Abstract
As one of the most clinically relevant human habitats, the human mouth is colonized by a set of microorganisms, including bacteria, archaea, fungi, and viruses. Increasing evidence has supported that these microbiota contribute to the two commonest oral diseases of man (dental caries and periodontal diseases), presenting significant risk factors to human health conditions, such as tumor, diabetes mellitus, cardiovascular diseases, bacteremia, preterm birth, and low birth weight in infants. It is widely accepted that oral microorganisms cause diseases mainly by a synergistic or cooperative way, and the interspecies interactions within the oral community play a crucial role in determining whether oral microbiota elicit diseases or not. Since a comprehensive understanding of the complex interspecies interactions within a community needs the knowledge of its endogenous residents, a plenty of research have been carried out to explore the oral microbial diversity. In this review, we focus on the recent progress in this field, including the oral microbiome composition and its association with human diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Only about 10 % of cells in our bodies are truly from the human host, and the rest are from human microbiota (Savage 1977; Wilson 2008). These commensal microorganisms help us resist pathogens, educate immune system, and provide some traits humans do not originally evolve with the body (Dethlefsen et al. 2007; Gill et al. 2006; Turnbaugh et al. 2007). For instance, the plant polysaccharides commonly consumed in the diet are rich in xylan-, pectin-, and arabinose-containing carbohydrate structures. Although the human genome lacks most of the enzymes required for degrading these compounds, the distal gut microbiota provides us with this capacity (Gill et al. 2006). In fact, the human genetic landscape is a blend of the human genome and the metagenome of microorganisms colonizing in/on the human bodies (Turnbaugh et al. 2007). Therefore, the genetic diversity of humans resides not only in the allele frequencies of shared Homo sapiens genes but also in the genes within our microbial communities (Bäckhed et al. 2005; Li et al. 2008). To fully understand the human genetic and physiological variations, the composition and structure of human microbiota in major parts (e.g., mouth, skin, and gut) of the body and their influencing factors must be characterized (Gill et al. 2006; Heijtz et al. 2011).
As one of the most clinically relevant microbial habitats, the oral cavity is colonized by a personalized set of microorganisms, including bacteria, archaea, fungi, and viruses. If the term “human microbiome” is used to describe the sum of microbes that live in symbiosis or commensalism with us and elicit various human diseases under certain conditions (Lederberg and McCray 2001), the “oral microbiome” is suitable to refer specifically to the microorganisms inhabiting the human mouth (Dewhirst et al. 2010). The oral microbiome not only greatly contributes to the two commonest human oral diseases (i.e., dental caries and periodontal diseases) but also has been proven to present a significant risk factor to human health, such as tumor (Farrell et al. 2011), diabetes mellitus (Löe 1993), cardiovascular diseases (Figuero et al. 2011), bacteremia (Bahrani-Mougeot et al. 2008), and preterm birth and low birth weight in infants (Mitchell-Lewis et al. 2001; Offenbacher et al. 2006). It is widely accepted that oral microorganisms cause diseases mainly by a synergistic or cooperative fashion (Darveau 2010; Griffen et al. 2012; Hajishengallis and Lamont 2012), which means, in addition to the host immune response, it is the dynamic balance of both synergistic and antagonistic interspecies interactions rather than the presence/absence of specific bacteria within the community that plays a crucial role in determining whether diseases occur or not (Kleinberg 2002; Marsh 2005; Medzhitov 2007; Socransky et al. 1998). Since a comprehensive understanding of the microbial interspecies interactions of a given community requires the full knowledge of its endogenous residents, a slew of investigations have been carried out to explore the microbial diversity, composition, and structure of oral microbial communities. For a long time, this research field has been impeded by the intrinsic limitation of the conventional culture-dependent methods. However, more than 50 % of the oral microorganisms are unable to be cultivated (Paster et al. 2006); culture-independent methods, such as reverse-capture checkerboard hybridization (Mager et al. 2003), fluorescence in situ hybridization (Gersdorf et al. 1993), terminal restriction fragment length polymorphism (Takeshita et al. 2008), denaturing/thermal gradient gel electrophoresis (Alves et al. 2009), microarrays (Lif Holgerson et al. 2011), and 16S rRNA clone library analysis (Aas et al. 2005), have be used to refine and redefine the knowledge of the microbial diversity in the different oral sites, substantially expanding the list of candidate pathogens associated with oral diseases. More importantly, in this decade, high-throughput DNA sequencing technologies, such as 454 pyrosequencing (Roche Applied Science, Basel, Switzerland), Illumina/Solexa Genome Analyzer (Illumina, San Diego, CA, USA), and SOLiD (Applied Biosystems, Foster City, CA, USA), have been used, dramatically increasing the resolution at which microbial communities can be analyzed. Here, we describe the recent progress in the field of oral microbiome. Since the majority of the research is concentrated on the domain Bacteria, we first discuss the bacterial diversity in different oral niches under health and disease conditions and the evidence confirming relationships between oral bacterial community shifts and some systemic diseases and health risk conditions, and then, a brief introduction of oral viruses, fungi, and archaea and their relationships with human health and diseases is presented.
Oral bacterial microbiome
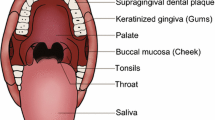
Over 700 bacterial species have been identified by culture-independent approaches in the human mouth, and more than 250 have been isolated, cultivated, and named (Paster et al. 2006). Undoubtedly, more novel species are expected to be identified (Belda-Ferre et al. 2012; Bik et al. 2010; Griffen et al. 2012; Keijser et al. 2008). Two types of surfaces are available in the human mouth for microbial colonization, including shedding (mucosa), and solid surfaces (teeth). In addition, microflora attaching to surfaces continuously shed into the saliva, making salivary microbiota the “fingerprint” of the oral microbiome inhabiting in the oral surface (Fábián et al. 2008). Since it has been well established that microorganisms colonizing the oral cavity display significant tropism for different inhabiting environments (Mager et al. 2003), we discuss the microbial composition of saliva, biofilms formed on the tooth surface, and mucosa separately.
Saliva
Each milliliter of saliva contains an average of 1.4 × 108 CFU bacteria, most of which belong to one of seven major phyla: Actinobacteria, Bacteroides, Firmicutes, Fusobacteria, Proteobacteria, Spirochaetes, and TM7 (The Human Microbiome Consortium 2012; Zaura et al. 2009). Although members of the population shared similar salivary organisms (The Human Microbiome Consortium 2012), there is an inter-individual difference in the salivary composition, although it is stable within a certain period for a given individual within a certain time frame (Lazarevic et al. 2010). However, salivary microbes do not show obvious geographical distribution characteristics, that is, the microbial composition does not indicate any strong influence of geography (Nasidze et al. 2009). The salivary microbiota may be used as biomarkers of disease diagnosis. For example, patients with dental caries (Yang et al. 2011a), periodontitis (Sakamoto et al. 2000), oral squamous cell carcinoma (Pushalkar et al. 2011), and pancreatic cancer (Farrell et al. 2011) show a different salivary bacterial composition and/or distribution from healthy populations.
Dental plaque
Dental plaque is a kind of biofilm building on the tooth surfaces (Yang et al. 2011b). It could be classified into two categories according to the location: supragingival dental plaque above the gingival margin and subgingival plaque below the gingival margin (Filoche et al. 2010). The supragingival plaque of healthy populations contains a rich variety of bacteria. Approximately 6,888 bacterial taxa in the supragingival ecology of 98 healthy adult subjects were identified with Firmicutes and Actinobacteria as the dominant groups (Keijser et al. 2008). The composition of supragingival plaque on the dental surfaces varies with the balance between health and disease conditions. Taking the development of dental caries as an example, the microbial composition of plaque on the surface of healthy enamel, white spot, and caries cavity was significantly different, and as caries progressed, the microbial diversity gradually decreased and dominant bacteria changed (Aas et al. 2008; Gross et al. 2010). Similar results were obtained from studies on root surfaces (Preza et al. 2008). The subgingival plaque is closely associated with the periodontal destruction. Microbiological studies of this niche mainly focus on changes during the development of periodontal diseases, which are described in the section “Periodontal diseases” of this review.
Oral mucosa
Compared with other oral niches, the microbes colonizing the oral mucosa are relatively limited. The tongue has attracted great attention due to its colonization by microbes associated with halitosis (i.e., unpleasant odor exhaled in breathing). The tongue dorsum of healthy populations is colonized by large amounts of Streptococcus salivarius, Rothia mucilaginosa, and an uncharacterized cultivable species of Eubacterium strain FTB41 (Kazor et al. 2003).
Oral bacteria related to oral diseases
Dental caries
Dental caries is one of the most prevalent oral diseases, and humans are susceptible to it during the whole life (Selwitz et al. 2007). Dental caries not only leads to tooth destruction but also causes pulp and periapical infection (Balakrishnan et al. 2000). When investigating the microbial diversity of dental caries, specimens are usually collected from supragingival plaque, saliva, and infected dentin of children with severe early childhood caries (S-ECC), caries-active adult patients, and elders suffering from root caries.
S-ECC is a kind of rampant caries involving multiple primary teeth, especially the maxillary anterior teeth (Drury et al. 1999). Streptococcus mutans has been studied intensively for its cariogenic properties and has even been regarded as a specific pathogen of caries. However, other bacteria, including species of Streptococcus, Veillonella, Actinomyces, Granulicatella, Leptotrichia, Thiomonas, Bifidobacterium, and Prevotella were also detected at higher frequencies in the plaque of children with S-ECC than that of caries-free children, and these bacteria are believed to be S-ECC associated (Becker et al. 2002; Kanasi et al. 2010; Ling et al. 2010; Tanner et al. 2011). Follow-up observation of S-ECC patients who underwent systemic treatments showed that children affected with S-ECC again had higher abundance of Prevotella nigrescens and Capnocytophaga in pre-treatment plaque compared with relapse-free patients (Tanner et al. 2011). In addition, if S-ECC is effectively controlled, the percentage of S. mutans in supragingival dental plaque would be reduced (Tanner et al. 2011).
The genera Streptococcus, Lactobacillus, Actinomycetes, Propionibacterium, and Veillonella were also detected with a high relative abundance in the plaque of caries-active adults (Belda-Ferre et al. 2012). In addition, Prevotella species including Prevotella sp., Prevotella histicola, and Prevotella shahii were not similarly distributed between healthy and caries-active hosts (Yang et al. 2011a). More significantly, genes related to acid production, DNA uptake, and stress responses were highly expressed in the plaque of caries-active patients (Belda-Ferre et al. 2012).
Root caries (RC) often occurs in elders with gingival recession. The genera Atopobium, Olsenella, Pseudoramibacter, Propionibacterium, and Selenomonas were found to be involved in the occurrence and development of root caries (Preza et al. 2008). A large number of Lactobacillus were detected in the biofilms formed on the healthy root surface of RC patients; however, no Lactobacillus were found in plaques formed on the root surface of caries-free populations (Preza et al. 2008, 2009).
From the clinical perspective, dental caries can be manifested as white spot with intact tooth structure and cavity. During the development of dental caries, microbial composition undergoes dynamic changes: the microbial diversity gradually decreases in the order of plaque from healthy enamel surfaces, white spot, plaque from cavity surfaces, and infected dentin (Gross et al. 2010). The predominant bacteria also changes during the progression, gradually transitioning from non-S. mutans, Streptococci, and Actinomycetes to S. mutans, Lactobacillus, and Bifidobacterium (Takahashi and Nyvad 2011). Interestingly, even at the same caries developmental stages, predominant bacteria vary between primary and permanent dentition. For example, the dominant bacteria in the white spot of both primary and permanent teeth are S. salivarius and Streptococcus parasanguis, whereas the bacteria most frequently detected from cavity surfaces are S. salivarius, S. parasanguis, Corynebacterium, and Actinomyces gerencseriae in primary dentition and S. salivarius, S. parasanguis, Campylobacter, and Selenomonas in permanent dentition. Furthermore, infected dentin contains a large number of acid-producing bacteria, such as S. mutans, Lactobacillus, and Propionibacterium (Aas et al. 2008).
Periapical infection
Periapical periodontitis is an infectious disease occurring on the tooth periapical tissue and it is the result of the interaction between the microbial community and the host immune response (José 2002; Siqueira and Rôças 2009a, b).
As demonstrated above, microbial colonization in the root canal system is the major pathogenic factor of periapical periodontitis (Siqueira 2011). A long ago, researchers found that root periapical infection was a multi-bacterial infection, by using culture-dependent technologies (Byström et al. 1987; Fabricious et al. 1982), which has been confirmed by studies with culture-independent molecular methods (Siqueira and Rôças 2009a, b). The most represented, abundant, and prevalent phyla in infected root canal were Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria at the phylum level (Siqueira 2011) and Olsenella uli, Prevotella baroniae, Porphyromonas endodontalis, Fusobacterium nucleatum, and Tannerella forsythia at the species level (Rôças et al. 2010). There are inter-individual variations in the microbial spectrum of infected root canals, that is, the microbial communities in infected root canals of different individuals are not exactly the same (Machado de Oliveira et al. 2007; Santos et al. 2011; Siqueira et al. 2008; Siqueira 2011). The microbial communities associated with root canal infection also differ according to the location of the affected tooth (Alves et al. 2009; Rôças et al. 2010). Even within a single infected root canal, the microbial composition in the apical and coronal regions differs: the former typically has a higher level of microbial diversity than the latter (Özok et al. 2012), and the dominant microbes are different (the apical area mainly contains obligate anaerobes) (Siqueira and Rôças 2005).
Although most of the periapical periodontitis can be controlled with root canal therapy, in some cases, the infection could be persistent or the filled canal could be re-infected. By using PCR-DGGE, Chugal et al. (2011) compared the microbiota residing at the apical portion of primary infected canals and root canals with failed treatments and found that the apical bacterial communities in primary infections were significantly more diverse, and different roots of the same teeth with primary infections contained almost identical bacterial composition while an equivalent sample collected from unsuccessfully treated tooth displayed low similarity.
In a small proportion of cases, bacteria colonized in the root canals could also develop communities on root surfaces outside the apical foramen and cause extraradicular infections, which is associated with refractory periapical periodontitis. Bacterial taxa detected in extraradicular infections belong to six phyla (Siqueira and Rôças 2009b). Species reported include Propionibacterium propionicum, Porphyromonas gingivalis, Prevotella intermedia, Prevotella oralis, Parvimonas micra, P. endodontalis, F. nucleatum, and T. forsythia (Noguchi et al. 2005; Siqueira and Rôças 2009b; Su et al. 2010).
The extensive application of molecular methods has not only dramatically increased the amount of available information related to root canal infection but has also, to some extent, led to changes in our understanding of the etiology of root canal infection (Siqueira and Rôças 2009a). For a long time, some Gram-negative bacteria were believed to be associated with specific symptoms of periapical periodontitis (Gomes et al. 1994). Because these Gram-negative bacteria were detected at similar frequencies in infected root canals without symptoms (Baumgartner et al. 1999; Rôças and Siqueira 2008; Siqueira et al. 2000), the overall structure of the microbial community are supposed to be closely associated with the symptoms instead (Sakamoto et al. 2006; Siqueira et al. 2004). For example, a much more diverse root canal microbial community was observed among periapical periodontitis patients with acute infection symptoms than these without clinic symptoms (Santos et al. 2011).
Periodontal diseases
Gingivitis is an inflammation limited in the gingiva. It is believed to result from the accumulation of plaque and the associated interactions between bacteria in dental plaque and gingival tissues (Moore et al. 1987). With the development of gingivitis, the dominant bacteria in subgingival plaque gradually shift from Streptococcus to Actinomycetes, Capnocytophaga, Campylobacter, Eikenella, Fusobacterium, and Prevotella (Zaura et al. 2009). Besides, the saliva microbiome of people with and without gingivitis showed significant differences (Huang et al. 2011).
Periodontitis is also a chronic inflammatory disease of the tooth-supporting tissues and the leading cause of tooth loss worldwide (Pihlstrom et al. 2005). Compared with gingivitis, it causes not only the progressive destruction of the gum but also periodontal membrane and alveolar bone. In contrast to previous results obtained using bacterial cultures, studies with methods based on 16S rRNA gene PCR/cloning/sequencing techniques have demonstrated that most dominant bacteria in sites affected by periodontitis are not Gram-negative species (Wade 2011). The reasons for the different findings might be that staining characteristics of Gram-positive anaerobic bacteria colonizing the subgingival plaque could vary and that excessively long periods of culture could also lead to negative gram staining (Kononen and Wade 2007). Another factor is that the culture technique has bias, and it only detects what is specific for the culturing conditions.
The subgingival microbial composition in patients with periodontitis undergoes extremely complex changes. More than 400 phenotypes have been detected from periodontal pockets (Wade 2011). Griffen et al. (2012) found that the relative abundances of 123 phenotypes in the subgingival plaque microbiome were increased in periodontitis, whereas the abundances of 53 others decreased. In addition to P. gingivalis, Treponema denticola, and T. forsythia, bacteria including Bacteroidetes species, Eubacterium saphenum, P. endodontalis, Prevotella denticola, Parvimonas micra, Peptostreptococcus species, Filifactor alocis, Desulfobulbus species, Dialister species, and Synergistetes species are closely related to periodontitis (Dahlén and Leonhardt 2006; Kumar et al. 2003, 2005; Paster et al. 2006). Streptococcus, Veillonella, Abiotrophia, Campylobacter, Capnocytophaga, Gemella, and Neisseria are considered as beneficial bacteria (Kumar et al. 2005). Recently, a study focusing on microorganisms associated with aggressive periodontitis yielded surprising results: Selenomonas was the dominant microbiota in subgingival plaque whereas Aggregatibacter actinomycetemcomitans, which was formerly believed to be closely associated with this disease (Schacher et al. 2007), was not detected (Faveri et al. 2008). However, this finding should be taken with cautious: one possibility is that A. actinomycetemcomitans is not present in aggressive periodontitis samples, and another possibility is that A. actinomycetemcomitans is below the detection of 16S rRNA gene clones. The subgingival plaque of refractory periodontitis patients contains a greater number of Parvimonas micra, Campylobacter gracilis, Eubacterium nodatum, Selenomonas noxia, T. forsythia, P. gingivalis, Prevotella species, Treponema species, and Eikenella corrodens compared to healthy subjects (Colombo et al. 2012, 2009).
As an important risk factor for periodontitis, smoking could affect the subgingival microbiota composition. In the subgingival environment, greater abundances of species belonging to Bacteroides, Campylobacter, Fusobacterium, Parvimonas, and Porphyromonas, and lower abundances of Veillonella, Neisseria, and Streptococcus have been detected in smokers with periodontitis than in never smokers (Shchipkova et al. 2010). Moreover, tobacco smoking also causes changes in the symbiotic and commensalistic relationships among the subgingival microbes. For example, never-smoking patients with high levels of Streptococci exhibited low levels Parvimonas while current smokers with high levels of Streptococci demonstrated high levels of Parvimonas (Shchipkova et al. 2010). Also, recent evidence suggests that Streptococci play an essential role in preventing colonization of periodontal ecology by pathogens (Stingu et al. 2008; Van Hoogmoed et al. 2008). It is possible that the protective function of Streptococci is impaired by tobacco, leading to a co-colonization pattern alteration. In addition, smoking cessation results in a decrease in the prevalence of P. endodontalis and Dialister pneumosintes and in the relative abundances of Parvimonas micra, Filifactor alocis, and Treponema denticola (Delima et al. 2010). An increase in the proportion of beneficial bacteria Veillonella parvula was also observed after quitting tobacco (Delima et al. 2010).
Halitosis
Halitosis (oral malodor) refers to unpleasant odor exhaled in breathing; it can be classified into intra-oral halitosis and extra-oral halitosis (Murata et al. 2002). Volatile sulfur compounds and malodorous fatty acids produced from the decomposition of sulfur-containing amino acids, peptides, and proteins by oral bacteria are considered to be the direct cause of intra-oral halitosis (Murata et al. 2002).
It has been gradually recognized that bacteria on the tongue dorsum, especially the posterior dorsum, are the main factor leading to intra-oral halitosis in people with complete dentition and healthy periodontal tissues (Allaker et al. 2008; Porter and Scully 2006). There are differences in the microbial diversity of the tongue dorsum between patients with intra-oral halitosis and healthy controls. The dominant bacteria found in the tongue dorsum of patients with intra-oral halitosis include Solobacterium moorei, Atopobium parvulum, and Eubacterium sulci, whereas S. salivarius, Rothia mucilaginosa, and an uncharacterized cultivable Eubacterium species are abundant in healthy controls (Kazor et al. 2003). It was also shown that Solobacterium moorei was only found in specimens from patients with halitosis (Haraszthy et al. 2007); these bacteria were once called the “arch-criminal of halitosis.” The oral distributions of S. salivarius in patients with halitosis and in healthy populations remain controversial. For example, Kazor et al. (2003) suggested that S. salivarius was the most dominant species on the tongue dorsum of healthy controls and was relatively rare or even absent on the tongue dorsum of halitosis patients. In contrast, Riggio et al. (2008) found that it was the dominant species in both halitosis patients and healthy persons with the same method (i.e., 16S rRNA gene cloning). Until today, there are still researchers who believe that S. salivarius represents as a benign commensal probiotic for the reduction of oral malodor (Masdea et al. 2012).
Oral bacteria and systemic diseases
Tumor
A tumor is a mass of tissue as a result of abnormal growth or division of cells (Cooper 1992). Microbe-induced inflammation is involved in 15–20 % of human tumors (Allavena et al. 2008). Researches in this area have focused on the correlation between the microbiome and tumor occurrence, microbiome shifts in cancer patients, the feasibility of identifying early diagnostic markers from the microbiome, and the effects of tumor treatments on the microbiome (Meurman 2010).
Oral squamous cell carcinoma (OSCC) is the most common malignant tumor in the oral cavity (Bagan et al. 2010). Levels of Capnocytophaga gingivalis, Prevotella melaninogenica, and Streptococcus mitis in the saliva of patients with OSCC significantly increased (Mager et al. 2005), and the salivary microbiomes of cancer patients were more similar to each other than the microbiomes of individuals in healthy populations (Pushalkar et al. 2011). The OSCC surface biofilm harbors increased aerobes and anaerobes, including Veillonella, Fusobacterium, Prevotella, Porphyromonas, Actinomyces, Clostridium, Haemophilus, Enterobacteriaceae, and Streptococcus species (Nagy et al. 1998). In addition, bacteria could be detected within the OSCC tissues. Hooper et al. (2007) found 52 different bacterial phylotypes from tumorous tissues, and the majority of species were saccharolytic and aciduric, including Proteobacteria, Fusobacterium, Streptococcus, Prevotella, and Veillonella, which is consistent with a previous study indicating the acidic and hypoxic microenvironment within tumor tissues (Raghunand et al. 2003).
The oral microbiome is also involved in or affected by tumors of distant organs (Ahn et al. 2012; Farrell et al. 2011). For example, the proportions of S. mitis and Neisseria elongata in the salivary microbiome were significantly lower in patients with pancreatic cancer than in healthy people (Farrell et al. 2011). As the authors discussed, the cross-sectional nature of this study has not enabled us to understand the mechanisms or the association, and whether these two types of bacteria can be called markers for early diagnosis of pancreatic cancer has yet to be investigated.
Surgery, radiotherapy, and chemotherapy are three primary approaches to cancer treatments. The changes of oral microbiome undergoing chemoradiotherapy have been confirmed by many approaches, including high-throughput sequencing (Hu et al. 2013; Meurman 2010; Napeñas et al. 2010; Shao et al. 2011; Xu et al. 2014). These studies suggested a shift to a more complex oral bacterial profile in patients undergoing cancer chemotherapy by a host-specific manner.
Diabetes mellitus
Diabetes mellitus (DM) is a clinical syndrome characterized by hyperglycemia due to a deficiency in the secretion of insulin and/or reduced insulin action (Alberti and Zimmet 1998). There are two main types: type 1 DM resulting from the body’s failure to produce insulin and type 2 DM resulting from insulin resistance. Both type 1 and type 2 DM show a three- to four-fold increased risk of periodontitis which is regarded as the sixth complication of DM (Löe 1993).
Some studies have explored the composition of subgingival dental plaque in diabetics compared with non-diabetics; however, no agreement has been reached regarding the effect of DM on periodontal subgingival microbiota. Hintao et al. (2007) found increased frequency of Treponema denticola, Streptococcus sanguinis, Prevotella nigrescens, Staphylococcus intermedius, and Streptococcus oralis in the supragingival plaque of type 2 diabetics compared with non-diabetics while no significant differences were found in subgingival plaque samples. Similar subgingival infection patterns were also observed between type 1 diabetics and non-diabetics after controlling of the periodontal severity (Lalla et al. 2006). In contrast, Ebersole et al. (2008) demonstrated that the periodontitis sites in type 1 DM patients showed a higher frequency of P. gingivalis, A. actinomycetemcomitans, and Campylobacter spp. A higher prevalence of P. gingivalis, Candida spp. (mainly Candida albicans and Candida dubliniensis), as well as a lower frequency of T. forsythia was also demonstrated in type 2 diabetics (Campus et al. 2005; Sardi et al. 2011). In a much more recent study using 16S rRNA gene sequencing, significant differences were observed in subgingival microbiota between type-2 DM and non-diabetic subjects: diabetic subjects presented higher abundance of total clones of TM7, Aggregatibacter, Neisseria, Actinomyces, Capnocytophaga, Gemella, Eikenella, Selenomonas, Fusobacterium, Veillonella, and Streptococcus and lower percentages of Synergistetes, Tannerella, Porphyromonas, Filifactor, Eubacterium, and Treponemas; moreover, some species, such as F. nucleatum, V. parvula, Veillonella dispar, and E. corrodens were detected significantly more often in diabetics (Casarin et al. 2013). Considering the elevated glucose content in subgingival microenvironment and altered or impeded immune responses of hosts (Ohlrich et al. 2010), there may indeed be differences in the subgingival microbiome in diabetic patients compared with non-diabetics.
Cardiovascular disease
Cardiovascular diseases are a set of diseases that include congestive heart failure, cardiac arrhythmias, valvular heart disease, and stroke and coronary artery disease (including atherosclerosis and myocardial infarction) (Ross 1999).
Atherosclerosis, a major component of cardiovascular diseases, is a condition caused by abnormal lipid metabolism and neurovascular dysfunction, in which yellow substances containing cholesterol and fat appear in the intima of large and medium arteries, often leading to thrombosis and ischemia (Ross 1999). Oral microbiota, including Streptococcus, Veillonella, P. gingivalis, F. nucleatum, T. forsythia, and Neisseria were detected from atherosclerotic plaques (Figuero et al. 2011; Ford et al. 2005; Koren et al. 2011; Pucar et al. 2007). The levels of Fusobacterium, Streptococcus, and Neisseria were found to be related to the risk factors for the disease, such as the plasma cholesterol level (Koren et al. 2011). These findings also indirectly support one of the pathogenic models of cardiovascular disease, namely, the infection model (the bacteria invade the bloodstream and subsequently get into the endothelium, resulting in endothelial dysfunction, inflammation, and atherosclerosis) (Seymour et al. 2007).
The atherosclerotic plaque rupture participating in thrombus formation and chronic inflammation may cause plaque instability (Libby et al. 2002). Chiu (1999) found P. gingivalis and S. sanguinis in unstable atherosclerotic plaques, and Ohki et al. (2012) detected A. actinomycetemcomitans, P. gingivalis, and Treponema denticola in thrombi of patients with acute myocardial infarction by PCR. These studies not only confirmed the presence of oral bacteria but also suggested that oral microbiota might have a role in plaque inflammation and instability.
Bacteremia
Bacteremia is an invasion of the bloodstream by bacteria. Invasive dental manipulation (tooth extraction for instance), as well as daily oral hygiene activities (brushing the teeth for example), can produce bacteremia (Poveda-Roda et al. 2008). According to the study of Bahrani-Mougeot et al. (2008), the most predominant species in the blood of following dental procedures were Streptococcus spp., followed by Peptostreptococcus micros, Veillonella dispar or V. parvula, and D. pneumosintes. Another research also confirmed that the most predominant Streptococcus species were S. mitis, S. oralis, and S. sanguinis (Forner et al. 2006). Although majority of such bacteremia is transient, it might also be a risk factor of distant site infections. For example, oral streptococci’s ability to aggregate platelets is a potential pathogenic factor in the development of endocarditis and formation of thrombi (Herzberg and Meyer 1996). A relationship has been reported between bacteremia caused by tooth brushing and the risk of cardiovascular diseases (Roberts 1999).
Other systemic diseases
The application of molecular biological methods continues to expand our understanding of the relationship between oral microorganisms and systemic diseases. For example, Docktor et al. (2012) utilized the human oral microbiome chip to reveal that tongue microbial diversity was lower in pediatric patients with Crohn’s disease than in healthy children and to show that changes in Fusobacteria and Firmicutes were among the most significant. Han et al. (2010) identified F. nucleatum with an oral origin in the uteri of miscarried women and preterm birth and low birth weight has been associated with high levels of T. forsythia, Campylobacter rectus, Prevotella intermedia, Prevotella nigrescens, and P. gingivalis (Mitchell-Lewis et al. 2001; Offenbacher et al. 2006). Goodson et al. (2009) even proposed that oral microorganisms might be involved in obesity.
Oral fungal, viral, and archaeal microbiome
Oral fungal microbiome
Fungi comprise a minor component of the oral microbiome, and they are collectively named oral fungal microbiome or mycobiome (Ghannoum et al. 2010). In a recent study aiming to obtain a comprehensive profile of the oral mycobiome, researchers identified 74 cultivable and 11 uncultivable fungal genera from oral rinse of 20 healthy individuals with pyrosequencing (Ghannoum et al. 2010). It was also found that each individual carried 9~23 fungal species, and the detection rate of Candida in the subjects was the highest, followed by Cladosporium, Aureobasidium, Saccharomyces, Aspergillus, Fusarium, and Cryptococcus (Ghannoum et al. 2010). The composition of fungal microbiome is, to some extent, associated with gender and race; it differs between white males and Asian males, but it is similar between white females and Asian females (Ghannoum et al. 2010).
Candida are the most frequently detected oral fungi (Ghannoum et al. 2010). The most common Candida species is C. albicans and less common species include Candida glabrata, Candida parapsilosis, Candida tropicalis, Candida krusei, Candida stellatoidea, Candida kefyr, and C. dubliniensis (Krishnan 2012). The frequency, intensity, species, and strains of oral Candida varied with age (Kleinegger et al. 1996), and the frequency of C. albicans decreased with increasing age (Qi et al. 2005). More interestingly, elders with high Candida load harbored a lower diverse salivary microbiome and had a distinct microbial composition toward dominance by Streptococci (Kraneveld et al. 2012). Candidiasis is the commonest infection caused by Candida (Cannon et al. 1995). In addition, this genus is considered to be involved in some other oral diseases. Recent evidence implies that the occurrence of caries in children was positively correlated with the frequency of oral candidal carriage (Raja et al. 2010; Yang et al. 2012), and that C. albicans is able to cause occlusal caries in rats at a high rate (Klinke et al. 2011), indicating the role of Candida in the cariogenic development. Actually, in vitro studies have proven that S. mutans could enhance the adherence of C. albicans and excrete lactate as a carbon source for yeast growth while the growth of yeast reduces oxygen tension to levels preferred by streptococci and provide growth stimulatory factors for the bacteria (Brogden and Guthmiller 2008; Metwalli et al. 2013). Moreover, Candida was also detected in periodontal pockets and C. albicans was observed to be highly associated with the severity of chronic periodontitis (Canabarro et al. 2013). Although still in its infancy, the research thus far strongly warrants that investigations on the microbiology of periodontitis should include yeasts.
Oral virome
The virome is the collective of viruses that populate an organism or ecosystem at any given time (Haynes and Rohwer 2011). The oral virome contains a range of viruses, the vast majority of which present homology with bacteriophages and their presence may be closely related to oral microbial diversity (Pride et al. 2012; Robles-Sikisaka et al. 2013). Comparisons of the salivary virome with respiratory and gut virome revealed that the habitat is an important selection factor for the human virome composition (Pride et al. 2012). The living environment of hosts has a role in shaping oral viral ecology, as demonstrated by Robles-Sikisaka et al. (2013) that a significantly greater fraction of shared viral homologous reads were observed among subjects from the same family than those from separate households.
Oral virome is primarily disease-associated, such as herpes simplex virus (pathogen of herpetic gingiva-stomatitis and herpes labialis), varicella zoster virus (pathogen of herpes zoster), and human papilloma virus (pathogen of papillomas) (Kumaraswamy and Vidhya 2011; Whitley and Roizman 2001). Herpes virus, including human cytomegalovirus (HCMV), Epstein–Barr virus (EBV) type 1–2, herpes simplex virus (HSV) type 1, and human herpes virus types 6–8, could also be detected in periodontal pockets of patients with chronic periodontitis (Imbronito et al. 2008), localized and generalized juvenile periodontitis (Imbronito et al. 2008), Papillon–Lefèvre syndrome periodontitis (Velazco et al. 1999), Down’s syndrome periodontitis (Hanookai et al. 2000), HIV-associated periodontitis (Contreras et al. 2001), and acute necrotizing ulcerative gingivitis (Contreras et al. 1997). Moreover, EBV-1 and HCMV are positively associated with the subgingival presence of some periodontal pathogens (Contreras et al. 1999) and the severity of periodontitis (Ling et al. 2004; Saygun et al. 2002). So, some oral microbiologists postulate that virus might play an important role in the pathogenesis of human periodontitis (Kubar et al. 2005; Saygun et al. 2002; Slots 2010) and herpesviruses may promote the process of periodontitis through releasing tissue-destructive cytokines, initiating cytotoxic or immunopathogenic event and boosting pathogenic periodontal bacteria growth (Slots and Contreras 2000). However, the role of oral virus in the periodontal pathogenesis is under debate. For instance, although an obvious association of the viruses with clinical samples was observed, Sunde et al. (2008) still believed that their presence might reflect that clinical samples contain more saliva or blood compared to healthy controls or an accumulation of lymphoid cells harboring virus in the inflamed tissue.
Oral archaeal microbiome
The Archaea are non-bacterial prokaryotes, and they are restricted to a small number of species/phylotypes. Human Archaea are mainly found in the gut and the oral cavity, majority of which are methanogens with few exceptions (Dridi et al. 2011). The diversity of oral Archaea is limited compared to bacteria domain and the reported oral Archaea include the genera Thermoplasmatales, Methanobrevibacter, Methanobacterium, Methanosarcina, and Methanosphaera (Dridi et al. 2011; Eckburg et al. 2003; Lepp et al. 2004; Nguyen-Hieu et al. 2013).
Studies have primarily focused on the role of oral Archaea in periodontal disease (Lepp et al. 2004; Li et al. 2009; Matarazzo et al. 2012; Vianna et al. 2008) and endodontic infections (Jiang et al. 2009; Vianna et al. 2006, 2009; Vickerman et al. 2007). These reports demonstrated a higher detection frequency of Archaea was observed in the infected population. The relative abundance of Archaea in subgingival plaque increased as the severity of chronic periodontitis increased (Lepp et al. 2004), and treated periodontitis sites showed a decrease in Archaea when the treatment was followed by improvement of lesions (Lepp et al. 2004; Lira et al. 2013). Although methanogenic archaea could promote periodontal tissue destruction, the archaea are not involved in the initiation of periodontal infection (Farrell et al. 2011). Furthermore, the presence of Archaea in infected root canal might be associated with clinical symptoms (Jiang et al. 2009). Research on Archaea has begun to extend to other oral infectious diseases, such as peri-implantitis (Faveri et al. 2011) and peri-coronitis (Mansfield et al. 2012). However, the role of Archaea in oral pathologies remains controversial and further studies are required to explore the potential mechanisms of these microorganisms (Nguyen-Hieu et al. 2013).
Conclusions
Although we can only partially annotate a portion of the completely sequenced oral microbiome, the vast amounts of data present an encrypted book available to researchers interested in the oral microbiome. Interpretation of the coded information within this encrypted book is sure to encourage and facilitate the investigation of microbial pathogenic mechanisms, drug development, and the identification of new diagnostic markers.
References
Aas JA, Paster BJ, Stokes LN et al (2005) Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43:5721–5732
Aas JA, Griffen AL, Dardis SR et al (2008) Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417
Ahn J, Chen C, Hayes R (2012) Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 23:399–404
Alberti KGMM, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Diabet Med 15:539–553
Allaker RP, Waite RD, Hickling J et al (2008) Topographic distribution of bacteria associated with oral malodour on the tongue. Arch Oral Biol 53(Supplement 1):8–12
Allavena P, Garlanda C, Borrello MG et al (2008) Pathways connecting inflammation and cancer. Curr Opin Genet Dev 18:3–11
Alves FRF, Siqueira JF Jr, Carmo FL et al (2009) Bacterial community profiling of cryogenically ground samples from the apical and coronal root segments of teeth with apical periodontitis. J Endod 35:486–492
Bäckhed F, Ley RE, Sonnenburg JL et al (2005) Host-bacterial mutualism in the human intestine. Science 307:1915–1920
Bagan J, Sarrion G, Jimenez Y (2010) Oral cancer: clinical features. Oral Oncol 46:414–417
Bahrani-Mougeot FK, Paster BJ, Coleman S et al (2008) Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol 46:2129–2132
Balakrishnan M, Simmonds RS, Tagg JR (2000) Dental caries is a preventable infectious disease. Aust Dent J 45:235–245
Baumgartner JC, Watkins BJ, Bae KS et al (1999) Association of black-pigmented bacteria with endodontic infections. J Endod 25:413–415
Becker MR, Paster BJ, Leys EJ et al (2002) Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40:1001–1009
Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R et al (2012) The oral metagenome in health and disease. ISME J 6:46–56
Bik EM, Long CD, Armitage GC et al (2010) Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4:962–974
Brogden KM, Guthmiller JM (2008) Polymicrobial diseases. ASM, Washington
Byström A, Happonen R-P, Sjögren U et al (1987) Healing of periapical lesions of pulpless teeth after endodontic treatment with controlled asepsis. Dent Traumatol 3:58–63
Campus G, Salem A, Uzzau S et al (2005) Diabetes and periodontal disease: a case–control study. J Periodontol 76:418–425
Canabarro A, Valle C, Farias MR et al (2013) Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J Periodontal Res 48:428–432
Cannon RD, Holmes AR, Mason AB et al (1995) Oral candida: clearance, colonization, or Candidiasis? J Dent Res 74:1152–1161
Casarin RCV, Barbagallo A, Meulman T et al (2013) Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J Periodontal Res 48:30–36
Chiu B (1999) Multiple infections in carotid atherosclerotic plaques. Am Heart J 138:534–536
Chugal N, Wang J, Wang R et al (2011) Molecular characterization of the microbial flora residing at the apical portion of infected root canals of human teeth. J Endod 37:1359–1364
Colombo APV, Boches SK, Cotton SL et al (2009) Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the Human Oral Microbe Identification Microarray. J Periodontol 80:1421–1432
Colombo AP, Bennet S, Cotton SL et al (2012) Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol 83:1279–1287
Contreras A, Falkler WA, Enwonwu CO et al (1997) Human Herpesviridae in acute necrotizing ulcerative gingivitis in children in Nigeria. Oral Microbiol Immunol 12:259–265
Contreras A, Umeda M, Chen C et al (1999) Relationship between herpesviruses and adult periodontitis and periodontopathic bacteria. J Periodontol 70:478–484
Contreras A, Mardirossian A, Slots J (2001) Herpesviruses in HIV-periodontitis. J Clin Periodontol 28:96–102
Cooper GM (1992) Elements of human cancer. Jones and Bartlett, Boston
Dahlén G, Leonhardt Å (2006) A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol Immunol 21:6–11
Darveau RP (2010) Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Micro 8:481–490
Delima SL, McBride RK, Preshaw PM et al (2010) Response of subgingival bacteria to smoking cessation. J Clin Microbiol 48:2344–2349
Dethlefsen L, McFall-Ngai M, Relman DA (2007) An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449:811–818
Dewhirst FE, Chen T, Izard J et al (2010) The human oral microbiome. J Bacteriol 192:5002–5017
Docktor MJ, Paster BJ, Abramowicz S et al (2012) Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis 18:935–942
Dridi B, Raoult D, Drancourt M (2011) Archaea as emerging organisms in complex human microbiomes. Anaerobe 17:56–63
Drury TF, Horowitz AM, Ismail AI et al (1999) Diagnosing and reporting early childhood caries for research purposes: a report of a workshop sponsored by the National Institute of Dental and Craniofacial Research, the Health Resources and Services Administration, and the Health Care Financing Administration. J Public Health Dent 59:192–197
Ebersole JL, Holt SC, Hansard R et al (2008) Microbiologic and immunologic characteristics of periodontal disease in Hispanic Americans with type 2 diabetes. J Periodontol 79:637–646
Eckburg PB, Lepp PW, Relman DA (2003) Archaea and their potential role in human disease. Infect Immun 71:591–596
Fábián TK, Fejérdy P, Csermely P (2008) Salivary genomics, transcriptomics and proteomics: the emerging concept of the oral ecosystem and their use in the early diagnosis of cancer and other diseases. Curr Genomics 9:11–21
Fabricious L, Dahlen G, ÖHman AE et al (1982) Predominant indigenous oral bacteria isolated from infected root canals after varied times of closure. Eur J Oral Sci 90:134–144
Farrell JJ, Zhang L, Zhou H et al (2011) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61:582–588
Faveri M, Mayer MPA, Feres M et al (2008) Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol Immunol 23:112–118
Faveri M, Gonçalves LFH, Feres M et al (2011) Prevalence and microbiological diversity of Archaea in peri-implantitis subjects by 16S ribosomal RNA clonal analysis. J Periodontal Res 46:338–344
Figuero E, Sánchez-Beltrán M, Cuesta-Frechoso S et al (2011) Detection of periodontal bacteria in atheromatous plaque by nested polymerase chain reaction. J Periodontol 82:1469–1477
Filoche S, Wong L, Sissons CH (2010) Oral biofilms: emerging concepts in microbial ecology. J Dent Res 89:8–18
Ford PJ, Gemmell E, Hamlet SM et al (2005) Cross-reactivity of GroEL antibodies with human heat shock protein 60 and quantification of pathogens in atherosclerosis. Oral Microbiol Immunol 20:296–302
Forner L, Larsen T, Kilian M et al (2006) Incidence of bacteremia after chewing, tooth brushing and scaling in individuals with periodontal inflammation. J Clin Periodontol 33:401–407
Gersdorf H, Pelz K, Göbel UB (1993) Fluorescence in situ hybridization for direct visualization of Gram-negative anaerobes in subgingival plaque samples. FEMS Immunol Med Microbiol 6:109–114
Ghannoum MA, Jurevic RJ, Mukherjee PK et al (2010) Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713
Gill SR, Pop M, DeBoy RT et al (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359
Gomes BPFA, Drucker DB, Lilley JD (1994) Association of specific bacteria with some endodontic signs and symptoms. Int Endod J 27:291–298
Goodson JM, Groppo D, Halem S et al (2009) Is obesity an oral bacterial disease? J Dent Res 88:519–523
Griffen AL, Beall CJ, Campbell JH et al (2012) Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185
Gross EL, Leys EJ, Gasparovich SR et al (2010) Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 48:4121–4128
Hajishengallis G, Lamont RJ (2012) Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419
Han YW, Fardini Y, Chen C et al (2010) Term stillbirth caused by oral Fusobacterium nucleatum. Obstet Gynecol 115:442–445
Hanookai D, Nowzari H, Contreras A et al (2000) Herpesviruses and periodontopathic bacteria in trisomy 21 periodontitis. J Periodontol 71:376–384
Haraszthy VI, Zambon JJ, Sreenivasan PK et al (2007) Identification of oral bacterial species associated with halitosis. J Am Dent Assoc 138:1113–1120
Haynes M, Rohwer F (2011) The Human virome. In: Nelson KE (ed) Metagenomics of the human body. Springer, New York, pp 63–77
Heijtz RD, Wang S, Anuar F et al (2011) Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 108:3047–3052
Herzberg MC, Meyer MW (1996) Effects of oral flora on platelets: possible consequences in cardiovascular disease. J Periodontol 67:1138–1142
Hintao J, Teanpaisan R, Chongsuvivatwong V et al (2007) The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral Microbiol Immunol 22:175–181
Hooper SJ, Crean SJ, Fardy MJ et al (2007) A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol 56:1651–1659
Hu YJ, Wang Q, Jiang YT et al (2013) Characterization of oral bacterial diversity of irradiated patients by high-throughput sequencing. Int J Oral Sci 5:21–25
Huang S, Yang F, Zeng X et al (2011) Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health 11:33
Imbronito AV, Okuda OS, Maria de Freitas N et al (2008) Detection of herpesviruses and periodontal pathogens in subgingival plaque of patients with chronic periodontitis, generalized aggressive periodontitis, or gingivitis. J Periodontol 79:2313–2321
Jiang YT, Xia WW, Li CL et al (2009) Preliminary study of the presence and association of bacteria and archaea in teeth with apical periodontitis. Int Endod J 42:1096–1103
José FS Jr (2002) Endodontic infections: concepts, paradigms, and perspectives. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 94:281–293
Kanasi E, Dewhirst FE, Chalmers NI et al (2010) Clonal analysis of the microbiota of severe early childhood caries. Caries Res 44:485–497
Kazor CE, Mitchell PM, Lee AM et al (2003) Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol 41:558–563
Keijser BJF, Zaura E, Huse SM et al (2008) Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87:1016–1020
Kleinberg I (2002) A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med 13:108–125
Kleinegger CL, Lockhart SR, Vargas K et al (1996) Frequency, intensity, species, and strains of oral Candida vary as a function of host age. J Clin Microbiol 34:2246–2254
Klinke T, Guggenheim B, Klimm W et al (2011) Dental caries in rats associated with Candida albicans. Caries Res 45:100–106
Kononen E, Wade (2007) Manual of clinical microbiology. ASM, Washington
Koren O, Spor A, Felin J et al (2011) Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A 108:4592–4598
Kraneveld EA, Buijs MJ, Bonder MJ et al (2012) The relation between oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS ONE 7:e42770
Krishnan PA (2012) Fungal infections of the oral mucosa. Indian J Dent Res 23:650–659
Kubar A, Saygun I, Özdemir A et al (2005) Real-time polymerase chain reaction quantification of human cytomegalovirus and Epstein-Barr virus in periodontal pockets and the adjacent gingiva of periodontitis lesions. J Periodontal Res 40:97–104
Kumar PS, Griffen AL, Barton JA et al (2003) New bacterial species associated with chronic periodontitis. J Dent Res 82:338–344
Kumar PS, Griffen AL, Moeschberger ML et al (2005) Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol 43:3944–3955
Kumaraswamy KL, Vidhya M (2011) Human papilloma virus and oral infections: an update. J Cancer Res Ther 7:120–127
Lalla E, Kaplan S, Chang SMJ et al (2006) Periodontal infection profiles in type 1 diabetes. J Clin Periodontol 33:855–862
Lazarevic V, Whiteson K, Hernandez D et al (2010) Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11:523
Lederberg J, McCray AT (2001) ‘Ome sweet’omics—a genealogical treasury of words. Scientist 15:2–8
Lepp PW, Brinig MM, Ouverney CC et al (2004) Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A 101:6176–6181
Li M, Wang B, Zhang M et al (2008) Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A 105:2117–2122
Li CL, Liu DL, Jiang YT et al (2009) Prevalence and molecular diversity of Archaea in subgingival pockets of periodontitis patients. Oral Microbiol Immunol 24:343–346
Libby P, Ridker PM, Maseri A (2002) Inflammation and atherosclerosis. Circulation 105:1135–1143
Lif Holgerson P, Harnevik L, Hernell O et al (2011) Mode of birth delivery affects oral microbiota in infants. J Dent Res 90:1183–1188
Ling LJ, Ho CC, Wu CY et al (2004) Association between human herpesviruses and the severity of periodontitis. J Periodontol 75:1479–1485
Ling Z, Kong J, Jia P et al (2010) Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol 60:677–690
Lira EAG, Ramiro FS, Chiarelli FM et al (2013) Reduction in prevalence of Archaea after periodontal therapy in subjects with generalized aggressive periodontitis. Aust Dent J 58:442–447
Löe H (1993) Periodontal disease: the sixth complication of diabetes mellitus. Diabetes Care 16:329–334
Machado de Oliveira JC, Siqueira JF, Rôças IN et al (2007) Bacterial community profiles of endodontic abscesses from Brazilian and USA subjects as compared by denaturing gradient gel electrophoresis analysis. Oral Microbiol Immunol 22:14–18
Mager DL, Ximenez-Fyvie LA, Haffajee AD et al (2003) Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 30:644–654
Mager D, Haffajee A, Devlin P et al (2005) The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med 3:27
Mansfield JM, Campbell JH, Bhandari AR et al (2012) Molecular analysis of 16S rRNA genes identifies potentially periodontal pathogenic bacteria and Archaea in the plaque of partially erupted third molars. J Oral Maxillofac Surg 70:1507–1514
Marsh PD (2005) Dental plaque: biological significance of a biofilm and community life-style. J Clin Periodontol 32:7–15
Masdea L, Kulik EM, Hauser-Gerspach I et al (2012) Antimicrobial activity of Streptococcus salivarius K12 on bacteria involved in oral malodour. Arch Oral Biol 57:1041–1047
Matarazzo F, Ribeiro AC, Faveri M et al (2012) The domain Archaea in human mucosal surfaces. Clin Microbiol Infect 18:834–840
Medzhitov R (2007) Recognition of microorganisms and activation of the immune response. Nature 449:819–826
Metwalli KH, Khan SA, Krom BP et al (2013) Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog 9:e1003616
Meurman JH (2010) Oral microbiota and cancer. J Oral Microbiol. doi:10.3402/jom.v2i0.5195
Mitchell-Lewis D, Engebretson SP, Chen J et al (2001) Periodontal infections and pre-term birth: early findings from a cohort of young minority women in New York. Eur J Oral Sci 109:34–39
Moore LVH, Moore WEC, Cato EP et al (1987) Bacteriology of human gingivitis. J Dent Res 66:989–995
Murata T, Yamaga T, Iida T et al (2002) Classification and examination of halitosis. Int Dent J 52:181–186
Nagy KN, Sonkodi I, Szöke I et al (1998) The microflora associated with human oral carcinomas. Oral Oncol 34:304–308
Napeñas JJ, Brennan MT, Coleman S et al (2010) Molecular methodology to assess the impact of cancer chemotherapy on the oral bacterial flora: a pilot study. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 109:554–560
Nasidze I, Li J, Quinque D et al (2009) Global diversity in the human salivary microbiome. Genome Res 19:636–643
Nguyen-Hieu T, Khelaifia S, Aboudharam G et al (2013) Methanogenic archaea in subgingival sites: a review. APMIS 121:467–477
Noguchi N, Noiri Y, Narimatsu M et al (2005) Identification and localization of extraradicular biofilm-forming bacteria associated with refractory endodontic pathogens. Appl Environ Microbiol 71:8738–8743
Offenbacher S, Lin D, Strauss R et al (2006) Effects of periodontal therapy during pregnancy on periodontal status, biologic parameters, and pregnancy outcomes: a pilot study. J Periodontol 77:2011–2024
Ohki T, Itabashi Y, Kohno T et al (2012) Detection of periodontal bacteria in thrombi of patients with acute myocardial infarction by polymerase chain reaction. Am Heart J 163:164–167
Ohlrich EJ, Cullinan MP, Leichter JW (2010) Diabetes, periodontitis, and the subgingival microbiota. J Oral Microbiol. doi:10.3402/jom.v2i0.5818
Özok AR, Persoon IF, Huse SM et al (2012) Ecology of the microbiome of the infected root canal system: a comparison between apical and coronal root segments. Int Endod J 45:530–541
Paster BJ, Olsen I, Aas JA et al (2006) The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000(42):80–87
Pihlstrom BL, Michalowicz BS, Johnson NW (2005) Periodontal diseases. Lancet 366:1809–1820
Porter SR, Scully C (2006) Oral malodour (halitosis). Brit Med J 333:632–635
Poveda-Roda R, Jimenez Y, Carbonell E et al (2008) Bacteremia originating in the oral cavity. A review. Med Oral Patol Oral Cir Bucal 13:E355–E362
Preza D, Olsen I, Aas JA et al (2008) Bacterial profiles of root caries in elderly patients. J Clin Microbiol 46:2015–2021
Preza D, Olsen I, Willumsen T et al (2009) Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis 28:509–517
Pride DT, Salzman J, Haynes M et al (2012) Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J 6:915–926
Pucar A, Milasin J, Lekovic V et al (2007) Correlation between atherosclerosis and periodontal putative pathogenic bacterial infections in coronary and internal mammary arteries. J Periodontol 78:677–682
Pushalkar S, Mane SP, Ji X et al (2011) Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol 61:269–277
Qi QG, Hu T, Zhou XD (2005) Frequency, species and molecular characterization of oral Candida in hosts of different age in China. J Oral Pathol Med 34:352–356
Raghunand N, Gatenby RA, Gillies RJ (2003) Microenvironmental and cellular consequences of altered blood flow in tumours. Br J Radiol 76:11–22
Raja M, Hannan A, Ali K (2010) Association of oral candidal carriage with dental caries in children. Caries Res 44:272–276
Riggio MP, Lennon A, Rolph HJ et al (2008) Molecular identification of bacteria on the tongue dorsum of subjects with and without halitosis. Oral Dis 14:251–258
Roberts GJ (1999) Dentists are innocent! “Everyday” bacteremia is the real culprit: a review and assessment of the evidence that dental surgical procedures are a principal cause of bacterial endocarditis in children. Pediatr Cardiol 20:317–325
Robles-Sikisaka R, Ly M, Boehm T et al (2013) Association between living environment and human oral viral ecology. ISME J 7:1710–1724
Rôças IN, Siqueira JF (2008) Root canal microbiota of teeth with chronic apical periodontitis. J Clin Microbiol 46:3599–3606
Rôças IN, Alves FRF, Santos AL et al (2010) Apical root canal microbiota as determined by reverse-capture checkerboard analysis of cryogenically ground root samples from teeth with apical periodontitis. J Endod 36:1617–1621
Ross R (1999) Atherosclerosis— an inflammatory disease. New Engl J Med 340:115–126
Sakamoto M, Umeda M, Ishikawa I et al (2000) Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol Immunol 44:643–652
Sakamoto M, Rôças IN, Siqueira JF et al (2006) Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol Immunol 21:112–122
Santos AL, Siqueira JF, Rôças JIN et al (2011) Comparing the bacterial diversity of acute and chronic dental root canal infections. PLoS One 6(11):e28088
Sardi JCO, Duque C, Camargo GACG et al (2011) Periodontal conditions and prevalence of putative periodontopathogens and Candida spp. in insulin-dependent type 2 diabetic and non-diabetic patients with chronic periodontitis—a pilot study. Arch Oral Biol 56:1098–1105
Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31:107–133
Saygun I, Şahin S, Özdemir A et al (2002) Detection of human viruses in patients with chronic periodontitis and the relationship between viruses and clinical parameters. J Periodontol 73:1437–1443
Schacher B, Baron F, Roßberg M et al (2007) Aggregatibacter actinomycetemcomitans as indicator for aggressive periodontitis by two analysing strategies. J Clin Periodontol 34:566–573
Selwitz RH, Ismail AI, Pitts NB (2007) Dental caries. Lancet 369:51–59
Seymour GJ, Ford PJ, Cullinan MP et al (2007) Relationship between periodontal infections and systemic disease. Clin Microbiol Infect 13:3–10
Shao ZY, Tang ZS, Yan C et al (2011) Effects of intensity-modulated radiotherapy on human oral microflora. J Radiat Res 52:834–839
Shchipkova AY, Nagaraja HN, Kumar PS (2010) Subgingival microbial profiles of smokers with periodontitis. J Dent Res 89:1247–1253
Siqueira JFJ (2011) Treatment of endodontic infections. Quintessence, London
Siqueira JF, Rôças IN (2005) Uncultivated phylotypes and newly named species associated with primary and persistent endodontic infections. J Clin Microbiol 43:3314–3319
Siqueira JF, Rôças IN (2009a) Community as the unit of pathogenicity: an emerging concept as to the microbial pathogenesis of apical periodontitis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 107:870–878
Siqueira JF, Rôças IN (2009b) Diversity of endodontic microbiota revisited. J Dent Res 88:969–981
Siqueira JF Jr, Rôças IN, Souto R et al (2000) Checkerboard DNA-DNA hybridization analysis of endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 89:744–748
Siqueira JF, Rôças IN, Rosado AS (2004) Investigation of bacterial communities associated with asymptomatic and symptomatic endodontic infections by denaturing gradient gel electrophoresis fingerprinting approach. Oral Microbiol Immunol 19:363–370
Siqueira JF, Rôças IN, Debelian GJ et al (2008) Profiling of root canal bacterial communities associated with chronic apical periodontitis from Brazilian and Norwegian subjects. J Endod 34:1457–1461
Slots J (2010) Human viruses in periodontitis. Periodontol 2000(53):89–110
Slots J, Contreras A (2000) Herpesviruses: a unifying causative factor in periodontitis? Oral Microbiol Immunol 15:277–280
Socransky SS, Haffajee AD, Cugini MA et al (1998) Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144
Stingu CS, Eschrich K, Rodloff AC et al (2008) Periodontitis is associated with a loss of colonization by Streptococcus sanguinis. J Med Microbiol 57:495–499
Su L, Gao Y, Yu C et al (2010) Surgical endodontic treatment of refractory periapical periodontitis with extraradicular biofilm. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 110:40–44
Sunde PT, Olsen I, Enersen M et al (2008) Human cytomegalovirus and Epstein–Barr virus in apical and marginal periodontitis: a role in pathology? J Med Virol 80:1007–1011
Takahashi N, Nyvad B (2011) The role of bacteria in the caries process. J Dent Res 90:294–303
Takeshita T, Nakano Y, Kumagai T et al (2008) The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J 3:65–78
Tanner ACR, Kent RL, Holgerson PL et al (2011) Microbiota of severe early childhood caries before and after therapy. J Dent Res 90:1298–1305
The Human Microbiome Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214
Turnbaugh PJ, Ley RE, Hamady M et al (2007) The Human Microbiome Project. Nature 449:804–810
Van Hoogmoed CG, Geertsema-doornbusch GI, Teughels W et al (2008) Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbiol Immunol 23:43–48
Velazco CH, Coelho C, Salazar F et al (1999) Microbiological features of Papillon-Lefèvre syndrome periodontitis. J Clin Periodontol 26:622–627
Vianna ME, Conrads G, Gomes BPFA et al (2006) Identification and quantification of Archaea involved in primary endodontic infections. J Clin Microbiol 44:1274–1282
Vianna ME, Holtgraewe S, Seyfarth I et al (2008) Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic Archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal diseases. J Bacteriol 190:3779–3785
Vianna ME, Conrads G, Gomes BPFA et al (2009) T-RFLP-based mcrA gene analysis of methanogenic archaea in association with oral infections and evidence of a novel Methanobrevibacter phylotype. Oral Microbiol Immunol 24:417–422
Vickerman MM, Brossard KA, Funk DB et al (2007) Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J Med Microbiol 56:110–118
Wade WG (2011) Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol 38:7–16
Whitley RJ, Roizman B (2001) Herpes simplex virus infections. Lancet 357:1513–1518
Wilson M (2008) Bacteriology of humans: an ecological perspective. Blackwell, Malden
Xu Y, Teng F, Huang S et al (2014) Changes of saliva microbiota in nasopharyngeal carcinoma patients under chemoradiation therapy. Arch Oral Biol 59:176–186
Yang F, Zeng X, Ning K et al (2011a) Saliva microbiomes distinguish caries-active from healthy human populations. ISME J 6:1–10
Yang L, Liu Y, Wu H et al (2011b) Current understanding of multispecies biofilms. Int J Oral Sci 3:74–81
Yang XQ, Zhang Q, Lu LY et al (2012) Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch Oral Biol 57:1048–1053
Zaura E, Keijser BJ, Huse SM et al (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259–271
Acknowledgments
This work was supported by the International Science and Technology Cooperation Program of China (grant number 2011DFA30940), the National Basic Research Program of China (“973 Pilot Research Program,” grant number 2011CB512108), project supported by the National Natural Science Foundation of China (grant numbers 30901689 and 81172579), and Sichuan Provincial Department of Science and Technology project (2013SZ0039).
Conflict of interests
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jinzhi He and Yan Li have equal contribution to this work.
Rights and permissions
About this article
Cite this article
He, J., Li, Y., Cao, Y. et al. The oral microbiome diversity and its relation to human diseases. Folia Microbiol 60, 69–80 (2015). https://doi.org/10.1007/s12223-014-0342-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-014-0342-2