Abstract
Cells are constantly exposed to a variety of mechanical perturbations and their response to these stimuli plays a vital role in their proper functioning. Here, we present a micro-mechanical device for providing a mechanical stimulus to cells cultured on it and observing the change in the deformation of the nucleus of the cell. Our device has the provision to stretch, in situ, single cells by different amounts through a single actuation based on their points of adhesion on the device. The device consists of folded beams that deform as in an accordion, which is actuated using a probe attached to an XYZ positioner. The device is microfabricated on glass coverslips using SU-8, which is transparent and allows for the visual measurement of the nucleus through high-magnification imaging during stretching. Many devices can be accommodated on a single coverslip and can be actuated independently. Growing cells on the device do not need any specialized technique: it is easily achieved by seeding cells at low density directly on the coverslip. Furthermore, the single-mask microfabrication process developed for the mechanism permits a range of stiffness by changing only one mask or the thickness of the structural layer. We demonstrate the utility of the device by culturing NIH 3T3 fibroblasts on the devices, stretching them in situ, and measuring the deformation of their nuclei using fluorescence imaging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Mechanical signals are being increasingly implicated as important regulators of cell function [1,2,3]. Both active and passive mechanical influences from the cell microenvironment affect important aspects of tissue function such as development [4], differentiation [5], etc. Mechanical signals have been hypothesized to be faster and longer range signaling mechanisms than chemical signals [6]. Some disease states are associated with improper mechanical signaling [7]. Hence, the study of the response of the cells to mechanical signals is deemed important. The present work describes design, fabrication, characterization and demonstration of a folded beam compliant device to study the response of cells to a mechanical stimulus. To show the utility of the device, we stretch individual cells grown on the device and quantify the deformation of the cell nuclei. We also describe the potential of using the device for measuring the stiffness of cells and the force exerted by cells. Compliant devices made of micromachined silicon beams have been shown to be useful for studying the mechanical response of cells in earlier studies [8, 9]. In this work, we focus on a micromachined compliant stretching device made of SU-8 polymer, which is transparent and hence amenable for visual inspection at high magnification. It also makes it possible to match the device stiffness to that of the cells. This can be done by changing either the in-plane widths of the beam segments of the mechanism or the thickness of the structural layer.
Studying the response of cells to mechanical stimuli involves the ability to induce measurable force or deformation on cells and to observe and quantify their response to such stimuli. Stretching is a commonly used mechanical stimulus to measure cell response. The response is tracked by fluorescence imaging of tagged cell components such as organelles, cytoskeletal filaments, proteins, etc. Growing cells on soft substrates and stretching the substrate induces stretching of the cells growing on them [10,11,12,13,14]. A large number of cells can be stretched by this technique but the stretching of an individual cell cannot be directly controlled. Furthermore, the measured response of cells to stretch contains contributions of cell-cell and cell-substrate interactions. High-resolution live imaging of the cell response is also usually limited because of the presence of the flexible substrates below the cells. Another popular approach is an optical stretcher [15,16,17,18]. In this technique, the contribution from other cells and the substrate is eliminated. However, this is an invasive technique because beads are attached to the cells and laser-based stretching might damage the cells. Various other techniques such as microplates [19,20,21], two-fingered microhand with microforce sensor [22] and some by means of Micro Electro Mechanical Systems (MEMS) [23,24,25,26] have also been developed to overcome these limitations. In these devices, individual cells are attached between two plates or members that can be moved away from each other using micro actuation techniques such as piezo, electrostatic comb drives, etc. Although, these systems, like the optical stretcher, have the ability to test individual cells with minimal cell substrate interaction, they can assess only one cell at a time and, hence, suffer from low throughput.
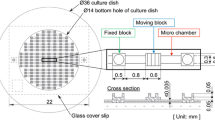
We present here a technique that meets the aforementioned requirements by proposing a microfabricated device made using folded beams. This device can perform uniaxial stretching of multiple individual cells with minimal substrate interaction. Our design consists of folded beams made of SU-8 (Figs. 1 and 2). The gap between the beams is less than the size of a cell, which is about 15 μm. When cells are seeded onto the device at sufficiently low density, we get individual cells attached randomly between the beams (Fig. 1). By pulling on the beams with a probe (Fig. 3), we can stretch these cells. The stretch on the cells depends on their location on the device. The cells toward the centre of the beams get stretched more than those at the ends of the beams. Hence, with a single actuation, we can observe the response to varying stretch on the cells (Fig. 1). These devices are fabricated on coverslips and hence are amenable for high-magnification live imaging. Furthermore, each device can be actuated independently without disturbing the cells on the other devices. The total footprint of a single device is approximately 1 mm × 0.5 mm and, hence, many (more than hundred) such devices can be microfabricated on a single coverslip. The devices are fabricated using a simple two-layer process without the need for alignment. It involves a base layer of OmniCoat that is preferentially removed from below the beams by a timed etch. Our stretchers have relatively easy fabrication (one mask, two-layer process without alignment), cell attachment (low density blanket cell seeding), and a simple actuation mechanism (micropipettes mounted on XYZ positioners). Since the stiffness of a folded beam mechanism varies by (1/l 3) and w 3 [27] where l and w are the length and width of the beams respectively, the stiffness of the device can be tuned based on the cell under study. Table 1 shows a comparison of the attributes the earlier mentioned techniques with the technique we have developed.
In a demonstrative study using our device, we observed the deformation of the nuclei of NIH 3T3 cells by fluorescence imaging during stretching. The shape of the nucleus has been shown to affect fundamental aspects of cell function such as gene expression and in turn protein expression [28, 29]. It is hypothesized that the nucleus plays an important role in mechanotransduction [30]. The forces on the cell membrane are transferred to the nucleus through the cytoskeleton [29]. The transmitted forces change the shape of the nucleus. Due to the change in the nuclear shape, the genes bound to lamin on the periphery of the nucleus may get dislodged leading to altered transcription [28]. Further, the mechanical properties of the nuclei have been shown to change from a positive Poisson’s ratio to a negative Poisson’s ratio as the cell exits pluripotency [31]. Hence, a device to deform a cell and observe the response of the nucleus to this stimulus will be valuable in understanding the mechanotransduction process.
In the following sections we describe the design of the device, the fabrication process, measurement of the stiffness of the device, comparison with finite element simulations, cell-seeding, culture, in situ stretching, and measuring the mechanical response.
2 Material and methods
2.1 Design of the device
The design was primarily driven by experimental convenience. Thus, it was decided to fabricate the device on coverslips to allow high-magnification imaging from underneath. We chose SU-8 for fabricating the device because it is transparent, elastic, and non-toxic to cells [32]. A folded beam suspension design was adapted for the device. The spacing between the folded beams was chosen based on the dimensions of cells in suspension. The spacing had to be such that suspended cells could sit between the beams without touching the coverslip below. The beams had to be sufficiently wide for the cells to attach. In its native environment the stiffness of the cell-substrate is comparable to the stiffness of the cell [33] and hence we wanted the cell stretcher to have stiffness comparable to that of the cell. With comparable stiffness, the cells are able to deform the mechanism and this can be used to compute the forces exerted by them [34]. Hence, the length and out-of-plane thickness of the folded beams was chosen such that the stiffness of the mechanism is of the same order of magnitude as compared to the stiffness of the cells. One end of the device was connected to a large pad to ensure firm adhesion to the surface. By using a timed development we could ensure preferential release of the beams while the pads remained attached to the coverslip. The device had a triangular frame at the other end to allow for actuation using a probe without disturbing the cells on the beams.
Previous studies have reported a modulus of elasticity of around 6 kPa for fibroblasts [33] and human Mesenchymal Stem Cells [35] and 2–5 kPa for human hepatocellular carcinoma [36]. For a cell having a modulus of elasticity in the 6 kPa range, this translates to a stiffness of approximately 0.024 N/m (assuming the cell to be a homogeneous cuboid of 20 × 10 × 2 μm and stretched along the 10 μm edge). In an earlier work, it was observed that for mechanisms with in-plane stiffness of around 0.042 N/m, the mechanisms tend to deform out-of-plane due to buoyancy [37] and are not amenable for easy in-situ experimentation. Hence, we wanted to design the mechanism with stiffness an order of magnitude greater than cell stiffness. Further, the dimensions of the device were constrained by the size of the cells, the material chosen, and the practical limits of the microfabrication process. The in-plane width of the beams was taken as 5 μm, which is a practical lower limit in our fabrication process. By choosing the beam length of 210 μm, an out-of-plane thickness of 1.5 μm, and by using SU-8 (Young’s modulus = 4.02 GPa [38]), simulations in COMSOL gave a stiffness of 0.178 N/m per folded beam, which meets our requirements. Table 2 shows the dimensions of the designed and fabricated device. Three such folded beams were connected in series by a 70 μm wide rectangular block. The rectangular block had to be wide enough so that it is relatively rigid compared to the beams. An XYZ positioner was used to actuate the devices. By having three folded beams in series, the minimum deformation that can be given to a cell is reduced to a third of the resolution of the XYZ positioner. Since three folded beams are connected in series, the stiffness of the device is a third of the stiffness of the folded beam (0.059 N/m).
The gap between the beams and that between two folded beams was taken as 10 μm, which is lower than the diameter of a cell in suspension (approximately 15–20 μm). When cells are seeded onto the device, the cells rest on top of the beams since their diameter is larger than the gap between the beams. It is likely that in some cases, the cells squeeze into the gaps between the beams as they grow. Indeed, it was observed in some cells but not all. Those cells that only spread laterally and not into the gap are considered for measurement.
The shape of the triangular frame used for actuation is equilateral and the length of the side of the outer triangle is 400 μm and that of the inner triangle is 225 μm. The size of the triangle was chosen such that a stiff probe (around 30–50 μm in diameter) can comfortably fit inside the inner triangle and safely actuate the device.
2.2 Microfabrication
Glass coverslips (22 mm circular or square) were cleaned by piranha cleaning. A thin layer for OmniCoat (Microchem) was formed by spin-coating at 3000 rpm for 40 s followed by a curing bake at 200 °C for one minute. Following this, a 1.5 μm thick layer of SU-8 2002 (Microchem), was created by spinning at 3000 rpm for one minute. The SU-8 was pre-baked at 95 °C for one min followed by UV exposure at an energy density of 35 mJ/cm2. A post-exposure bake at 95 °C of 2 min was done followed by development for 15 min in SU-8 developer (Microchem). The coverslip was then dipped in acetone to quench the developing solution and then dipped in OminCoat developer (MF-26A) for 2–3 s to only release the beams and the triangular regions of the device and not the pads. Since the dimensions of the triangular region and the beams are lower than that of the pad regions these regions will release faster than the pad regions. We conducted timed trials until we could get only the regions we needed released. The Omni Coat developer was quenched in DI water and the coverslips were allowed to rest and dry to remove all the water. A schematic representation of the fabrication protocol has been included as Online Resource 1.
2.3 Device characterization
The dimensions of the device were obtained using a Scanning Electron Microscope (SEM) and the thickness was obtained using an optical profilometer (Veeco, WYKO NT1100).
To determine the in-plane stiffness of the device, we used an optical fibre-based force transducer [39]. The force transducer consists of a thin optical fibre (diameter 22 μm) of length 9.5 mm. The fibre is held vertical and one end of the fibre is firmly held by a clamp while the other end is placed inside the triangular frame of the device. The device as well as the optical fibre and clamp assembly are kept on top an optical microscope. The clamp is pulled horizontally by a piezo actuator and the displacement of the clamp is noted. A laser is passed through the optical fibre. The displacement of the laser spot emitted at the tip is recorded and measured through the microscope using a position sensitive photo detector with a resolution of 35 nm. The difference between the displacements of the piezo actuator and the end on the device gives the deformation of the optical fibre. The force exerted on the device is calculated from the deformation of the optical fibre. For these dimensions of the optical fiber, the transducer had a force resolution of 0.805 nN. The displacement of the device is equal to the displacement of the laser spot. The force vs. displacement curves for three devices were obtained. The stiffness was calculated as the slope of a linear fit to this data.
Using the dimensions from the SEM and profilometer, a CAD model of the fabricated device was created in SolidWorks (www.solidworks.com) and analyzed in COMSOL (www.comsol.com). The material properties of SU-8 were obtained from the literature [26]. The end of the device attached to the pads was assumed to be completely fixed and a force was applied at the apex point of the triangle. The geometry was meshed with tetrahedral elements. The displacement was interpolated from the nodes of the mesh using a quadratic scheme.
2.4 Cell culture
NIH 3T3 fibroblasts cells (ATCC R CRL_1658TM) were used for experiments. The cell lines were cultured in T25 flasks (NEST-25 cm2 cell culture flask, canted neck) in high glucose Dulbecco’s Modified Eagle Medium (Sigma Aldrich, Cat. No. D5648-1 l) with 10% fetal bovine serum (Origin: South America, Gibco, Invitrogen, Ref-10270-106). The cells were cultured in an incubator maintained at 37 °C and 5% CO2. The device was coated with 20 μg/mL fibronectin (Sigma Aldrich, Cat. No. F2006) for an hour before seeding cells for the experiment. Cells were trypsinized using Trypsin-EDTA solution (Sigma Aldrich, Cat. No. T3924), seeded on the devices at a concentration where individual cells attach on the devices and allowed to grow for 12 h before experiments were conducted.
2.5 Cell stretching
The fabricated devices were sterilized under UV in a biosafety hood prior to cell seeding. For ease of experiments, the coverslips were attached to custom-fabricated punched Petri dishes. The experiments were conducted on a fully motorized Leica DMI 6000 B, inverted, fluorescence microscope (Fig. 4) with a live cell stage that maintained the samples at 37 °C, 5% CO2 and 90% humidity during the experiment. A custom frame was designed and fabricated using a biocompatible material on a 3D printer (Objet Connex 260). It allows the micropipette holder to enter the live cell chamber without loss of heat and CO2. It had two slots cut into it and the slots were covered by a slit rubber sheet which allowed the pipette holder inside. We used MP-285 (Sutter Instrument Co.), which gives us a movement resolution of 40 nm. A glass micropipette (Borosilicate glass with filament OD 1 mm, ID 0.50 mm, BF 100-5–10) was made using a micropipette puller (Sutter Instrument Co. – Flaming brown Micropipette puller model P-97). The pulled pipette was cut and polished to a tip diameter or around 50 μm by a Micro Forge (Narishige-Micro Forge MF-900). The large diameter and polishing was necessary since it needed to be capable of deforming the device without itself bending or damaging the device.
The micropipette tip was used to probe the device by keeping the tip on the inside of the triangular section of the device and moving it (Fig. 3) to deform the beams. To visualize the cells and nuclei during stretching, Calcein AM (Life Technologies-L3224, kit) and Hoechst (Invitrogen molecular probes 134,406 component B, Hoechst 33,342) were used.
2.6 Fluorescence and confocal imaging
Since there is no material below the coverslips, we were able to image with high-NA objectives (20X 0.7 NA dry objective) to observe general cell/nucleus deformation during stretching and confocal imaging with a 63X, 1.4 NA oil immersion objective during the experiments.
For confocal imaging, cells were fixed in a 4% paraformaldehyde/PBS solution followed by permeabilisation using 0.5% Triton X-100 in PBS. Actin fibres in the cells were visualized using Rhodamine Phalloidin (Molecular Probes; R415) and the nucleus using the Hoechst which was added prior to imaging the deformations. Samples were finally imaged with a confocal microscope (Leica Microsystems, TCS SP5 II) using a 63X oil immersion objective.
3 Results and discussion
3.1 Experimentation and modelling
The in-plane dimensions of the fabricated device were measured from SEM images and the out-of-plane thickness was measured using an optical profilometer. The width of the beams was 7 μm, length was 212 μm and the out-of-plane thickness was 1.5 μm (Table 2). The gap between the beams was 9 μm. The beam width is slightly larger than what was designed. The exact dimensions can be obtained by slightly reducing the energy of the UV exposure or by increasing the SU-8 development time. In spite of the slight differences in dimensions, these devices were found be compliant enough for the cells to deform them.
The stiffness of the device was measured using the optical fibre-based force transducer [39]. The clamped end of the optical fibre was displaced by the piezo actuator in steps of 10 μm. The displacement of the other end which was actuating the device (Fig. 5a) was obtained from the image of the laser spot on the microscope. The end of the optical fibre on the device was allowed to stabilise. After stabilising, many readings (around hundred) of the force applied by the optical fibre and the displacement of the laser spot are recorded for each step of the piezo actuator. The forces and displacements were averaged and plotted against each other (Fig. 5c). The stiffness was obtained by measuring the slope of a line fitted to the force and displacement data. The force vs displacement curve was almost linear with an R2 value > 0.997 for all the devices. Stiffness was obtained for three devices (Fig. 5c). The mean and standard deviation of the stiffness is 0.438 ± 0.085 N/m.
Mechanical characterization of the stretcher. a Stretcher being deformed by the optical fiber for measuring the stiffness. b Finite element Simulation showing the deformation of the stretcher c Sample force vs displacement curve obtained experimentally (red) and force vs displacement curve from the finite (black) element simulation
Using finite element simulations, the displacement of the apex of the device was obtained for various forces applied at the same point (0.5, 1, 1.5, 2 μN) (Fig. 5b). The force vs displacement data obtained was fit to a linear curve (Fig. 5c). The stiffness was calculated as the slope of this line. The stiffness at the actuation point obtained was 0.159 N/m (Fig. 5b). The stiffness of the device determined experimentally is approximately three times larger than what is obtained from the simulations. The deformation of the device (Fig. 3) shows that the beams closer to the actuation point deform more than the ones away from it.
The discrepancy can be attributed to a few factors that include: (a) uncertainty in the material properties, (b) friction and adhesion between the device and the glass coverslip beneath, (c) drag of the liquid, and (d) added mass effect of the liquid as the beams move apart. We argue that the difference in Young’s modulus cannot explain a discrepancy of a factor of three. Friction and adhesion are also not likely the major causes because they would only shift the force-displacement curve up rather than change the slope in the linear regime. We also rule out the effect of drag as the speed of stretch was low (about 1 μm/s). This leaves the added mass effect as a possible cause of the large discrepancy. By “added mass”, we mean the additional force required to stretch the device as the liquid enters and moves along with the beam as the beams move apart. This means that the beams move the liquid even as they move themselves. Modleing the additional force to move a solid submerged in a liquid [40] is beyond the scope of this paper. In the absence of such modelling, this device can be used to induce a desired deformation on the cells. The response of the cells can be obtained for various amounts of stretch rather than various magnitudes of forces. In fact, the focus of this paper is on observing the nuclear deformation due to stretching the cells.
The stiffness of the device is only an order of magnitude larger than that of the cells and hence can be deformed by the cells as can be seen in Online Resource 1. Once the adhesion/friction is modelled, the deformation of the device can be used to compute the force applied on the cells by the mechanism [34]. The stiffness of the cell can be calculated by noting the deformation of the cell, obtained from the images, and the computed forces. The method for obtaining the stiffness of the cell from the deformation of the device is detailed in an earlier work [37]. While that micro-mechanical gripper was designed for compressing cells in suspension, our device is capable of in situ stretching attached cells.
3.2 Nuclear deformation under stretching
The cells attach between the beams (Fig. 6) and proliferate on the mechanism (Online Resource 1). The actin staining (Fig. 6c and g) showed that cells attach to different points on the device and spread well. The SEM pictures (Fig. 6i and j) show that the cells are on top of the device and attached to the beams.
Cells on the stretcher. a, b, c, d shows confocal images – brightfield, nucleus, actin and merged respectively, of a cell on an undeformed folded beam. e, f, g, h shows confocal images – brightfield, nucleus, actin and merged respectively, of a cell on an actuated beam. i, j SEM images of cells attached to the beams
Figure 7a–c show the nucleus prior to stretching the cell and Fig. 7aI, bI, cI show the deformed nuclei after stretching. The red arrows (Fig. 7a–c) points to the nucleus of the cell subjected to deformation. To quantify the effect of stretching the cells on the nucleus, fluorescence images of the nucleus taken before and after actuation of the stretcher were analyzed using ImageJ software (ImageJ, U. S. National Institutes of Health). All nuclei analyzed were of cells attached between the beams and having their major axis aligned along the length of the beams. The images were thresholded manually to clearly identify the nucleus. An ellipse was fit to the nucleus and its major and minor axes were calculated. The direction of stretching is perpendicular to the major axis. Since the cells used were terminally differentiated fibroblasts, the nucleus can be assumed to be having a positive Poisson’s ratio [31]. Hence there should be a decrease in the length of the major axis and an increase in the length of the minor axis on stretching along the minor axis. Table 3 shows the data of the major and minor axis before and after stretching for five cells. We can see that there is a decrease in the length of the major axis and a corresponding increase along the minor axis. The variability in the deformation of the nucleus is due to varying stretch applied to the cells. We have been able to stretch the nuclei to a deformation of up to 11%. For some of the cells, the cell detaches from the device at larger stretches and the nucleus relaxes to an unstressed configuration. Hence all nuclei could not be stretched to the same extent.
Nuclear deformation during cell stretching of three cells, cell 1 (a, a I), cell 2 (b, b I) and cell 3 (c, c I) with a, b, c showing the unstretched nuclei and a I, b I, c I showing the stretched nuclei. The red arrows point to the nuclei in each image. For images a, b, c, a I, b I, c I, Scale bar = 25 μm. The overlaid image of the nucleus (blue), cell (green) in fluorescence and brightfield (greyscale) is shown in (d). For (d) Scale bar = 100 μm
Using confocal microscopy, we can study the change in the volume of the nucleus upon cell stretching. This device can also be used to study the response of the other cellular components linked to mechanotransduction by fluorescently labelling them and observing them during stretching.
4 Conclusions
We have developed a micro-mechanical device for stretching single cells and observing the changes in real time. Our device is simple to fabricate, easy to use and requires only a single XYZ positioner to actuate. The device is transparent and fabricated on coverslips. Hence, they are amenable for high-magnification imaging during operation. A large number of these devices can be microfabricated on a single coverslip and independently actuated. Stretching cells on one of the devices does not disturb the cells on others. This allows for multiple experiments such as different stretches, stretch rates, etc. to be performed on a single coverslip. Furthermore, we can adjust the stiffness of the device by varying in-plane widths of beam segments or the thickness of the structural layer.
Attachment of the cells on the device is accomplished by coating the entire coverslip with Extra Cellular Matrix (ECM) proteins and seeding cells on it. We do not need to pattern the ECM proteins nor place the cells individually on the device. By fluorescent labelling of specific cell components, we can observe their response to mechanical stimuli. This device can be a simple and useful tool to study the effects of mechanical stimuli on cells. As a demonstration, we have cultured NIH 3T3 fibroblasts on the device, stretched them in situ, and measured the deformation of the nucleus as the cell was stretched. We show that cells are capable of deforming the device in Online Resource 1 suggesting that this device can be used to measure the forces exerted by cells. We accept that there is a possibility that the cells attach to the surface of the glass coverslip. To avoid this, in future, we plan to have the same mechanism as an overhanging structure and eliminate any surface interaction.
References
Wang H, Dembo M, Wang Y et al (2000) Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Phys 279(5):1345–1350
Paszek MJ, Zahir N, Johnson KR et al (2005) Tensional homeostasis and the malignant phenotype. Cancer Cell 8:241–254. https://doi.org/10.1016/j.ccr.2005.08.010
Discher DE, Janmey P, Wang Y-L (2005) Tissue cells feel and respond to the stiffness of their substrate. Science 310:1139–1143. https://doi.org/10.1126/science.1116995
Farge E (2003) Mechanical induction of twist in the drosophila foregut / stomodeal primordium. Curr Biol 13:1365–1377. https://doi.org/10.1016/S0960-9822(03)00576-1
Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126:677–689. https://doi.org/10.1016/j.cell.2006.06.044
Wang N, Tytell JD, Ingber DE (2009) Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 10:75–82. https://doi.org/10.1038/nrm2594
Suresh S (2007) Biomechanics and biophysics of cancer cells. Acta Mater 55:3989–4014. https://doi.org/10.1016/j.actamat.2007.04.022
Rajagopalan J, Tofangchi A, Saif MTA (2010) Drosophila neurons actively regulate axonal tension in vivo. Biophys J 99:3208–3215. https://doi.org/10.1016/j.bpj.2010.09.029
Rajagopalan J, Tofangchi A, Saif MTA (2010) Linear high-resolution BioMEMS force sensors with large measurement range. J Microelectromech Syst 19:1380–1389. https://doi.org/10.1109/JMEMS.2010.2076780
Potter SM, DeMarse TB (2001) A new approach to neural cell culture for long-term studies. J Neurosci Methods 110:17–24. https://doi.org/10.1016/S0165-0270(01)00412-5
Wang JHC, Goldschmidt-Clermont P, Yin FCP (2000) Contractility affects stress fiber remodeling and reorientation of endothelial cells subjected to cyclic mechanical stretching. Ann Biomed Eng 28:1165–1171. https://doi.org/10.1114/1.1317528
Moraes C, Chen J-H, Sun Y, Simmons CA (2010) Microfabricated arrays for high-throughput screening of cellular response to cyclic substrate deformation. Lab Chip 10:227–234. https://doi.org/10.1039/b914460a
Wang Q, Zhang X, Zhao Y (2013) Micromechanical stimulator for localized cell loading: fabrication and strain analysis. J Micromech Microeng 23:15002. https://doi.org/10.1088/0960-1317/23/1/015002
Huang Y, Nguyen NT (2013) A polymeric cell stretching device for real-time imaging with optical microscopy. Biomed Microdevices 15:1043–1054. https://doi.org/10.1007/s10544-013-9796-2
Guck J, Ananthakrishnan R, Mahmood H et al (2001) The optical stretcher: a novel laser tool to micromanipulate cells. Biophys J 81:767–784. https://doi.org/10.1016/S0006-3495(01)75740-2
Dao M, Lim CT, Suresh S (2003) Mechanics of the human red blood cell deformed by optical tweezers. J Mech Phys Solids 51:2259–2280. https://doi.org/10.1016/j.jmps.2003.09.019
Rørth P (2012) Fellow travellers: emergent properties of collective cell migration. EMBO Rep 13:984–991. https://doi.org/10.1038/embor.2012.149
Sugiura T, Miyoshi H, Nishio T, Honda A (2012) Cell palpation with an optically trapped particle. J Micro-Nano Mechatronics 7:131–136. https://doi.org/10.1007/s12213-012-0051-3
Thoumine O, Ott A, Cardoso O, Meister JJ (1999) Microplates: a new tool for manipulation and mechanical perturbation of individual cells. J Biochem Biophys Methods 39:47–62. https://doi.org/10.1016/S0165-022X(98)00052-9
Thoumine O, Ott A (1997) Time scale dependent viscoelastic and contractile regimes in fibroblasts probed by microplate manipulation. J Cell Sci 110:2109–2116
Sun Y, Liu X, Wang W, Lansdrop BM (2007) Vision-based cellular force measurement using an elastic microfabricated device. J Micromech Microeng 17:1281–1288
Ohara K, Kawakami D, Takubo T et al (2012) Dextrous cell diagnosis using two-fingered microhand with micro force sensor. J Micro-Nano Mechatronics 7:13–20. https://doi.org/10.1007/s12213-012-0040-6
Serrell DB, Law J, Slifka AJ et al (2008) A uniaxial bioMEMS device for imaging single cell response during quantitative force-displacement measurements. Biomed Microdevices 10:883–889. https://doi.org/10.1007/s10544-008-9202-7
Fior R, Maggiolino S, Lazzarino M, Sbaizero O (2011) A new transparent bio-MEMS for uni-axial single cell stretching. Microsyst Technol 17:1581–1587. https://doi.org/10.1007/s00542-011-1325-8
Antoniolli F, Maggiolino S, Scuor N et al (2014) A novel MEMS device for the multidirectional mechanical stimulation of single cells: preliminary results. Mech Mach Theory 78:131–140. https://doi.org/10.1016/j.mechmachtheory.2014.03.009
Sato K, Kamada S, Minami K (2010) Development of microstretching device to evaluate cell membrane strain field around sensing point of mechanical stimuli. Int J Mech Sci 52:251–256. https://doi.org/10.1016/j.ijmecsci.2009.09.021
Wai-Chi W, Azid AA, Majlis BY (2010) Formulation of stiffness constant and effective mass for a folded beam. Arch Mech 62:405–418
Fedorchak GR, Kaminski A, Lammerding J (2014) Cellular mechanosensing: getting to the nucleus of it all. Prog Biophys Mol Biol 115:76–92. https://doi.org/10.1016/j.pbiomolbio.2014.06.009
Dahl KN, Ribeiro AJS, Lammerding J (2009) Nuclear shape, mechanics, and mechanotransduction. Circ Res 102:1307–1318. https://doi.org/10.1161/CIRCRESAHA.108.173989
Isermann P, Lammerding J (2013) Nuclear mechanics and mechanotransduction in health and disease. Curr Biol. https://doi.org/10.1016/j.cub.2013.11.009
Pagliara S, Franze K, Mcclain CR et al (2014) Transition from pluripotency in embryonic stem cells distinguished by an auxetic nucleus. Nat Mater 13:638–644. https://doi.org/10.1038/nmat3943.
Xue P, Bao J, Chuah YJ et al (2014) Protein covalently conjugated SU-8 surface for the enhancement of mesenchymal stem cell adhesion and proliferation. Langmuir 30:3110–3117. https://doi.org/10.1021/la500048z
Solon J, Levental I, Sengupta K et al (2007) Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J 93:4453–4461. https://doi.org/10.1529/biophysj.106.101386
Reddy AN, Ananthasuresh GK (2008) On computing the forces from the noisy displacement data of an elastic body. Int J Numer Methods Eng 76:1645–1677. https://doi.org/10.1002/nme.2373
Tee SY, Fu J, Chen CS, Janmey PA (2011) Cell shape and substrate rigidity both regulate cell stiffness. Biophys J 100:L25–L27. https://doi.org/10.1016/j.bpj.2010.12.3744
Zhu X, Zhang N, Wang Z, Liu X (2016) Investigation of work of adhesion of biological cell (human hepatocellular carcinoma) by AFM nanoindentation. J Micro-Bio Robot 11:47–55. https://doi.org/10.1007/s12213-016-0089-8
Bhargav SDB, Jorapur N, Ananthasuresh GK (2015) Micro-scale composite compliant mechanisms for evaluating the bulk stiffness of MCF-7 cells. Mech Mach Theory 91:258–268. https://doi.org/10.1016/j.mechmachtheory.2015.04.002
Lorenz H, Despont M, Fahrni N et al (1997) SU-8: a low-cost negative resist for MEMS. J Micromech Microeng 7:121–121. https://doi.org/10.1088/0960-1317/7/3/010
Seshagiri Rao RV, Kalelkar C, Pullarkat PA (2013) Optical fiber-based force transducer for microscale samples. Rev Sci Instrum. https://doi.org/10.1063/1.4824198
Dong RG (1978) Effective mass and damping of submerged structures. UCRL-52342, April, Lawrence Livermore Laboratory, U. California, Livermore, CA, USA
Acknowledgements
Authors would like to acknowledge Prof. Pramod Pullarkat and his student Sushil Dubey, Raman Research Institute for helping with the stiffness measurements and Anitha Shiva, Center for Nano Science and Engineering, Indian Institute of Science for helping with the microfabrication and Gowri Balachandran for helping take the SEM images of the device. This work was supported by the “Bioengineering and Biodesign Initiative” grant from the Department of Biotechnology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Online resource 1
(AVI 2619 kb)
Rights and permissions
About this article
Cite this article
Kollimada, S., Balakrishnan, S., Malhi, C.K. et al. A micro-mechanical device for in-situ stretching of single cells cultured on it. J Micro-Bio Robot 13, 27–37 (2017). https://doi.org/10.1007/s12213-017-0102-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12213-017-0102-x