000. In vitro selection and characterization of a drought clone of Tagetes minuta. Plant Science 159: 213–222.
Peuke, A.D., C. Scharml, W. Hartung, and H. Rennenberg. 2002. Identification of drought-sensitive beech ecotypes by physiological parameters. New Phytologist 154: 373–387.
Price, A.H., K.A. Steele, B.J. Moore, P.B. Barraclough, and L.J. Clark. 2000. A combined RFLP and AFLP linkage map of upland rice (Oryza sativa L.) used to identify QTLs for root-penetration ability. Theoretical and Applied Genetics 100: 49–56.
Quarrie, S.A. 1996. New molecular tools to improve the efficiency of breeding for increased drought resistance. Plant Growth Regulators 20: 167–178.
Ribaut, J.M., C. Jiang, D. Gonzáles de León, G.O. Edmeades, and D.A. Hoisington. 1997. Identification of quantitative trait loci under drought conditions in tropical maize. II. Yield components and marker-assisted selection strategies. Theoretical and Applied Genetics 94: 887–896.
Rossouw, F.T., and J. Waghmarae. 1995. The effect of drought on growth and yield of two South African potato cultivars. South African Journal of Science 91: 149–150.
Rouppe van der Voort, J., P. van Zandvoort, H. van Eck, R.T. Folkertsma, R.C.B. Hutten, J. Draaistra, F.J. Gommers, E. Jacobsen, J. Helder, and J. Bakker. 1997. Use of allele specificity of comigrating AFLP markers to align genetic maps from different potato genotypes. Molecular Genetics and Genomics 255: 438–447.
Sakthivelu, G., M.K. Akitha Devi, P. Giridhar, T. Rajasekaran, G.A. Ravishanker, T. Nedev, and G. Kosturkova. 2008. Drought-induced alterations in growth, osmotic potential and in vitro regeneration of soybean cultivars. General and Applied Plant Physiology 34(1–2): 103–112. Special issue.
Sari-Gorla, M., P. Krajewski, N. Di Fonzo, M. Villa, and C. Frova. 1999. Genetic analysis of drought tolerance in maize by molecular markers. II. Plant height and flowering. Theoretical and Applied Genetics 99: 289–295.
Schaefer-Pregl, R., E. Ritter, L. Concilio, J. Hesselbach, L. Lovatti, B. Walkemeier, H. Thelen, F. Salamini, and C. Gebhardt. 1998. Analysis of quantitative trait loci (QTLs) and quantitative trait alleles (QTAs) for potato tuber yield and starch content. Theoretical and Applied Genetics 97: 834–846.
Schafleitner, R., R.O.G. Rosales, A. Gaudin, C.A.A. Aliaga, G.N. Martinez, L.R.T. Marca, L.A. Bolivar, F.M. Delgado, R. Simon, and M. Bonierbale. 2007. Capturing candidate drought tolerance traits in two native Andean potato clones by transcription profiling of field grown plants under water stress. Plant Physiology and Biochemistry 45: 673–690.
Stam, P. 1993. Construction of integrated genetic linkage maps by means of a new computer package: Joinmap. The Plant Journal 3: 739–744.
Tanksley, S.D. 1993. Mapping polygenes. Annual Review of Genetics 27: 205–233.
Tuberosa, R., M.C. Sanguineti, P. Landi, S. Salvi, E. Casarini, and S. Conti. 1998. RFLP mapping of quantitative trait loci controlling abscisic acid concentration in leaves of drought-stressed maize (Zea mays L.). Theoretical and Applied Genetics 97: 744–755.
van den Berg, J.H., E.E. Ewing, R.L. Plaisted, S. McMurry, and M. Bonierbale. 1996a. QTL analysis of potato tuberization. Theoretical and Applied Genetics 93: 307–316.
van den Berg, J.H., E.E. Ewing, R.L. Plaisted, S. McMurry, and M. Bonierbale. 1996b. QTL analysis of potato tuber dormancy. Theoretical and Applied Genetics 93: 317–324.
Van Eck, H., J. Rouppe van der Voort, J. Draaistra, P. van Zandvoort, E. van Enckevort, B. Segers, J. Peleman, E. Jacobsen, J. Helder, and J. Bakker. 1995. The inheritance and chromosomal localization of AFLP markers in a non-inbred potato offspring. Molecular Breeding 1: 397–410.
Walbot, V., and C. Warren. 1988. Regulation of Mu element copy number in maiz line with an active or inactive mutator transposonable element system. Molecular Genetics and Genomics 211: 27–34.
Watanabe, S., K. Kojima, Y. Ide, and S. Sasaki. 2000. Effects of Saline and osmotic stress on praline and sugar accumulation in Populus euphratica in vitro. Plant Cell, Tissue and Organ Culture 63: 199–206.
Wu, S.J., L. Ding, and J.K. Zhu. 1996. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. The Plant Cell 8: 617–627.
Yadav, R., B. Courtois, N. Huang, and G. McLaren. 1997. Mapping genes controlling root morphology and root distribution on a double-haploid population of rice. Theoretical and Applied Genetics 94: 619–632.
Acknowledgements
Present publication was realized with the support of the project TÁMOP-4.2.2/B-10/1-2010-0025 and NKTH-TECH-09-A3-2009-0210. The experiments comply with the current laws of Hungary in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gorji, A.M., Matyas, K.K., Dublecz, Z. et al. In Vitro Osmotic Stress Tolerance in Potato and Identification of Major QTLs. Am. J. Potato Res. 89, 453–464 (2012). https://doi.org/10.1007/s12230-012-9268-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-012-9268-x