Abstract
Twenty-four acid- and bile-tolerant lactobacilli isolates from dairy products were identified and further in vitro characterized for the presence of functional traits potentially useful for probiotic applications, which included desirable and undesirable traits, such as biofilm formation, ability to inhibit intestinal pathogens, antibiotic susceptibility, and enzyme activity. The majority of examined strains were susceptible to certain antimicrobial agents (streptomycin, gentamicin, clindamycin, erythromycin, tetracycline, quinupristin–dalfopristin), except for three strains of Lactobacillus rhamnosus with minimal inhibitory concentration levels for streptomycin higher than the microbiological breakpoints (≥32 μg/mL), which are considered as resistant. Undesirable traits such as α-chymotrypsin or N-acetyl-β-glucosaminidase activities were not detected, but low β-glucuronidase, and moderate and high β-glucosidase activities were recorded in nine strains, which were eliminated from further examination together with three isolates showing unsuitable antibiotic resistance. Of the remaining 12 isolates, 4 (Lactobacillus fermentum 202, Lactobacillus gallinarum 7001, L. rhamnosus 183, and Lactobacillus plantarum L2-1) manifested an outstanding potential to inhibit selected intestinal pathogens in an agar spot test, including Escherichia coli and Salmonella spp., and simultaneously demonstrated strong biofilm-forming capacity. In conclusion, the results of our in vitro experiments showed that the above four strains had a potential probiotic value and met the criteria to be identified as a possible probiotic microorganism, with the necessity of verification through well-designed in vivo experimental, clinical, and technological studies before the strains can be used as probiotics or as starter probiotic cultures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The concept of using Lactobacillus species for disease treatment and prevention as well as health restoration and maintenance is not new. In the recent times, there has been a renewal of interest in the use of probiotics (as distinct from antibiotics), driven in large part by consumers, the lay press, and the rapid emergence of antibiotic-resistant pathogenic strains. Dairy products, together with the contents in the gastrointestinal tract, are the main sources for isolation of novel potential probiotic organisms (Ambadoyiannis et al. 2005). Lactobacillus and other beneficial strains originally isolated from dairy products are probably the most suitable candidates for inclusion into these foods as probiotics, because they are well adapted to the conditions and may therefore be more competitive than probiotics from other sources. These isolates do require further in vitro and in vivo investigations and specific studies on their technological properties in dairy fermentation. Moreover, before the suitable strains can be eventually used as probiotic starter cultures in dairy products, it is necessary to screen their potential beneficial effects without occurrence of unacceptable properties, such as harmful biochemical activities (Heavey and Rowland 2004), and transmissible antibiotic resistances (Salyers et al. 2004). The testing of their in vivo efficacy is expensive and time-consuming. For this reason, reliable in vitro methods are required for selecting new strains of probiotic bacteria to obtain profitable in vivo effects. The aim of the present study was therefore to apply in vitro tests (namely biofilm formation, ability to inhibit intestinal pathogens, antibiotic susceptibility, and enzyme activity) for validation of the probiotic potential of Lactobacillus strains isolated from dairy sources, and to select candidate probiotic strains that meet the established criteria and could therefore be potentially used as novel probiotics.
Materials and methods
Bacterial isolates and identification
The Lactobacillus isolates analyzed were obtained from the Institute of Biotechnology and Food Science, Faculty of Chemical and Food Technology (Slovak University of Technology, Bratislava, Slovakia) and were recovered from dairy products such as sheep’s milk (n = 4), cow’s milk (n = 6), cow’s cheese (n = 6), and sheep cheese (n = 8). Isolates were identified using protein “fingerprints” determined by MALDI-TOF mass spectrometry (Bruker Daltonics MALDI Biotyper) as described by Bessède et al. (2011). They belonged to the seven following species: Lactobacillus rhamnosus (n = 12), Lactobacillus fermentum (n = 5), Lactobacillus acidophilus (n = 1), Lactobacillus helveticus (n = 1), Lactobacillus gallinarum (n = 1), Lactobacillus plantarum (n = 2), and Lactobacillus casei (n = 2). All isolates were routinely grown in MRS medium (Oxoid Ltd., England) by anaerobic incubation at 37°C for 48 h.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed according to Kushiro et al. (2009) by using MIC strip tests (Liofilchem, Italy).
Enzyme activities were assayed following the method of Arora et al. (1990) using the API-ZYM system (bioMérieux, France). The inocula of 65 μL of the McFarland standard 1 suspension were deposited in each well. Enzyme activity readings were taken after 4 h of anaerobic incubation at 37°C and after addition of Zym A and Zym B reagents. Color intensity values from 0 to 5 and their relevant value in nanomoles were assigned for each reaction according to a color chart enclosed with the kit.
Biofilm formation
The strains were inoculated to the MRS agar (Oxoid) and incubated for 24 h at 37°C. One colony was transferred to 5 mL of MRS broth (Oxoid) and incubated for 24 h at 37°C. Then the culture was centrifuged (10,000×g, 10 min), and the sediment was resuspended in phosphate-buffered saline (PBS) to reach the McFarland standard 1 suspension that corresponded to 1.5–3 × 108 CFU/mL. A volume of 100 μL of the culture was inoculated in a well of a polystyrene microtitre plate (Nunc, Denmark) and incubated for 20 h at 37°C.
Crystal violet assay
A modified version of a previously described method (Toledo-Arana et al. 2001) was used. The biofilm formed in the well of the microtitre plate was washed five to six times with 200 μL of PBS and dried for 30 min at 37°C in an inverted position. A volume of 50 μL of a 1 % (W/V) solution of crystal violet (Merck) in ethanol was added and incubated for 15 min at 25°C. The dye solution was aspirated away, and the well was washed with 5 × 400 μL of distilled water. After removing water and drying for 10 min at 25°C, 200 μL of the ethanol–acetone (80:20, V/V) mixture was added. The absorbance at 570 nm of the dye solutions was measured in Synergy HT Multi-Mode Microplate Reader (BioTek, USA).
Inhibition of pathogens
The capacity of the strains to inhibit the representative groups of intestinal pathogens was determined using the agar spot test described by Jacobsen et al. (1999). The assayed strains included Escherichia coli NB2007 (vt1, vt2 positive), E. coli T12 (ESBLs), and Salmonella spp. H9812. Inhibition zones were read after finishing the incubation according to the following criteria: (−) = no inhibition, (+/−) = inhibition but no clear-cut zone, (+) = zone of inhibition between 2 and 5 mm around spots, and (++) = zone of inhibition bigger than 5 mm.
Bacteriocin, hydrogen peroxide production
Bacteriocin production was determined according to Aslim et al. (2005). Analytical Merckoqant peroxide test strips (Merck) were used to measure hydrogen peroxide production by selected lactobacilli on detection scale between 0 and 25 mg/L. The test was prepared and evaluated according manufacturer’s recommendations.
Results and discussion
Antibiotic susceptibility
The majority of our isolates were susceptible to tetracycline, with minimum inhibitory concentration (MIC) values ranging from 1 to 8 μg/mL, to gentamicin (2 to 4 μg/mL), to erythromycin (0.25 to 0.5 μg/mL), to quinupristin/dalfopristin (0.5 to 2 μg/mL), to clindamycin (0.125 to 0.5 μg/mL), and to streptomycin (8 to 32 μg/mL), except for three L. rhamnosus strains 173 (≥64 μg/mL), 123 (≥128 μg/mL), and 161 (≥64 μg/mL) with MIC levels for streptomycin higher than 32 μg/mL, which are considered as resistant according to the microbiological breakpoints given by the Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2008). From a safety point of view, this phenotypic result underlines the necessity to elucidate the resistance mechanisms at the molecular level, and in particular, the presence of antibiotic resistance genes should be carefully evaluated. In spite of this, these strains were considered as potentially hazardous and were discarded as prospective candidate probiotics.
Enzyme activities of the 21 lactobacilli strains are shown in Table 1. Alkaline phosphatase, trypsin, lipase (C14), and α-mannosidase activities, and also undesirable activities such as α-chymotrypsin or N-acetyl-β-glucosaminidase were not recorded. Weak β-glucuronidase activity (5 nmol of substrate hydrolyzed) was detected in the L. rhamnosus G1, G4, LC705, 81, and ALC01 strains. The L. rhamnosus ALC01, L. casei 163, and L. fermentum G6 showed moderate β-glucosidase activity (10–20 nmol of substrate hydrolyzed), and L. rhamnosus G1, G4, LC705, 81, and 7039 and L. helveticus G11 showed high β-glucosidase activity (30–40 nmol of substrate hydrolyzed). α-Chymotrypsin, β-glucuronosidase, β-glucosidase, and N-acetyl-β-glucosaminidase activities may have negative effects in the colon; all have been associated with intestinal diseases (Heavey and Rowland 2004). Therefore, another nine lactobacilli isolates with β-glucuronosidase and β-glucosidase activities were eliminated from further screening. The therapeutic and preventive properties of probiotics are primarily based on their metabolic potential. Bacteria used in probiotic supplements are found to be the producers of various metabolites including enzymes, antimicrobial peptides and/or proteins, and other biologically active substances. Among the enzymes perhaps the most significant is β-galactosidase which helps in lactose digestion and ameliorates the disorders associated with lactose intolerance (Hussain et al. 2008). Except for L. rhamnosus 7039 with weak β-galactosidase activity, the remaining isolates showed predominantly high β-galactosidase activity.
Biofilm formation
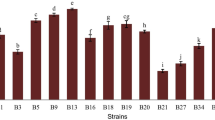
The majority of the 12 remaining lactobacilli isolates displayed a medium ability to form biofilms on an abiotic surface, except for L. acidophilus G9, which showed a very low capacity (A 570 lower than 0.1), and L. gallinarum 7001, L. plantarum L2-1, and L. fermentum G3 and 202, which displayed a very strong biofilm-forming capacity (A 570 higher than 0.2 or 0.3; Table 2).
To our knowledge, the ability to adhere to abiotic surfaces is not routinely tested in probiotic strains, even though the potential role of this trait in host health and disease is relevant. Biofilm formation is a multifactorial process that confers resistance of bacteria to mechanical or biotic stresses and to antibiotics; hence, it is not a desirable trait in pathogens. On the other hand, this property allows commensal bacteria to colonize the gut environment, remain more active, and compete with pathogens for surface colonization (Probert and Gibson 2002). Usually, the competition depends on the ability to produce antagonistic substances, which can change physical and mechanical environmental conditions or block the available surface receptors, leading to growth inhibition of some bacteria or even to their total elimination (Koninkx and Malago 2006). The probiotic strategies for the prevention and treatment of disease may require discovery and development of strains that form effective biofilms.
Inhibition of pathogens
The 12 mentioned strains were analyzed by an agar spot test for their capacity to inhibit representative diarrheal pathogens (Table 3). The best competence to suppress all of the selected pathogens on a high or moderate level was shown by L. gallinarum 7001, L. plantarum L2-1, L. rhamnosus 183, and L. fermentum 202. It is known that lactobacilli suppress pathogenic bacteria by producing inhibitory substances (Bernet-Camard et al. 1997; Dunne et al. 2001). For specification of the inhibition mechanism, our lactobacilli were assayed for the production of bacteriocin-like substances and hydrogen peroxide. It was observed that inhibitory activities of two strains (L. fermentum 202, 2 mg/L, and L. rhamnosus 183, 5 mg/L) were due to the production of hydrogen peroxide, while L. gallinarum 7001 and L. rhamnosus 183 exhibited antibacterial activity in neutralized (pH 6.5–7.0) and catalase-treated culture supernatants (bacteriocin-like compound producers). At present, the nature of the inhibitory potential of L. plantarum L2-1 remains unknown.
Conclusion
Our results imply that four lactobacilli strains (L. fermentum 202, L. gallinarum 7001, L. rhamnosus 183, and L. plantarum L2-1) comply with in vitro tests recommended by the Food and Agriculture Organization of the United Nations and the World Health Organization (FAO/WHO 2002), such as high antimicrobial activity and capacity to biofilm formation, and absence of undesirable properties such as unsuitable antibiotic resistance and enzymatic activity. All four may therefore be considered appropriate probiotic candidates; however, it is necessary to confirm their safety in in vivo experiments before allowing their inclusion in foods and/or food supplements.
References
Ambadoyiannis G, Hatzikamari M, Litopoulou-Tzanetaki E, Tzanetakis N (2005) Probiotic and technological properties of enterococci isolates from infants and cheese. Food Biotechnol 18:307–325
Arora G, Lee BH, Lamoureux M (1990) Characteristics of enzyme profiles of Lactobacillus casei species by a rapid API–ZYM system. J Dairy Sci 73:264–273
Aslim B, Yuksekdag ZN, Sarikaya E, Beyatli Y (2005) Determination of the bacteriocin-like substances produced by some lactic acid bacteria isolated from Turkish dairy products. LWT- Food Sci Technol 38:691–694
Bernet-Camard MF, Lievin V, Brassart D, Neeser JR, Servin AL, Hudault S (1997) The human Lactobacillus acidophilus strain LA1 secretes a non-bacteriocin antibacterial substance(s) active in vitro and in vivo. Appl Environ Microbiol 63:2747–2753
Bessède E, Angla-gre M, Delagarde Y, Sep Hieng S, Ménard A, Mégraud F (2011) Matrix-assisted laser-desorption/ionization biotyper: experience in the routine of a University hospital. Clin Microbiol Infect 17:533–538
Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O’Sullivan GC, Shanahan F, Collins JK (2001) In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 73:386S–392S
FAO/WHO (2002) Guidelines for the evaluation of probiotics in food. Report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. London, Ontario (April 30 and May 1, 2002)
Heavey PM, Rowland IR (2004) Microbial-gut interactions in health and disease. Gastrointestinal cancer. Best Pract Res Clin Gastroenterol 18:323–336
Hussain M, Khan NT, Wajid A, Rasool SA (2008) Technological characterization of indigenous enterococcal population for probiotic potential. Pak J Bot 40:867–875
Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Møller PL, Michaelsen KF, Pærregaard A, Sandstrom B, Tvede M, Jakobsen M (1999) Screening of probiotics activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol 65:4949–4956
Koninkx JFJG, Malago JJ (2006) The protective potency of probiotic bacteria and their microbial products against enteric infections—review. Folia Microbiol 53:189–194
Kushiro A, Chervaux CH, Cools-Portier S, Perony A, Legrain-Raspaud S, Obis D, Onoue M, Moer A (2009) Antimicrobial susceptibility testing of lactic acid bacteria and bifidobacteria by broth microdilution method and E-test. Int J Food Microbiol 132:54–58
Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2008) Technical guidance on the update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J 732:1–15
Probert HM, Gibson GR (2002) Bacterial biofilms in the human gastrointestinal tract. Curr Issues Intest Microbiol 3:23–27
Salyers AA, Gupta A, Wang Y (2004) Human intestinal bacteria as reservoirs for antibiotics resistance genes. Trends Microbiol 12:412–416
Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penadés JR, Lasa I (2001) The enterococcal surface protein, Esp, is involved in Enterocccus faecalis biofilm formation. Appl Environ Microbiol 67:4538–4545
Acknowledgments
The work was financially supported by the ERDF project no. 26220220152. We would like to thank Assoc. Prof. Greiffová from the Institute of Biotechnology and Food Science (Faculty of Chemical and Food Technology, Slovak University of Technology, Bratislava, Slovakia) for kindly providing the lactobacilli strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bujňáková, D., Kmeť, V. Functional properties of Lactobacillus strains isolated from dairy products. Folia Microbiol 57, 263–267 (2012). https://doi.org/10.1007/s12223-012-0121-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-012-0121-x