Abstract
Most important during probiotic selection are gastric acid and bile tolerance, the adhesion to the luminal epithelium to colonize the lower gastrointestinal tract of a human and safety for human consumption. The aim of this study was to evaluate the selected probiotic in vitro properties of Lactobacillus spp. Strains isolated from traditional fermented food. A total 38 strains were isolated from the pickled samples and 14 were identified as Lactobacillus spp. The survival of almost all strains after incubation at pH 2.5 did not change markedly, and remained at above 90 % (109 CFU/mL). The strains also exhibited a high survival rate at pH 3.5 (>90 %), whereas pH 1.5 all were died. Just four strains could survive 90 min. at pH 1.5 (<39 %). The incubation with 0.2 % bile salt solution resulted in a survival rates of 81–94 % after 24 h, whereas after incubation in 2 and 4 % bile salt solution it was 59–94 %. All tested strains showed very good and good resistance to 0.4 % phenol addition, however only Lb. johnsonii K4 was able to multiply. The hydrophobic nature of the cell surface of the tested strains was moderated recording hydrophobicity of Lb. johnsonii K4 and Lb. rhamnosus K3 above 60 %. Safety evaluation excluded four of tested strains as candidate probiotics, according to antibiotic resistance patterns and certain metabolic activities. On the basis on the results 10 of the selected Lactobacillus strains are safe and can survive under gastrointestinal conditions, which requires them to future in vitro and in vivo probiotic studies.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Lactic acid bacteria (LAB) are characterized by their production of lactic acid and are predominant participants in many industrial and artisanal plant, meat, and dairy fermentations. LAB group include mainly cocci and rods belonging to the genera: Streptococcus, Leuconostoc, Lactococcus, Oenococcus, Carnobacterium, Lactobacillus, Pediococcus, Enterococcus, Tetragenococcus, Vagococcus i Weisella. The Bifidobacterium genus some authors include also to LAB group due to the reason that produce lactic acid and present in a similar environment, while Bifidobacterium belongs to the Actinomycetes [30]. In general, LAB occur in habitats with a rich nutrition supply. Carbohydrates are used as carbon and energy source for these bacteria by a homofermentative or heterofermentative pathway. LAB are characterized by the production of 0.6–3 % lactic acid as a major catabolic end product from glucose. Some strains can also produce acetate, ethanol, and formate from pyruvate under low substrate concentrations and strictly anaerobic conditions [2].
LAB, particularly Lactobacillus genera, have recently received much attention due to their “generally recognized as safe” (GRAS) status and to their potential health-promoting effects as probiotics. According to FAO/WHO Report [19] probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host” and they should meet certain requirements in vitro and in vivo. Two of the currently most widely used in vitro tests are resistance to gastric acidity and bile salts, as based on both survival and growth studies. Approximately 2.5 l of gastric juice [10] and 1 l of bile [5] are secreted into the human digestive tract every day. Thus, it is essential for the bacteria to have protection systems to withstand the low pH in the stomach, digestive enzymes and bile of the small intestine [28, 65]. Other functional properties used to characterize probiotics are the production of antimicrobial compounds, the ability to modulate immune responses and bacterial adherence to intestinal epithelial cells and/or mucus [9, 33, 37, 54]. The mechanisms of adhesion is not fully understood, however the process appears to be multifactorial as adhesion cannot be attributed to one component [27] and includes electrostatic interactions, hydrophobic interactions and specific bacterial structures such as external appendages [57]. Regarding the safety of probiotic strains, FAO/WHO Working Group recommends some of the following tests: determination of antibiotic resistance patterns and assessment of certain metabolic activities [19]. It was found that probiotic strains may harbor resistance genes which may be transferred to pathogenic bacteria [65], as well as enzyme activities which may have negative effects in the colon [23].
Strains of Lactobacillus spp. occur naturally in the human intestine, and for this reason, such strains are also preferentially developed for commercial use as probiotics. However, some authors report that bacteria isolated from traditional fermented food products also possess potential probiotic properties and could be developed as a commercial starter cultures in food industry [1, 6, 13, 31, 34, 35, 40, 50, 63, 65].
Currently, much attention is paid to the obtaining new probiotic strains of bacteria with satisfactory technological properties in the food industry [26, 46, 47, 64]. Researches effort allowed to obtain sufficient numbers of well-characterized probiotic strains available for commercial use around the world [18, 52, 55]. However, the isolation and characterization of new strains are still needed, especially in developing countries, mainly to the design of new probiotic foods, where it is still limited access to probiotic strains, especially among small manufactories, such as dairies [64].
The aim of this study was identification of Lactobacillus spp. bacteria isolated from plant material (pickled cucumbers and cabbage) and evaluation of the safety and behavior of bacteria under gastrointestinal track conditions as selected probiotic properties.
Materials and Methods
Isolation of Bacterial Strains
Six samples of cucumber pickles and cabbage pickles were obtained from household of Poland central region. Samples were collected aseptically in pre-sterile poly-bags kept in an icebox and transported to the laboratory for analyses. 10 g of sample were weighed aseptically and homogenized for 2 min in stomacher (Stomacher 80 Biomaster, Seward) containing 90 ml of peptone water. Briefly, the homogenized samples were serially diluted with sterile peptone water and pour-plated on MRS (de Man, Rogosa and Sharpe) agar (Merck) and were incubated at 30 °C for 72 h. The colonies were selected randomly. Purity of the isolates was checked by streaking twice again on fresh MRS agar plates and further test.
38 strains of presumptive LAB strains were finally selected from these media. Stock cultures of each strain were transferred frozen at −80 °C in 20 % (w/v) glycerol for further studies.
Phenotypic Characterization
The selected strains were examined for cell morphology, Gram staining and catalase test. Strains were characterized phenotypically by testing their growth in MRS broth at 10, 15, and 45 °C and at pH 3.9 and 9.6. 17 of isolated strains identified as Lactobacillus spp. have been tested to carbohydrate fermentation profiles using the API 50CH identification system (BioMérieux).
Genotypic Characterization
Molecular identification was carried out by amplification of 16S rDNA gene of all identify phenotypically Lactobacillus strains using polymerase chain reaction (PCR). Bacterial DNA was isolated according to manufacturer’s description (Isolate Genomic DNA Kit, Bioline). The primer combination 27F/1492R (5′-AGA GTT TGA TCC TGG CTC AG-3′/5′- GGT TAC CTT GTT ACG ACT T-3′) was used for the amplification of 16S rDNA. 16S rDNA sequencing was performed with the superior strain screened from the study by Illumina technique. The homology search was carried out by BLAST software.
Probiotic Control Strain, Culture Media, and Growth Conditions
The well-studied commercial probiotic strain Lactobacillus plantarum 299v was used as a reference strain. All bacterial strains used in this study are listed in Table 1. Strains were cultivated in MRS broth using a 1 % inoculum, incubation was provided at 30 °C for 72 h.
Studies on Resistance to Gastrointestinal Conditions
Resistance to the Low pH Conditions
Cultures of all Lactobacillus strains were cultivated for 24 h in MRS medium, centrifuged, washed twice by PBS buffer and re-suspended to final concentration 108–109 CFU/mL in PBS. Bacteria were incubated in 0.85 % NaCl at pH 1.5, 2.5 and 3.5 for 2 h at 37 °C to study the impact of pH [11]. The bacteria were then spread-plated (100 µl aliquots) onto MRS agar medium and incubate for 72 h at 30 °C. Lactobacillus survival (in %) was calculated as follows:
Ni/Nx × 100, where Ni = log CFU/mL after incubation, Nx = log CFU/mL before incubation.
Resistance to High Concentration of Bile Salts
Cultures of all Lactobacillus strains were cultivated for 24 h in MRS medium than 1 % of cultures were transferred into fresh MRS broth with 0.2, 2 and 4 % (w/v) bile salt addition (Ox gall powder; Sigma Aldrich). Effect of the bile was estimated after 24 and 48 h treatment [11]. The bacteria were then spread-plated (100 µl aliquots) onto MRS agar medium and incubate for 72 h at 30 °C. Lactobacillus survival (in %) was calculated as follows:
Ni/Nx × 100, where Ni = log CFU/mL after incubation, Nx = log CFU/mL before incubation.
Bile Salt Hydrolase (BSH) Activity
BSH activity of the cultures was detected using the plate screening procedure described by Du Toit et al. [15] and Franz et al. [20]. Overnight cultures were spotted onto MRS agar plates containing 0.5 % (w/v) sodium taurodeoxycholate (Sigma Aldrich) and 0.37 g/l CaCl2. Plates were incubated anaerobically, in an anaerobic jar (BioMérieux) at 37 °C. Colonies with precipitation zones were considered as BSH-positive.
Resistance to 0.4 % (v/v) Phenol
The ability of all Lactobacillus strains to grow in the presence of phenol was tested by inoculating cultures (1 % of overnight culture) in MRS broth with and without 0.4 % phenol. Serial dilutions were spread-plated (100 µl aliquots) onto MRS agar at time 0 and after 24 h of incubation at 37 °C to enumerate surviving bacteria [67].
Bacterial Adhesion to Hydrocarbons
The bacterial adhesion to hydrocarbons (BATH) test was performed according to Reniero et al. [53]. Bacterial cells were washed with PBS buffer and resuspended in the same buffer. Absorbance was adjusted to 0.25 ± 0.05 to standardize the number of bacteria (108 CFU/mL) at 600 nm. Then, equal proportions of viable bacterial suspension and solvent (xylene) were mixed by vortexing for 5 min. The aqueous phase was removed after 1 h of incubation at room temperature and its absorbance was measured. Results were reported as percentages according to the formula:
BATH % = [(Ao−A)/Ao]*100, where Ao and A are absorbance before and after mixing with xylene, respectively.
Safety Evaluation
The selected strains were investigated for their antibiotic resistance profile using the E test (bioMérieux) using MRS agar and anaerobic incubation conditions. The strains were considered resistant when they showed MIC values higher than the MIC breakpoints established by the EFSA [16]. Enzyme activities were examined by API ZYM kit (bioMérieux) according to manufacturer’s instruction.
Statistical Analysis
All the experiments were performed in triplicate. Error bars on graphs show the standard deviation. The data were analyzed by analysis of variance (ANOVA) and Student t test. Differences were considered significant at P ≤ 0.05.
Results
Characterization and Identification of Bacterial Strains
A total 38 strains were isolated from the pickled samples and 17 were identified as Lactobacillus spp. according to phenotypic characterization. All the strains were Gram-positive, catalase-negative. Most tested strains were able to growth at 15 and 45 °C, just 9 of total 38 have tolerated temperature 10 °C. The growth at pH 9.6 was characteristic of most strains. At pH 3.9, just 7 of total 38 tested strains were able to growth well, and other strains represent moderate or slight growth. According to this tests 17 strains were identified, which were rod shaped and exhibited characteristics typical for Lactobacillus. These selected strains were identified to the species level also by sugar fermentation investigations (Table 1).
Almost all strains exhibited the ability to ferment glucose, fructose, mannose, mannitol, maltose, sacharose, and turanose. 12 of the total 17 strain were able to reduce lactose. Three strains were not identified according to API test and did not future investigated.
The nucleotide BLAST analysis showed that all of 14 isolates belong to the phylum Firmicutes and family Bacillaceae. Phylogenetic analyses of the 16S rDNA region identified strains O12, O13, O14, O16, O18 as Lb. casei, and K3 as Lb. rhamnosus, while strains O19, O20, O21, O23, K1 as Lb. plantarum. 2 strains (O22 and O24) were identified as Lb. brevis and strain K4 as Lb. johnsonii. All strains showed 97–100 % similarity in the nucleotide sequence of the 16S rDNA gene of the identify Lactobacillus type strain.
Studies on Resistance to Gastrointestinal Conditions
The ability of survival under simulates gastrointestinal conditions of 14 strains of lactic acid bacteria was examined. This examination included the resistance to low pH of the environment, high concentration of bile salts and 0.4 % phenol addition.
The first step in determining probiotic properties (in vitro tests) of lactic acid bacteria was the study of resistance to low pH of the environment. The effect of the successive incubations in low pH solutions (1.5, 2.5, and 3.5) on the viability of the strains are shown in the Table 2.
Individual strains performed different degrees of sensitivity toward gastric acidity. In the solution at pH 1.5 the highest level of survival (more than 98 %) was observed after 30 min of Lb. johnsonii K4 strain incubation. However after 60 min of incubation, other strains selected on the basis of high tolerance to low pH–strains Lb. casei O18 and Lb. plantarum O23 (levels of survival with 57–68 % cells). After 90 min of incubation only 4 isolates–Lb. casei O13 and O18 and Lb. plantarum O21 and O23-performed high tolerance to pH 1.5. These strains performed the survival rate of 23–38 %. None of the strains survive at pH 1.5 for 120 min of incubation.
The survival of all tested strains after incubation at pH 2.5 and 3.5 did not change markedly and remained at above 90 % (109 CFU/mL) and what is interesting some strains were able to multiply.
If strains survive the synthetic gastric juice, the next major challenge is the survival in the artificial small intestine. The effects of the successive incubations through synthetic bile salts on the viability of the strains are shown in the Fig. 1.
All of isolates were capable of growth with the concentration of 0.2 % bile salts (levels of survival with 81–93 % cells after 24 h of incubation and 57–86 % cells after 48 h of incubation). The strains Lb. brevis O22, Lb. plantarum O23 and K1 tolerated bile salts at concentration of 2 % with levels of survival with more than 85 % of cells after 24 h and 70 % after 48 h of incubation. Strains capable of growth in the presence of up to 4 % of bile salts were Lb. brevis O22 and Lb. plantarum K1 (more than 84 % cells after 24 and 48 h of incubation).
All Lactobacillus strains were tested for their ability to hydrolyse the sodium salt of taurodeoxycholic acid. In almost all cases, we did not observe deconjugation of bile salts. Just one strain Lb. johnsonii K4 showed BSH activity, while two strains Lb brevis O22 and O24 possessed weak bile salt hydrolase activity (Table 3).
Resistance to phenol was tested as an indicator for survival under intestinal conditions for all 14 Lactobacillus spp. strains isolated form pickled vegetables and the reference strain. The results are shown on Fig. 2. All tested strains showed very good and good resistance to this compound. Strains Lb. johnsonii K4, Lb. casei O16 and O18, Lb. rhamnosus K3 and Lb. plantarum K1 were distinguished from the other strains, as being the most resistant 0.4 % phenol, what indicate that these strains tolerate stress intestinal conditions. However, just one strain Lb. johnsonii K4 was able to growth in the presence of phenol during 24 h incubation at 37 °C.
Cell Surface Hydrophobicity
The hydrophobic nature of the cell surface the tested strains was measured photometrically using BATH assay. The results are listed in Table 3. We have found that two of Lactobacillus strains (Lb. rhamnosus K3 and Lb. johnsonii K4) showed quite high, recording above 60 % hydrophobicity values. However 12 of 14 tested strains showed moderate hydrophobicity character (Table 3).
Safety Evaluation
All tested strains were sensitive to ampicillin and except two strains (Lb. plantarum O23 and K1) to tetracycline (Table 4). However, all these strains were considered as resistant to the antibiotics streptomycin, gentamicin (except Lb. rhamnosus K3), and vancomycin, when using the MIC breakpoint values as suggested by EFSA [16].
None of the tested isolates produced enzymes such as alkaline phosphatase, esterase lipase (C8), lipase (C14), trypsin, α-chymotrypsin, acid phosphatase, α-galactosidase, α-mannosidase, and α-fucosidase (Table 5). Also the carcinogen enzyme, β-glucuronidase was not produced by any of the isolated lactobacilli. However, esterase (C4), leucine arylamidase, valine arylamidase, cystine arylamidase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, α and β-glucosidase, and N-acetyl-β-glucosaminidase were detected.
Discussion
Traditional fermented food can be considered as open ecosystems that contain undefined LAB with high competitive potential. Several of these traditional foods are kept in unrefrigerated conditions and are rich sources for new LAB strains with unprecedented antimicrobial activity as well as probiotic characteristics [51]. According to Holzapfel [25], there is an increasing need to select new strains of bacteria with functional properties for commercial production and for improvement of quality of existing traditional fermented food.
The lactic acid bacteria isolated and identified in our pickled cucumber and cabbage samples have been also reported in other investigation of fermented foods. Lactobacillus plantarum has been predominantly isolated from natural fermented cabbage and cucumber in earlier studies. Other authors isolate also such species as Lactobacillus brevis, Lactobacillus buchneri, Lactococcus lactis subsp. lactis, and Lactobacillus acidophilus [4, 17, 21, 36, 48, 49, 60, 61, 66, 69]. The dominance of Lb. plantarum in pickled vegetables has been attributed to its high acid tolerance [3, 43].
Ability to overcome physical and chemical barriers such as gastric acidity and bile toxicity in the gastrointestinal tract and adherence to mucus or human epithelial cell lines are among the in vitro tests that are frequently suggested for the evaluation of the probiotic potential of the bacterial strain [7, 38, 56].
Stresses to microorganisms begin in the stomach, with pH between 1.5 and 3.0 and in the upper intestine which contains bile [58]. Stomach pH can be as low as 1.0 [68]. However, survival of ingested bacteria in the stomach obviously is influenced by the buffering capacity of food matrix, that is why in most in vitro assays pH 3.0 has been preferred [56]. Del Piano et al. [14] tested seven of Lactobacillus plantarum strains for their resistance to both simulated gastric juice and human gastric juice withdrawn on an empty stomach from healthy individuals. It was noted that less than 20 % of the bacteria survived after an hour of exposure to simulated gastric juice, while human gastric juice allowed a survival rate between 15 and 45 %.
Survival of bacterial strains in low pH conditions is a more accurate indication of the ability of strains to survive passage through the stomach. For this reason, in vitro survival tests in three different low pH (1.5, 2.5, 3.5) values were conducted. Analysis of results in this study suggest that all tested strains are able to survive in great number of cells under the conditions simulated gastric juice at pH 2.5 and 3.5. The applied in the present study experimental conditions are the simulation of the human gastric juice environment, not quite adequately, but are usually used by many researchers to quickly screening of potential probiotic properties of the bacteria [3, 38, 56, 65]. The other researchers have obtained similar results, noting that the survival rate of bacteria of the genus Lactobacillus in a low pH ranges from 30–100 %. In the studies conducted by Casey et al. [7] LAB isolates were examined for ability to survive with simulated gastric juice at pH 1.85. Only one of the strains (Lactobacillus johnsonii 4) was able to survive after 30 min of incubation. Also in similar studies Kumar et al. [31] observed that only four strains of Lactobacillus (out of 34) could survive up to 2 h at pH 1.85. Schillinger et al. [56] suggested that Lb. casei group were more susceptible to gastric acidity than strains of the Lb. acidophilus group. On the other hand Mathara et al. [38] observed that Lb. fermentum strains showed better survival when exposed to pH 2.0 and 2.5 solution than Lb. acidophilus strains, while Lb. plantarum represent poor viability [40].
Bile acids are synthesized in the liver from cholesterol and are secreted from the gall bladder into the duodenum in the conjugated form. The physiological concentration of bile acids in the small intestine is between 5.000 and 20.000 lM [24]. The relevant physiological concentration of human bile ranges from 0.3 to 0.5 %.
Many authors accented that there is still no consensus about concentration of bile salts which are used in simulating small intestine [14, 65]. Studies in bile salt must be used with at least 0.15 % concentration [56].
In our selection of Lactobacillus strains, we have used the criteria (pH and bile resistance) similar to those had published by Kurman [32]. According to the author the use of 4 % bile salt concentration is a good indicator of the characteristics of probiotics. It has been also suggested that LAB strains vegetable origins are generally less bile-resistant [42]. Mathara et al. [38] have found that strains of Lb. acidophilus and Lb. rhamnosus group showed highest bile salt tolerance, compared to other lactobacilli.
A recent study reported that the strains isolated from pickled vegetables represent good survival under bile salt stress conditions. All numbers of cells of tested strains decrease after 24 and 48 h of incubation. Most resistant to these conditions were Lb. brevis O22, Lb. plantarum K1, and O23. Suskovic et al. [59] reported that Lb. plantarum strains isolated from fermented olives have been tolerance to low pH and high bile salts concentration. Also Lb. plantarum strains isolated from fermented milk products were resistant to 0.1–0.5 % concentration of bile salts [39].
It is considered that BSH enzyme might be a detergent shock protein that enables bacteria to survive the intestinal bile stress [12]. While Moser & Savage [41] have proposed that BSH activity facilitates the survival of lactobacilli in the gastrointestinal tract. It is likely that bile salt hydrolase activity may contribute to resistance of lactic acid bacteria to the toxicity of conjugated salts in the small intensity. On the other hand, some data indicate that BSH activity and resistance to bile salts are unrelated in lactobacilli [41], what was also showed in our study. Moreover, most BSH activity relates to strains that have been isolated from the intestines or feces of mammals-rich environment, and non-conjugated bile acids. Strains of other environments, such as milk or vegetables-environment in which bile salts are absent, usually do not possess bile salt hydrolase activity [62] as we have also observed in this study.
Phenols may be formed in the intestine as a result of the bacterial deamination of some aromatic amino acids derived from dietary and endogenous proteins and are believed to act as co-carcinogens [8, 59]. Some LAB create the anti-carcinogenic mechanisms allowing for tolerance to phenols. Vizoso Pinto et al. [65] reported that Lb. plantarum strains isolated from children’s faeces tolerate phenol at 0.4 % for 24 h as their numbers did not decrease from an initial inoculum, but Lb. johnsonii strains were in a great extent inhibited. On the other hand in our present study Lb. johnsonii K4 strain showed the best resistant to the phenol from the tested strains. Also Kumar et al. [31] have found that 4 strains isolated form vegetables pickles showed different degrees of sensitivity toward this compound. On the other hand Xanthopoulos et al. [67] reported, that most of the tested strains Lb. paracasei subsp. paracasei, Lb. rhamnosus, Lb. acidophilus, Lb. gasseri, and Lb. reuteri isolates from new-born infants grew, although at lower levels, in the presence of phenol at 0.4 %, during incubation for 24 h. In the present study, the investigated lactobacilli strains survival and growth rates were slightly affected by phenol during 24 h incubation, so it could be supposed that bacteria are able to grow and live in the presence of this compound in colon during the transit time.
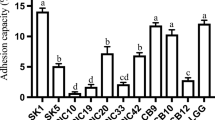
Having survived the acid and bile challenges of the gastrointestinal tract, it may be advantageous for LAB to adhere to the luminal epithelium and colonize the lower gastrointestinal tract of a human. High cell surface hydrophobicity may favor the colonization of mucosal surfaces and play a role in the adhesion of bacteria to epithelial cells [22]. On the other hand Mathara et al. [39] did not find any correlations between hydrophobicity and bacterial adhesion. Thus, hydrophobicity may be helpful for adhesion, but it is obviously not a prerequisite for a strong adherence capacity [56]. All isolates examined in this study demonstrated high degrees of adherence to hydrocarbons, but we found the considerable differences between the strains. Strains of Lb. johnsonii K4 and Lb. rhamnosus K3 and also Lb. brevis O22 and O24 exhibit higher hydrophobicity values as compared to Lb. plantarum and Lb. casei strains. The other strains represent moderate and low character of hydrophobicity (Table 3). The high cell surface hydrophobicity of the Lb. rhamnosus K3 and Lb. johnsonii K4 strains could play a significant role in interaction with other organic mucin layer of the gut. It needs to be investigated.
The other authors have observed that Lb acidophilus and Lb. fermentum groups obtained better hydrophobicity values than Lb. plantarum and Lb. rhamnosus [38, 55]. Studies on the cell surface characteristic of the Lb. casei group suggest a relatively hydrophilic nature of these strains [45].
The methods used in this study do not give comprehensive information about the survival bacteria under gastrointestinal conditions. The in vitro assays are quite different from the in vivo conditions, because the human gut food matrix pays the protective role for the bacteria [5]. However, the in vitro tests provide important information about species and strain differences and are very helpful and powerful tools especially for quick screening the bacteria for probiotic activity.
One of the safety considerations in probiotic studies is the verification that a potential probiotic strain does not harbor acquired and transferable antibiotic resistance genes. Studies on the antibiotic resistance of lactobacilli indicated that they were usually resistant to major classes of antibiotics such as β-lactams, aminoglycosides, cephalosporins, and quinolones [29]. In the present study, all strains were resistant to streptomycin, vancomycin, and except Lb. rhamnosus K3 to gentamicin. However, according to previous studies [31, 40, 65] such antibiotic resistance observed for Lactobacillus strains in this work, were considered to be intrinsic or natural resistances and, therefore, non-transmissible. The resistance of Lactobacillus strains to tetracycline has also been previously observed and often considered as potential transferable acquired resistances [44]. In this study only 2 strains (Lb. plantarum O23 and K1) were resistant to tetracycline and for this reason, at this stage, they were excluded as candidate probiotics. Future research should focus on the location and potential transferability of antibiotic resistance genes.
Safety assessment must also include the lack of harmful activities, such as β-glucuronidase activity. β-glucuronidases liberate toxins and mutagens that have been glucuronated in the liver and excreted into the gut with the bile. This can lead to high local concentrations of carcinogenic compounds within the gut, thus increasing the risk of carcinogenesis. Also α-chymotrypsin, β-glucosidase, and N-acetyl-β-glucosaminidase activities may have negative effects in the colon [23]. In present study, all tested strains have not possess β-glucuronidase and α-chymotrypsin activity, however 3 of Lactobacillus strains (Lb. casei O14, Lb. plantarum O21 and K1) produce β-glucosidase, and N-acetyl-β-glucosaminidase, which excludes them from further probiotic investigations. Among the enzymes perhaps the most significant is β-galactosidase which helps in lactose digestion and ameliorates the disorders associated with lactose intolerance. Fewer (11 of 14 tested strains) possess β-galactosidase activity which suggests the possibility of using the strains in the production of fermented milk products.
Conclusions
In this study, the in vitro tests were used to screening the most important probiotic activity of bacteria isolated from pickled cucumber and cabbage. The findings revealed that strains of Lactobacillus spp. investigated at this study were characterized by the high survival under simulates gastrointestinal conditions. Strains Lb. johnsonii K4, Lb. brevis O22 and Lb. plantarum K1 tolerate gastric and small intestinal conditions very well, and also strain Lb. johnsonii K4 has a high adhesion capacity. Furthermore, strains group Lactobacillus plantarum and Lactobacillus brevis tolerate gastric-intestinal conditions better than Lactobacillus casei group.
All tested strains probably could be able to enter the intestinal tract alive, which requires them to future studies on in vitro and in vivo probiotic properties. However, four of tested strains (Lb. casei O14, Lb. plantarum O21, O23, K1) have been excluded as probiotic candidate according to the antibiotic resistant and enzymatic profile study.
We are currently characterizing the antagonistic activity to the pathogenic bacteria and bacteriocins produced by these cultures. On the basis of the results 10 of the Lactobacillus spp. strains isolated from pickled cucumber and cabbage have the potential for use in developing probiotic food as a starter culture.
References
Ashenafi M, Busse M (1991) Growth of Bacillus cereus in fermenting tempeh made from various beans and its inhibition by Lactobacillus plantarum. J Appl Bacteriol 70:329–333
Axelsson L (2004) Lactic Acid Bacteria: Classification and Physiology. In: Dekker.M (ed) Lactic Acid Bacteria Microbiological and Functional Aspects, 3rd edn, pp.19–85
Bao Y, Zhang Y, Zhang Y, Liu Y, Wang S, Dong X, Zhang H (2010) Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 21:695–701
Barrangou R, Yoon SS, Breidt F, Fleming HP, Klaenhammer TR (2002) Identification and characterization of Leuconostoc fallax strains isolated from an industrial sauerkraut fermentation. Appl Environ Microbiol 68:2877–2884
Begley M, Gahan CGM, Hill C (2005) The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651
Botes A, Todorov SD, von Mollendorff JW, Botha A, Dicks LMT (2007) Identification of lactic acid bacteria and yeast from boza. Process Biochem 42:267–270
Casey P, Casey G, Gardiner G, Tangney M, Stanton C, Ross R, Hill C, Fitzgerald G (2004) Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett Appl Microbiol 39:431–438
Chung KT, Fulk GE, Slein MW (1975) Tryptophanase of fecal flora as a possible factor in the etiology of colon cancer. J Natl Cancer Inst 554:1073–1078
Collado MC, Isolauri E, Salminen S, Sanz Y (2009) The impact of probiotic on gut health. Curr Drug Metab 10:68–78
Cotter PD, Hill C (2003) Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67:429–453
Cukrowska B, Motyl I, Kozáková H, Schwarzer M, Górecki RK, Klewicka E, Śliżewska K, Libudzisz Z (2009) Probiotic Lactobacillus strains: in vitro and in vivo studies. Folia Microbiol 54:533–537
De Smet I, Van Hoorde L, Vande Woestyne M, Christianes H, Verstraete W (1995) Significance of bile salt hydrolytic activities of lactobacilli. J Appl Microbiol 79:292–301
De Valdez GF, De Giori GS, Garro M, Mozzi F, Oliver G (1990) Lactic acid bacteria from naturally fermented vegetables. Microbiol Aliments Nutr 8:175–179
Del Piano M, Morelli L, Strozzi G, Allesina S, Barba M, Deidda F, Lorenzini P, Ballare M, Montino F, Orsello M, Sartori M, Garello E, Carmagnola S, Pagliarulo M, Capurso L (2006) Probiotics: from research to consumer. Digest Liver Dis 31:248–255
Du Toit M, Franz CMAP, Dicks LMT, Schillinger U, Haberer P, Warlies B, Ahrens F, Holzapfel WH (1998) Characterisation and selection of probiotic lactobacilli for a preliminary minipig feeding trial and their effect on serum cholesterol levels, faeces pH, and faeces moisture content. Int J Food Microbiol 40:93–104
EFSA (2008) Technical guidance. update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. prepared by the panel on additives and products or substances used in animal feed. The EFSA J 732:1–15
Enitan A, Adeyemo J, Ogunbanwo ST (2011) Influence of growth conditions and nutritional requirements on the production of hydrogen peroxide by lactic acid bacteria. Afr J Microbiol Res 5:2059–2066
Ezendam J, van Loveren H (2006) Probiotics: immunomodulation and evaluation of safety and efficacy. Nutr Rev 64:1–14
FAO/WHO (2002) Guidelines for the Evaluation of Probiotics in Food. Report a Joint FAO/WHO Working Group, London, pp. 1–11
Franz C, Specht I, Haberer P, Holzapfel WH (2001) Bile salt hydrolase activity of enterococci isolated from food: screening and quantitative determination. J Food Prot 64:725–729
Gündoğdu AK, Karahan AG, Çakmakçi ML (2005) Production of nitric oxide (NO) by lactic acid bacteria isolated from fermented products. Eur Food Res Technol 223:35–38
He X, Lux R, Kuramitsu HK, Anderson MH, Shi W (2009) Achieving probiotic effects via modulating oral microbial ecology. Adv Dent Res 21:53–56
Heavey PM, Rowland IR (2004) Gastrointestinal cancer. Best Pract Res Cl Ga 18:323–336
Hofmann A (1991) Enterohepatic circulation of bile acids. In: Schultz SG, Forte JG, Rauner BB (eds) Handbook of physiology. Section 6: The gastrointestinal system, vol 3, pp 567–580
Holzapfel WH (2002) Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol 75:197–212
Hoque MZ, Akter F, Hossain KM, Rahman MSM, Billah MM, Islam KMD (2010) Isolation, identification and analysis of probiotic properties of Lactobacillus spp. from selective regional yoghurts. World J Dairy & Food Sci 5:39–46
Izquierdo E, Horvatovich P, Marchioni E, Aoude-Werner D, Sanz Y, Ennahar S (2009) 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis 30:949–956
Jensen H, Grimmer S, Naterstad K, Axelsson L (2012) In vitro testing of commercial and potential probiotic lactic acid bacteria. Int J Food Microbiol 53:216–222
Kaur IP, Chopra K, Saini A (2002) Probiotics: potential pharmaceutical applications. Eur J Pharm Sci 15:1–9
König H, Fröhlich J (2009) Lactic acid bacteria. Biology of Microorganisms on Grapes. In: Must and in Wine, Springer, Berlin, pp. 3–29
Kumar M, Ghosh M, Ganguli A (2012) Mitogenic response and probiotic characteristics of lactic acid bacteria isolated from indigenously pickled vegetables and fermented beverages. World J Microbiol Biotechnol 28:703–711
Kurman JA (1988) Starters with selected intestinal bacteria. Bulletin IDF 227:41–55
Lebeer S, Vanderleyden J, De Keersmaecker SC (2008) Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 72:728–764
Lee HM, Lee Y (2006) Isolation of Lactobacillus plantarum from kimchi and its inhibitory activity on the adherence and growth of Helicobacter pylori. J Microbiol Biotechnol 16:1513–1517
Lei V, Jakobsen M (2004) Microbiological characterization and probiotic potential of koko and koko sour water, African spontaneously fermented millet porridge and drink. J App Microbiol 96:384–397
Liu P, Shen S, Ruan H, Zhou Q, Ma L, He G (2011) Production of conjugated linoleic acids by Lactobacillus plantarum strains isolated from naturally fermented Chinese pickles. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 12:923–930
Marco ML, Pavan S, Kleerebezem M (2006) Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol 17:204–210
Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Holzapfel WH (2004) Isolation, identification and characterisation of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int J Food Microbiol 94:269–278
Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Guigas C, Franz C, Holzapfel WH (2008) Functional properties of Lactobacillus plantarum strains isolated from maasai traditional fermented milk products in kenya. Curr Microbiol 56:315–321
Mathara JM, Schillinger U, Guigas C, Franz C, Kutima PM, Mbugua SK, Shin H-K, Holzapfel WH (2008) Functional characteristics of Lactobacillus spp. from traditional maasai fermented milk products in kenya. Int J Food Microbiol 126:57–64
Moser SA, Savage DC (2001) Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl Environ Microbiol 67:3476–3480
Mourad K, Nour-Eddine K (2006) In vitro preselection criteria for probiotic Lactobacillus plantarum strains of fermented olives origin. Int J Probiotics Prebiotics 1:27
Muyanja CMBK, Narvhus JA, Tremo J, Langsrud T (2003) Isolation, characterisation and identification of lactic acid bacteria from bushera a Ugandan traditional fermented beverage. Int J Food Microbiol 80:201–210
Nawaz M, Wang J, Zhou A, Ma C, Wu X, Moore JE, Millar BC, Xu J (2011) Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr Microbiol 62:1081–1089
Pelletier C, Bouley C, Cayuela C, Bouttier S, Bourlioux P, Bellon-Fontaine MN (1997) Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei and Lactobacillus rhamnosus strains. Appl Environ Microbiol 63:1725–1731
Pennacchia C, Ercolini D, Blaiotta G, Pepe O, Mauriello G, Villani F (2004) Selection of Lactobacillus strains from fermented sausages for their potential use as probiotics. Meat Sci 67:309–317
Pennacchia C, Vaughan EE, Villani F (2006) Potential probiotic Lactobacillus strains from fermented sausages: further investigations on their probiotic properties. Meat Sci 73:90–101
Pérez-Borla O, Davidovich LA, Roura SI (2010) Isolation and characterization of proteolytic microorganisms from fresh and fermented cabbage. Food Sci Technol-Leb 43:298–301
Plengvidhya V, Breidt FZ, Fleming HP (2007) DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl Environ Microbiol 73:7697–7702
Psani M, Kotzekidou P (2006) Technological characteristics of yeast strains and their potential as starter adjuncts in Greek-style black olive fermentation. World J Microbiol Biotechnol 22:1329–1336
Ramadan MM, Tantawy EA, Shehata MS (2005) Effect of gamma rays on seed germination and seedling growth of some timber trees. Annals of Agric Sci Moshtohor 43:869–883
Reid G, Jass J, Sebulsky MT, McCormick JK (2003) Potential uses of probiotics in clinical practice. Clin Microbiol Rev 16:658–672
Reniero R, Cocconcelli P, Bottazzi V, Morelli L (1992) High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J Gen Microbiol 138:763–768
Saarela M, Mogensen G, Fondén R, Mättö J, Mattila-Sandholm T (2000) Probiotic bacteria: safety, functional and technological properties. J Biotechnol 84:197–215
Santosa S, Farnworth E, Jones PJH (2006) Probiotics and their potential health claims. Nutr Rev 64:265–274
Schillinger U, Guigas C, Holzapfel W (2005) In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int Dairy J 15:1289–1297
Servin AL, Coconnier M-H (2003) Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Cl Ga 17:741–754
Shin HS, Huang EJ, Park BS, Sakai T (1999) The effects of seed inoculation on the rate of garbage composting. Environ Technol 20:293–300
Suskovic J, Brkic B, Matosic S, Maric V (1997) Lactobacillus acidophilus M 92 as potential probiotic strain. Milchwissenschaft 52:430–435
Tamang JP, Tamang B, Schillinger U, Franz C, Gores M, Holzapfel WH (2005) Identification of predominant lactic acid bacteria isolated from traditionally fermented vegetable products of the eastern himalayas. Int J Food Microbiol 105:347–356
Tamminen M, Joutsjoki T, Sjöblom M, Joutsen M, Palva A, Ryhänen E-L, Joutsjoki V (2004) Screening of lactic acid bacteria from fermented vegetables by carbohydrate profiling and PCR–ELISA. Lett Appl Microbiol 39:439–444
Tanaka H, Doesburg K, Iwasaki T, Mierau I (1999) Screening of lactic acid bacteria for bile salt hydrolase activity. J Dairy Sci 82:2530–2535
Todorov SD, Botes M, Guigas C, Schillinger U, Wiid I, Wachsman MB, Holzapfel WH, Dicks LMT (2008) Boza, a natural source of probiotic lactic acid bacteria. J Appl Microbiol 104:465–477
Vinderola B, Capellini F, Villarreal S, Viviana QA, Jorge R (2008) Usefulness of a set of simple in vitro tests for the screening and identification of probiotic candidate strains for dairy use. Food Sci Technol-Leb 41:1678–1688
Vizoso Pinto M, Franz C, Schillinger U, Holzapfel W (2006) Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int J Food Microbiol 109:205–214
Wang C-Y, Lin P-R, Ng C-C, Shyu Y-T (2010) Probiotic properties of Lactobacillus strains isolated from the feces of breast-fed infants and Taiwanese pickled cabbage. Anaerobe 16:578–585
Xanthopoulos V, Litopoulou-Tzanetaki E, Tzanetakis N (2000) Characterization of Lactobacillus isolates from infant faeces as dietary adjuncts. Food Microbiol 17:205–215
Xanthopoulos V, Litopoulou-Tzanetaki E, Tzanetakis N (eds) (1997) In vitro study of Lactobacillus species strains on bile tolerance and cholesterol removal. In: Lactic acid bacteria-Lactic 97. Presses Universitaires de Caen, Caen
Xiong T, Song S, Huang X, Feng C, Liu G, Huang J, Xie M (2013) Screening and identification of functional Lactobacillus specific for vegetable fermentation. J Food Sci 78:84–89
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zielińska, D., Rzepkowska, A., Radawska, A. et al. In Vitro Screening of Selected Probiotic Properties of Lactobacillus Strains Isolated from Traditional Fermented Cabbage and Cucumber. Curr Microbiol 70, 183–194 (2015). https://doi.org/10.1007/s00284-014-0699-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0699-0