Abstract
A marker-saturated linkage map of potato was used to genetically map a locus involved in the resistance against wart disease Synchytrium endobioticum race 1. The locus mapped on the long arm of chromosome 4 and is named Sen1-4 in contrast to a Sen1 locus on chromosome 11. The AFLP markers from the Sen1-4 interval enabled the isolation of BAC clones from an 11 genome equivalent BAC library. This was achieved via fingerprinting of BAC pools with the AFLP primer pairs that resemble the genetic marker loci. With non-selective AFLP primers, fingerprints of individual BAC clones were generated to analyse the overlap between BAC clones using FPC. This resulted in a complete contig and a minimal tiling path of 14 BAC clones enclosing the Sen1-4 locus. The BAC contig has a genetic length of ~6 cM and a physical length of ~1 Mb. Our results demonstrate that map-based cloning of Sen1-4 can be pursued on the basis of a strategy of marker saturation alone. Genetic resolution achieved by screening large numbers of offspring for recombination events may not be required. Together with the construction of the BAC contig, a physical map with the position of the markers is accomplished in one step. This provides proof of concept for the utility of the marker saturation that is offered by the ultra dense AFLP map of potato for gene cloning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Map-based cloning has proven to be a successful method for the isolation of, for example, resistance genes in several plant species such as tomato (Martin et al. 1993; Dixon et al. 1996), Arabidopsis (Bent et al. 1994; Mindrinos et al. 1994; Grant et al. 1995), sugar beet (Cai et al. 1997) and barley (Büschges et al. 1997). Most map-based cloning projects start with a bulked segregant analysis (BSA) (Michelmore et al. 1991) to find bulk-specific markers. Subsequently, these markers are mapped in a segregating population to determine the order and distance relative to the trait of interest. Using a graphical display of the marker scores in the offspring genotypes, new pools can be composed using plants with recombination events in the proximity of the target locus for a second cycle of BSA. This second BSA will narrow the window to be saturated, and thus improve the chance to identify the markers that are physically sufficient close to the gene of interest to allow chromosome landing (Tanksley et al. 1995) in order to find the BAC-clone containing the gene of interest. A level of marker saturation, which is equivalent to a physical spacing of markers that is less than the average insert size of the genomic library (e.g. BAC or PAC library) to be used, offers an alternative to the screening of offspring for recombinants. In fact high-resolution mapping only results in genetic distances (cM) and not in physical distances (kb). Moreover, it seems that recombination is not a random event. Chromosomal intervals rather show an alternation of hotspots and coldspots for recombination. Therefore, the assumption of random distribution of AFLP markers (in fact the distribution of EcoRI sites) is safer than assuming the random distribution of recombination events.
The process of a targeted saturation with markers for BAC landing and/or contig construction is commonly performed for each map-based cloning project separately. This is labourious and time consuming in view of the required efforts and resources. The combination of BSA and AFLP fingerprinting (Vos et al. 1995) is an efficient approach to scan thousands of loci. Nevertheless, the numbers of AFLP primer combinations employed for a BSA are usually more than 1,000 combinations (Büsches et al. 1997; Harkins et al. 1998). Subsequently, a sufficiently large number of bulk-specific, putatively linked markers have to be tested using a mapping population. An alternative for the former approach could be a once and for all, genome-wide saturation with markers loci that are ready to be used for map-based cloning. In the case the locus-specific AFLP technique (Rouppe van der Voort et al. 1997a) is used to generate marker loci, then the same selective AFLP primers can be employed to recognise the AFLP amplification products within fingerprints of (pools of) BAC clones, without the need to store probes or locus-specific PCR primer information.
To construct a genome-wide saturated marker map of the potato genome, and to avoid many separate locus-specific marker saturation efforts with BSA, an EU project was started (FAIR5-PL97-3565; Isidore et al. 2003; http://potatodbase.dpw.wau.nl/UHDdata.html), aiming to reach the required level of marker saturation. It was assumed that a single linkage map with >10,000 AFLP marker loci should offer such a level of saturation of the 850 Mb potato genome that BAC landing becomes feasible with markers within a BAC length distance of the target gene. Therefore, in potato a diploid mapping population was established (Rouppe van der Voort et al. 1997b) in which several interesting traits and genes are segregating, including a locus involved in the resistance against Synchytrium endobioticum. This population with >10,000 marker loci should be an ideal starting point for map-based cloning of this resistance gene, and comparable to earlier work on this material (Bakker et al. 2004; Huang et al. 2004). This specific form of ‘BAC landing’ has also been termed ‘contig seeding’ or ‘contig initiation’ (Bryan et al. 2002).
The fungus S. endobioticum, the causal agent of potato wart disease, is an obligate soil-borne pathogen, producing persistent resting spores that can survive for many years (Hampson 1996). Salaman and Lesley (1923) already found that immunity against S. endobioticum was dominant and suggested that two dominant genes could induce immunity independently. The research of Lunden and Jørstad (1934) and Maris (1973) subscribe the model of dominant genes that are inherited in a Mendelian manner, but also reported influences of minor genes that modify the activity of the resistance genes. Hehl et al. (1999) reported the genetic mapping of a single dominant resistance gene against S. endobioticum race 1 (Sen1) on chromosome XI, using DNA sequence homology between the Sen1 locus and the N gene for resistance to TMV.
In this paper we report the mapping of Sen1-4, a gene on chromosome IV that confers resistance to race 1 of S. endobioticum. The position of Sen1-4 on the ultra dense map of potato indicates a number of co-segregating AFLP markers that have been used to identify the corresponding BACs from a BAC library. Based on the resulting contig containing the Sen1-4 locus, the general usefulness of the ultra dense potato map for map-based cloning is discussed.
Materials and methods
Plant material
The F1 mapping population descended from a cross between the diploid parents SH83-92-488 and RH89-039-16 (Rouppe van der Voort et al. 1997b; Isidore et al. 2003). The 120 genotypes of this mapping population are maintained in a screen house by annual propagation of seed tubers. This population segregated for resistance to S. endobioticum race 1. The maternal clone SH83-92-488 is a donor of the resistance, and the paternal clone is susceptible.
Wart disease test
Genotypes from this mapping population were evaluated for resistance to S. endobioticum using the Spieckermann test (Spieckermann and Kotthoff 1924). This test was performed within the quarantine facilities of the potato-breeding company Averis, Valthermond. Briefly, the Spieckermann bioassay involves inoculation of excised eye pieces from tubers with winter spores of S. endobioticum, followed by an incubation period of several weeks, after which wart disease symptoms are scored. The test was performed twice using 8 eye pieces per genotype, resulting in 16 tested eye pieces. A potato clone was found susceptible if at least one sample expressed clear disease symptoms.
Genetic mapping of the resistance gene against S. endobioticum race 1
Diploid potato is an obligate out-breeder, and therefore genetic maps are constructed using marker loci that segregate in the F1 progeny. The marker loci used in this study have been obtained in the framework of an EU project aiming at the construction of an ultra dense map of potato (Isidore et al. 2003; http://potatodbase.dpw.wau.nl/UHDdata.html). The maternal map consists of 4,187 segregating marker loci (Aa×aa) that are heterozygous in the pistilate parent SH83-92-488. The paternal map has 3,431 marker loci (aa×Aa) segregating from the staminate parent RH89-039-16, and 2,765 bridge markers (Aa×Aa) allow to connect the maternal and paternal maps. The co-segregating marker loci are grouped in the so-called bins, where each bin differs from its neighbouring bin(s) by one recombination event in the mapping population (Isidore et al. 2003). Within a bin, there is no information about the order of the markers, but linkage phase is known. The 4,187 markers of the maternal linkage map of SH83-92-488 reside in 989 bins divided over 12 linkage groups. The segregation for resistance/susceptibility for wart disease was added as a marker locus to the maternal genetic map data, after which the location of Sen1-4 relative to the 989 bins was determined using JoinMap2.0 software (Stam 1993).
BAC-library and plasmid isolation
A HindIII BAC-library of the resistant parent (SH83-92-488) was available (Rouppe van der Voort et al. 1999). This ~11 genome equivalent BAC library comprises almost 98,000 BAC clones with an average insert size of 100 kb. The BAC library was stored in 255 plates containing 384 BAC clones per plate. For the identification of the BACs enclosing the Sen1-4 locus, DNA was isolated from plate-pools, row/column pools of single plates and single BAC clones (see pooling strategy below). To prepare plate-pool DNA, the BAC clones of single BAC library plates were stamped on a plate with solid LB medium, using a 384-pin replicator. The bacterial colonies were grown overnight, and washed off using liquid LB medium. Plasmid DNA was isolated from these pooled cells using the alkaline lysis protocol (Heilig et al. 1997). The plasmid DNA isolation of row/column pools and single BAC clones was performed according to the protocol described by Klein et al. (1998). The row/column pools were constructed before plasmid isolation, but after the clones were grown in liquid LB medium (Sambrook et al. 1989).
Pooling strategy and library
Our BAC-pooling strategy is composed of two steps: (1) combining 255 library plate pools into 64 super pools to identify the library plate containing a BAC of interest. (2) From each positive library plate, the 16 rows and 24 columns were pooled in pairs of 2, resulting in 8 row pools and 12 column pools, to detect the single BAC of interest (Table 1). DNA of library plate pools was used to generate EcoRI/MseI AFLP template (Vos et al. 1995) using 100 ng plasmid DNA. The super pools were assembled with aliquots of +1/+1 selectively amplified AFLP template of the plate pools. The library plates were arrayed in rectangles of eight rows by eight columns. This was repeated four times to accommodate the 255 library plates. Every row and column was pooled into a super pool, representing (384×8×100 kb/840 Mb) 0.4 potato genome equivalents of DNA. After diluting the template of the super pools 20 times, the mixture was used for selective amplification using the AFLP primer combinations of the marker loci of the bins enclosing Sen-1. The putatively positive plate pools from this analysis have been confirmed, using the AFLP template from the single plate pools. The confirmed positive library plates were used to inoculate 384-well plates with LB medium (Sambrook et al. 1989) to grow single colonies overnight. Approximately 50 ng plasmid DNA of these plate-specific row and column pools was used for AFLP template preparation. Subsequently, pre-amplification and selective amplification was performed as described for the super pools. This screening resulted in four putatively positive BAC clones at a specific row–column intersection. To find the correct BAC clone, each of the four BACs was tested separately with the AFLP markers.
All selective AFLP amplifications were performed using IR dye labelled EcoRI primers (Biolegio BV, Malden, The Netherlands) after which the products were visualised on a denaturing polyacrylamide gel using a NEN® IR2 DNA analyser (LI-COR® Biosciences, Lincoln, NE, USA).
Contig construction
Fingerprint patterns of individual BAC clones were made by non-selective amplification of AFLP template. This template was generated using the enzyme combination HindIII/TaqI because HindIII was also used for partial digestion of potato genomic DNA for BAC-library construction of clone SH83-92-488. When HindIII is used in combination with a frequent cutter enzyme to construct AFLP template, the outermost restriction fragments of the inserted potato DNA are included and restriction fragments combining potato and vector DNA sequence are excluded. Moreover, those most distal HindIII/TaqI amplicons enable the detection of the smallest possible overlap between the single BACs. HindIII/TaqI template from single BACs was prepared using the standard AFLP protocol (Vos et al. 1995) adjusted for the restriction temperature of TaqI. Fragments were generated using the HindIII and TaqI primers without selective nucleotides after which the generated fragments were separated on a NEN® IR2 DNA analyser using standard conditions. The generated fragments were scored by the image interpretation software CrossChecker (Buntjer 2000; http://www.dpw.wau.nl/pv/pub/CrossCheck/download.html). The length of the fragments was estimated using a 10-bp ladder (SequaMark). The data set of AFLP band mobilities was multiplied with ten to produce a functional data set for FPC (Soderlund et al. 2000) which was used for contig construction of the selected BAC clones. A tolerance of 3 and a cutoff value of 1e−04 were used.
Results
Genetic localisation of Sen1-4
Tubers from 80 out of 135 individuals of the F1 mapping population were tested for the resistance to S. endobioticum race 1 using the Spiekermann test. The female parent SH83-92-488 was resistant to S. endobioticum, whereas the male parent RH89-039-16 was susceptible. The offspring segregated for resistance in a 35:45 (S–R) ratio, not significantly different from a 1:1 ratio (P=0.26). This suggests a single locus involved in resistance, heterozygous in the diploid female parent SH83-92-488. The phenotypic segregation for resistance was used to map the locus relative to the 989 bins of the ultra dense map parent SH83-92-488. This resulted in the identification of a region on chromosome IV ranging from bin 37 to bin 41 (see http://potatodbase.dpw.wau.nl/locishow.php?lowbin=SH04B037&highbin=SH04B042). All genetic positions within this interval have an equal likelihood to accommodate the R gene due to the absence in the disease test of specific plants that carry a recombination in this interval (Plant #13 recombination defines the border between bin 37 and bin 38, Plant #159 recombination defines the border between bin 38 and bin 39, Plants #164 and 55 define the border between bin 39 and bin 41). In six plants (#57, 27, 34, 48, 88 and 95) the flanking markers did not support the observed resistant phenotype, suggesting escapes. In offspring clone 67 the flanking markers indicated a false susceptible observation. These conflicting observations do not affect the accuracy of mapping because plants, with false positive or false negative observations, did not originate from a female gamete with a recombined chromosome 4. On the basis of these observations we postulate an R gene, tentatively called Sen1-4, involved in a monogenic qualitative resistance against S. endobioticum race 1. The name of this locus is an abbreviation of the pathogen, the race and its chromosomal position. Half tetrad analysis (data not shown) indicates that the region is located on the long arm of chromosome 4 approx. 5 cM below the centromere (bin 31). After removal of the <ab×ab> type markers, as well as the PstI/MseI or SacI/MseI markers, 13 mapped <ab×aa> segregating EcoRI/MseI AFLP markers remained which were used for BAC landing (Table 2).
To ensure the construction of the BAC contig beyond the interval, another three markers from the proximal bin 35 (EACAMCTC_127, EAAGMCGA_373, EAGCMATG_134) and the only one marker from the distal bin 42 (EAGTMACC_252) were also used (Table 2).
BAC identification
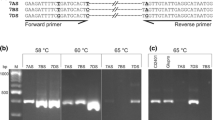
The SH83-92-488 BAC library was screened for BACs that carried AFLP markers from the interval comprising Sen-1. To this end 64 super pools were fingerprinted with the 13 AFLP primer combinations that were used to amplify the marker loci, listed in Table 2. The 0.4 genome equivalent templates showed an average of 40 fragments per lane, upon selective PCR amplification with (Eco/Mse+3/+3) AFLP primers extended with three selective nucleotides (Fig. 1). Deconvolution of super pool fingerprints for the markers of interest indicated a group of putatively positive plates. To verify these positive plates, the presence of the AFLP markers was confirmed by fingerprinting the single plate pools with the appropriate AFLP primer combinations. Eventually we obtained for the AFLP markers between three and seven positive plates (Table 2), with an average of 4.7 plates per marker. This is in close agreement with the expected number of five and a half positives when using an 11 genome equivalent BAC library in combination with heterozygous AFLP-markers. The specific plate pools that were retrieved with AFLP markers are already indicative for the physical distance between certain marker loci. Marker EAACMCAT_307 and EAGTMACC_252 both landed on a BAC in pool 115, suggesting a physical distance of less than 100 kb (the average BAC insert size).
Row and column pools were constructed by pooling single well cell cultures of the library plates. Plasmid DNA was isolated from these pools to prepare AFLP-template and to test the row and column pools for the presence of the AFLP marker. This procedure identified a set of putatively positive single BACs, which were then tested separately to determine the AFLP marker containing BACs.
Contig construction
For the construction of a contig enclosing the Sen 1-4 interval, fingerprints of individual BAC clones were produced by non-selective AFLP HindIII and TaqI primer amplification (Fig. 2). Analysis of fingerprints from non-overlapping BAC clones can be used as control experiment to gain an impression of the chance of co-incidental co-migration of non-homologous PCR fragments. This was observed in a few cases among very short fragments, which is consistent with the relative abundance of small-sized fragments (below ≈150 bases). To ensure homology between co-migrating bands only the amplified fragments larger than 200 nucleotides were used during contig construction. The AFLP band mobilities were multiplied with ten to produce a functional dataset for FPC (Soderlund et al. 2000) which was used for contig construction of the selected BAC clones. This resulted in two contigs (Fig. 3) of which the first enclosed the region starting with BAC 092E05 (positive for AFLP marker EACA/MCTC-127 which genetically mapped in bin 35) through BAC 247A20 (positive for AFLP marker EAGG/MCTT-403 mapped in bin 41). The second contig starts with BAC 202N21 (positive for AFLP marker EAAC/MCAT-307 mapped in bin 41) and ends with BAC 115N09 (positive for marker EAGT/MACC-252 mapped in bin 42). This last contig appeared to be very short, and is comprised of five BAC clones. All five BACs were positive for the AFLP-marker EAAC/MCAT-307 (clones 115N09, 122 K18, 138N09, 202N21, 246B04), and among these, one BAC clone (115N09) was also positive for EAGT/MACC-252. This shows that the second contig includes the genetic position of distal recombination event of the Sen1-4 interval, between bin 41 and bin 42.
The genetic and physical map of the Sen1-4 interval. In the top part (a) the marker-saturated genetic map is shown (map units are indicated with bin numbers). The genetic positions of recombination events are indicated with an X. Two adjacent recombination events result in an empty bin (not shown). In the middle part (b) the genetic and physical order of the AFLP markers from this region is shown. In the bottom part (c) the BAC contig is shown. Vertical arrows below the marker names indicate the BAC clones that were positive for the AFLP marker. The order and overlap of the BAC clones were determined by, with FPC, using BAC fingerprints with the HindIII/TaqI AFLP primer pair without selective nucleotides
With FPC the order of the BACs and the individual bands obtained by BAC fingerprinting could be determined. The gap between the ‘bin35/41-contig’ and the ‘bin41/42-contig’ was located within bin 41, between marker EAGG/MCTT-403 and EAAC/MCAT-307, each landing on five BAC clones. Closer examination of the fingerprinting patterns showed one fragment of ~208 nucleotides long, which was common to BAC clones 247A20 and 202N21. According to the ‘bands file’ of FPC this shared fragment was indeed located at the farthest end in both contigs. This small overlap is insufficiently significant for FPC to join both contigs with the parameters used. Nevertheless, the locus specificity of co-migrating AFLP fragments and the distal placement of the band in either contig offers sufficient confidence to conclude that the gap between the ‘bin35/41-contig’ and the ‘bin41/42-contig’ is connected by BAC clones 247A20 and 202N21.
Discussion
Inheritance of wart disease resistance
In this paper we report the genetic mapping of a resistance gene against S. endobioticum race 1 on potato chromosome IV, and therefore propose the name Sen 1-4. The observation of a monogenic inherited resistance is seemingly in contrast to earlier genetic models. Earlier studies on the inheritance of wart disease race 1 by classical genetics have resulted in genetic models comprising at least two genes (Salaman and Lesley 1923; Lunden and Jørstad 1934; Maris 1973). However, the approach taken by Hehl et al. (1999) as well as our approach implies the correlation of phenotypic segregation in a bioassay relative to loci on a linkage map. Both mapping studies show that a single locus is involved in resistance, albeit on different chromosomes (chromosome XI, Hehl et al. 1999; chromosome IV, this paper). As such linkage mapping can only display heterozygous loci segregating in the mapping population, whereas the earlier genetic models were based on multiple crosses. Intercrosses between parental clones from this study and the study by Hehl et al. (1999) may allow to reconcile the classical genetic models and loci currently mapped. A duplicate gene model (AaBb×AaBb=15:1), or epistatic interactions such as complementary genes (AaBb×AaBb=9:7) or recessive suppression (AaBb×AaBb=13:3) can be inferred from intercrosses between parents such as SH83-92-488 × H80.577/1 (resistant × resistant), SH83-92-488 × H80.576/16 (resistant × susceptible), RH89-039-16 × H80.577/1 (susceptible × resistant) and, RH89-039-16 × H80.576/16 (susceptible × susceptible), as well as intercrosses of offspring clones. Comparing of the pedigrees of both mapping populations (data not shown; Dr. C. Gebhardt, personal communication) did not show any parents in common. In addition, it should be noted that the resistance against S. endobioticum race 1 is commonly observed in approx. 60% the modern potato cultivars. Therefore the relation between the two loci currently mapped and the classical genetic models remains inconclusive.
Utility of the marker saturation offered by the ultra dense map of potato
For potato a marker dense linkage map was constructed consisting of a maternal map comprising 4,187 segregating marker loci and a paternal map comprising 3,413 marker loci. The co-segregating marker loci are grouped in the so-called bins, where each bin differs from its neighbouring bin(s) by one recombination event in the mapping population (Isidore et al. 2003). The marker density between the bins of the linkage map varies considerably, ranging from 0 up to 531 markers per bin, with the most marker dense bins occurring at the presumed centromeric regions of the chromosomes. The mapping of the resistance gene against S. endobioticum race 1 was performed by using this ultra dense genetic map of potato and adding the disease test results as a single marker locus. Sen1-4 was located between bin 35 and 42 of linkage group 4. All genetic positions within this interval have an equal likelihood to accommodate the R gene due to the absence of specific plants in the disease test. Although the average number of markers used was less than two per bin for this region, which is lower than the average of 4.2 markers per bin, it was possible to construct a contig. In fact the level of marker saturation is even more positive because neither the AFLP markers of the PstI/MseI and SacI/MseI primer combinations, nor the 3:1 segregating AFLP markers have been exploited.
This contig is spanning a genetic distance of ~6 cM (seven recombination events in 120 offspring) and a physical distance of ~1 Mb, including the minimal overlap between BAC clones 247A20 and 202N21 located within bin 41. This confirms that the level of marker saturation is adequate and equivalent to a marker spacing that is less than the average insert size of this BAC library. This allows BAC landing and contig construction at a low genetic resolution (≈100 offspring). The unknown order of the markers within a bin is not an obstacle for BAC landing.
The BAC library may contain more BAC clones to reinforce the contig. This may apply not only to the 247A20 and 202N21 connection, but also to the other connections that do not have the expected ~5-fold deep coverage. Since the super pools of the BAC library have been surveyed only with AFLP markers, the best coverage is observed around the position where the marker lands on the BACs. The poorest coverage is observed between the marker positions where the end of a cluster of BACs meet the end of the next cluster.
The results also describe the co-retention of AFLP markers in super pools and plate pools. The joint presence or absence of markers in the 64 super pools allows physical mapping of the markers, because the level of co-retention is proportional to the physical distance of the markers (Borm et al. 2003; de Boer et al. 2004).
The below average number of markers per bin (including an empty bin 40) already suggested the limited physical size and, in connection to this, an elevated recombination frequency in this region. This is in agreement with the findings of Ganal et al. (1989), Young and Tanksley (1989) and King et al. (2002) that there is no direct relation between the genetic linkage intensities and the physical distance between markers over the genome. At this moment a genome-wide physical map is constructed using markers generated with non-selective EcoRI/MseI AFLP fingerprinting of the potato clone RH89-039-16. In combination with the AFLP markers mapped in the ultra dense genetic map, these contigs will be linked to the genetic map of potato. When this physical map is finished more can be said about the physical clustering and the genetic/physical ratio per region of EcoRI/MseI markers in the potato mapping population.
Efficient identification of BACs via BAC pools
Most BAC libraries include ten–thousands of BAC clones to provide sufficient genome coverage to ensure the presence of the desired fragment of the genome. To identify a single BAC clone of interest among all these BACs, without making extreme efforts, several pooling strategies have been developed. Generally speaking, we recognise two systems: (1) the step-wise screening of library plates, and rows and columns of within positive plates; (2) the assembling of pools in three or more dimensions. De-convolution of positive pool signals then leads directly to the BAC clone of interest (Klein et al. 2000; Rogel-Gaillard et al. 2001; Whisson et al. 2001). This might be efficient for genome-wide physical map construction (Klein et al. 2000), where all information gained can be used to construct contigs and a large numbers of markers are used to screen all the pools. However, for local contig construction with only a few markers across a small interval, the assembling of multi-dimensional BAC pools will take too much effort relative to the number of markers by which the pools are going to be screened. In this study we have used a combination of the step-wise and the dimensional pooling strategy. At the level of library plates we constructed two-dimensional row–column super-pools, because the low number of positive plates to be expected (5.5 per 255 plates) allows to obtain a reduction of the number of PCR steps (255 to 64), without a severe trade-off of gaining many false positives. If multiple super pools (rows and columns) were positive, the de-convolution will result in a few false positives, which could be verified at the plate pool level easily. Similarly, combining of two rows and two columns will result in fewer PCR reactions to identify the BAC clone within a plate. With four additional PCR reactions the single positive BAC clone can be distinguished easily from the three false positives, from the four putatively positive clones suggested by the row and column pools. Although it is not easy to provide quantitative figures, we suggests that it is more efficient to use this step-by-step pooling design and verify false positives than investing into a pooling strategy by which no false positives arise, but always a very large number of PCR reactions have to be tested.
Towards cloning of Sen1-4
So far, we have a genetic map of ~6 cM and a physical contig of ~1 Mb, comprising the Sen1-4 locus, without the resolution that could be gained from the analysis of recombinants from large numbers of offspring. With this, we have demonstrated that the concept of BAC landing (Tanksley et al. 1995) does not require the genetic resolution of many recombinants, but only the marker saturation as provided by our dense linkage map (Isidore et al. 2003). For future research we have selected a minimal tiling path of 14 BAC clones. These BAC clones could be used for direct transformation, if it were not possible to postulate candidate genes. However, for the cloning of a disease resistance gene, it is more obvious to screen the 14 BACs for R gene like sequences via Southern analysis or with primers based on the highly conserved NBS motif (e.g. Van der Linden et al. 2004). BACs that contain such resistance gene analogs (RGA’s) can be subcloned and sequenced. Functional open reading frames can be used for complementation studies as well, if BAC transformation is deemed to be problematic. Complete DNA sequencing of the minimal tiling path is not inconceivable either. In conclusion, the added value of the genetic resolution gained by a priori screening many offspring for recombinants is doubtful. Moreover, it will require two growing seasons to generate sufficient numbers and sizes of potato tubers from those recombinants for the wart disease bio-assay. Only when large numbers of RGA’s are detected across many BACs, the resolving power of genetic recombination should be employed to narrow the interval and the number of candidate RGA’s. Irrespective of the procedure chosen, the material required for the bio-assay to confirm cloning of the correct Sen1-4 resistance gene will take at least several years.
By using a marker-saturated linkage map of potato, it was possible to genetically map the resistance gene against S. endobioticum race 1 and to detect sufficient BAC clones originating from this region. By performing non-selective AFLP the overlap between different BAC clones could be determined resulting in a contig and a minimal tiling path enclosing the Sen1-4 locus. The method of ‘BAC-landing’ used in this paper provides an illustration of the concept of ‘contig seeding’ or ‘contig initiation’ (Bryan et al. 2002) and underlines the utility of the ultra dense map of potato.
References
Bakker E, Achenbach U, Bakker J, Van Vliet J, Peleman J, Segers B, Van der Heijden S, Van der Linde P, Graveland R, Hutten R, Van Eck HJ, Coppoolse E, Van der Vossen E, Bakker J, Goverse A (2004) A high-resolution map of the H1 locus harbouring resistance to the potato cyst nematode Globodera rostochiensis. Theor Appl Genet 109(1):146–152
Bent AF, Kundel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265:1856–1860
De Boer JM, Borm TJA, Brugmans B, Bakker ER, Bakker J, Visser RGF, Van Eck HJ (2004) Construction of a genetically anchored physical map of the potato genome. Plant and animal genomes XII, San Diego. http://www.intl-pag.org/pag/12/abstracts/W54_PAG12_245.html
Borm T, Brugmans B, De Boer J, Van der Vossen E, Bakker J, Visser R, Van Eck HJ (2003) Bac-pool mapping: a method for physical distance estimation. Plant and animal genome XI, Town & Country Convention Center, San Diego, 11–15 January 2003. http://www.intl-pag.org/11/abstracts/P2d_P130_XI.html
Bryan G, Milbourne D, Isidore E, McNicoll J, Tierney I, Purvis A, Williamson S, Ramsay L, McLean K, Waugh W (2002) Application of a potato UHD genetic linkage map for BAC landing and contig initiation in a region of linkage group V. S.C.R.I. Ann. Rep 2001/2002. http://bitrws400.scri.sari.ac.uk/Document/AnnReps/02Indiv/22UHD.pdf
Buntjer JB (2000) Cross checker: computer assisted scoring of genetic AFLP data. Plant and animal genome VIII, San Diego, 9–12 January 2000. http://www.intl-pag.org/pag/8/abstracts/pag8664.html
Büschges R, Hollricher K, Panstruga R, Simons G, Wolters M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schultze-Lefert P (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Cai DG, Kleine M, Kifle S, Harloff HJ, Sandal NN, Marcker KA, Klein-Lankhorst R, Salentijn EMJ, Lange W, Stiekema WL, Wyss U, Grundler FMW, Jung C (1997) Positional cloning of a gene for nematode resistance in sugar beet. Science 275:832–834
Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84:451–459
Ganal MW, Young ND, Tanksley SD (1989) Pulsed-field gel electrophoresis and physical mapping of large DNA fragments in the Tm-2a region of chromosome 9 in tomato. Mol Gen Genet 215:395–400
Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269:843–846
Hampson MC (1996) A quantitative assessment of wind dispersal of resting spores of Synchytrium endobioticum, the causal agent of wart disease of potato. Plant Dis 80:779–782
Harkins DM, Johnson GN, Skaggs PA, Mix AD, Dupper GE, Devey ME, Kinloch BB, Neale DB (1998) Saturation mapping of a major gene for resistance to white pine blister rust in sugar pine. Theor Appl Genet 97:1355–1360
Hehl R, Faurie E, Hesselbach J, Salamini F, Whitham S, Baker B, Gebhardt C (1999) TMV resistance gene N homologues are linked to Synchytrium endobioticum resistance in potato. Theor Appl Genet 98:379–386
Heilig JS, Lech K, Brent R (1997) Large-scale preparation of plasmid DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 1.7.1–1.7.3
Huang S, Vleeshouwers VGAA, Werij JS, Hutten RCB, Van Eck HJ, Visser RGF, Jacobsen E (2004) The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. MPMI 17(4):428–335
Isidore E, Van Os H, Andrzejewski S, Bakker J, Barrena I, Bryan GJ, Buntjer J, Caromel B, Van Eck HJ, Ghareeb B, De Jong W, Van Koert P, Lefebvre V, Milbourne D, Ritter E, Rouppe van der Voort JNAM, Rousselle-Bourgeois F, Van Vliet J, Waugh R (2003) Toward a marker-dense meiotic map of the potato genome: lessons from linkage group I. Genetics 165(4):2107–2116
King J, Armstead IP, Donnison IS, Thomas HM, Jones RN, Kearsey MJ, Roberts LA, Thomas A, Morgan WG, King IP (2002) Physical and genetic mapping in the grasses Lolium perenne and Festuca pratensis. Genetics 161:315–324
Klein RR, Morishige DT, Klein PE, Dong J, Mullet JE (1998) High throughput BAC DNA isolation for physical map construction of sorghum (sorghum bicolor). Plant Mol Biol Rep 16:351–364
Klein PE, Klein RR, Cartinhour SW, Ulanch PE, Dong J, Obert JA, Morishige DT, Schlueter SD, Childs KL, Ale M, Mullet JE (2000) A high-troughput AFLP-based method for constructing integrated genetic and physical maps: progress toward asorghum genome map. Genome Res 10:789–807
Lunden AP, Jørstad J (1934) Investigations on the inheritance of immunity to wart disease (Synchytrium endobioticum (Schilb.)Perc.) in the potato. Genetics 29:375–385
Maris B (1973) Studies with potato dihaploids on the inheritance of resistance to wart disease. Potato Res 16:324
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Mindrinos M, Katagiri F, Yu GL, Ausubel FM (1994) The Arabidopsis thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78:1089–1099
Rogel-Gaillard C, Piumi F, Billault A, Bourgeaux N, Save J, Urien C, Salmon J, Chardon P (2001) Construction of a rabbit bacterial artificial chromosome (BAC) library: application to the mapping of the major histocompatibility complex to position 12q1.1. Mamm Genome 12:253–255
Rouppe van der Voort JNAM, Van Zandvoort P, Van Eck HJ, Folkertsma RT, Hutten RCB, Draaistra J, Gommers FJ, Jacobsen E, Helder J, Bakker J (1997a) Use of allele specificity of comigrating AFLP markers to align genetic maps from different potato genotypes. Mol Gen Genet 255(4):438–447
Rouppe van der Voort JNAM, Wolters P, Folkertsma RF, Hutten RBC, Van Zandvoort P, Vinke H, Kanyuka K, Bendahmane A, Jacobsen E, Janssen R, Bakker J (1997b) Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers. Theor Appl Genet 95:874–880
Rouppe van der Voort J, Kanyuka K, Van der Vossen E, Bendahmane A, Mooijman P, Klein-Lankhorst R, Stiekema W, Baulcombe D, Bakker J (1999) Tight physical linkage of the nematode resistance gene Gpa2 and the virus resistance gene Rx on a single segment introgressed from the wild species Solanum tuberosum subsp. andigena CPC 1673 into cultivated potato. MPMI 12:197–206
Salaman RN, Lesley MA (1923) Genetic studies in potatoes; the inheritance of immunity to wart disease. Genetics 13:177–186
Sambrook J, Fritsch EF, Maniatis TT (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Soderlund C, Humphray S, Dunham A, French L (2000) Contigs built with fingerprints, markers and FPC V4.7. Genome Res 10:1772–1787
Spieckermann A, Kotthoff P (1924) Die Prüfung von Kartoffeln auf Krebsfestigkeit. Deut Landw Presse 51:114–115
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3:739–744
Tanksley SD, Ganal MW, Martin GD (1995) Chromosome landing: a paradigm for map-based gene cloning in plants with large genomes. Trends Genet 11:63–68
Van der Linden CG, Wouters DCAE, Mihalka V, Kochieva EZ, Smulders MJM, Vosman B (2004) Efficient targeting of plant disease resistance loci using NBS profiling. Theor Appl Genet 109(2):384–393
Vos P, Hogers R, Bleeker M, Rijans M, Van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Whisson SC, Van der Lee T, Bryan GJ, Waugh R, Govers F, Birch PR (2001) Physical mapping across an avirulence locus of Phytophthora infestans using a highly representative, large-insert bacterial artificial chromosome library. Mol Genet Genomics 266(2):289–295
Young ND, Tanksley SD (1989) RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor Appl Genet 77:353–359
Acknowledgements
We gratefully acknowledge the breeding company Averis Seeds BV, Valthermond for the wart disease bio-assay on the parents and offspring clones of the mapping population. This research was supported by the Dutch Technology Foundation STW http://www.stw.nl/projecten/W/wpb5283.html (grant WPB.5283).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Wenzel
Rights and permissions
About this article
Cite this article
Brugmans, B., Hutten, R.G.B., Rookmaker, A.N.O. et al. Exploitation of a marker dense linkage map of potato for positional cloning of a wart disease resistance gene. Theor Appl Genet 112, 269–277 (2006). https://doi.org/10.1007/s00122-005-0125-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0125-x