Abstract
Introduction
The pathophysiological increase in microvascular permeability plays a well-known role in the onset and progression of diseases like sepsis and atherosclerosis. However, how interactions between neutrophils and the endothelium alter vessel permeability is often debated.
Methods
In this study, we introduce a microfluidic, silicon-membrane enabled vascular mimetic (μSiM-MVM) for investigating the role of neutrophils in inflammation-associated microvascular permeability. In utilizing optically transparent silicon nanomembrane technology, we build on previous microvascular models by enabling in situ observations of neutrophil-endothelium interactions. To evaluate the effects of neutrophil transmigration on microvascular model permeability, we established and validated electrical (transendothelial electrical resistance and impedance) and small molecule permeability assays that allow for the in situ quantification of temporal changes in endothelium junctional integrity.
Results
Analysis of neutrophil-expressed β1 integrins revealed a prominent role of neutrophil transmigration and basement membrane interactions in increased microvascular permeability. By utilizing blocking antibodies specific to the β1 subunit, we found that the observed increase in microvascular permeability due to neutrophil transmigration is constrained when neutrophil-basement membrane interactions are blocked. Having demonstrated the value of in situ measurements of small molecule permeability, we then developed and validated a quantitative framework that can be used to interpret barrier permeability for comparisons to conventional Transwell™ values.
Conclusions
Overall, our results demonstrate the potential of the μSiM-MVM in elucidating mechanisms involved in the pathogenesis of inflammatory disease, and provide evidence for a role for neutrophils in inflammation-associated endothelial barrier disruption.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increased permeability of the microvasculature to both leukocytes and plasma components are hallmarks of inflammation. Alterations in microvascular barrier integrity are implicated in a variety of inflammatory diseases, including sepsis.10 Despite the fact that both leukocyte and plasma ‘permeability’ have been known since the observations of Cohnheim,9,23,24 the extent to which leukocyte engagement contributes to microvascular permeability remains unclear.46 The common view that these two forms of permeability are uncoupled is based on many examples of leukocyte adhesion without accompanying permeability and vice versa (reviewed in Refs. 19 and 29). However, there are counter examples both in vivo53 and in vitro13,17 where the engagement of endothelium by leukocytes is the only plausible stimulus responsible for a subsequent increase in molecular permeability. Developing a better understanding of the specific role neutrophils play in vascular barrier disruption will facilitate the development of novel anti-inflammatory therapeutics capable of modulating this off-target response.
Live cell movies of leukocytes transmigrating across endothelium clearly show short-lived disruptions of endothelial junctions.48 The fact that the junctions quickly reseal following the leukocyte exit is cited as an explanation for the ability of endothelial monolayers to maintain a barrier to molecular transmission despite leukocyte transmigration,19,29 however the same images suggest that some local molecular leakage will likely accompany transmigration events. Thus, the controversy in the literature may be a matter of degrees: experimental circumstances involving a large number of simultaneous leukocyte engagements will show concurrent molecular transmission in bulk permeability assays, while the same assays will not be sensitive to less numerous and/or asynchronous events. Indeed, one clear example of neutrophil-induced monolayer disruption is provided by Gautum et al., who demonstrated both a decrease in transendothelial electrical resistance (TEER) and an increase in the albumin permeability of unstimulated human umbilical vein endothelial cells (HUVECs) when N-Formylmethionyl-leucyl-phenylalanine (fMLP) was introduced in the trans compartment of a membrane-based chemotactic chamber.17 Because fMLP is a powerful neutrophil chemoattractant, more than 200,000 transmigration events were estimated to occur within the hour of observation—a number equal to the estimated number of endothelial cells (ECs) plated on the Transwell™ filter.
While these observations argue that aggressive, synchronous leukocyte transmigration should be accompanied by a detectable molecular permeability rise in bulk assays, modern era microphysiological systems (MPS) (also tissue chip or organ-on-a chip) should be able to detect an increase in molecular permeability with more modest physiological levels of leukocyte engagement. Ideally, these systems will integrate assays for molecular transport in close proximity to the culture, overcoming dilutions and transfer losses that make bulk assays less sensitive. Thus far, vascular barrier MPS models have successfully integrated electrodes for TEER measurements.3,55 However, by employing conventional polymeric track-etched membranes, these devices sacrifice live cell imaging, operate with a high baseline resistance to transport, or both.5,37 Thus, while MPS devices have been used to measure the molecular permeability of the endothelium3,55 and have demonstrated leukocyte transmigration,21 they have not been used—nor have they been equipped—to investigate the relationship between the two.
We have recently introduced the µSiM (a microphysiological system enabled by a silicon-based membrane) for the construction of dual chamber (defined apical and basal compartments) MPS that enable live cell imaging. We applied the µSiM concept to create models of the cerebral vascular barrier37 and the lacuna-canalicular network of cortical bone.34 The work builds on more than a decade of basic studies establishing the optical clarity,1,5,35 biocompatibility,1,14,35,40 and extraordinary permeability22,28,49,51 of ultrathin (15 nm–400 nm thick) silicon-based nano and microporous ‘nanomembranes.’ In the current study we apply the µSiM concept to create a microvascular mimetic (µSiM-MVM) with the capacity for in situ measurements of electrical impedance and transmembrane molecular diffusion. We then use the µSiM-MVM to confirm that an MPS with these integrated permeability assays can be used to detect the impact of leukocyte transmigration on the barrier properties of an otherwise unstimulated endothelial monolayer. Ultimately, we observed a decrease in endothelial barrier integrity during neutrophil transmigration, that was partially attenuated following the blocking of neutrophil-expressed β1 integrins.
Materials and Methods
Nanomembranes
The nanomembrane utilized for the μSiM-MVM was 400 nm thick silicon dioxide (SiO2) membranes with hexagonally packed 3 µm holes spaced 9 µm apart (GFLAT300-1Q-3.0LP; SiMPore, Inc., West Henrietta, NY). These pore characteristics give the membranes a porosity of ~ 5% and an area density of ~ 8 × 105 cm−2 or ~ 16 per cell once a monolayer is confluent. The 3 µm pore dimension enables complete transmigration of neutrophils into the basal compartment of the μSiM-MVM. At this density however, microscale pores do compromise cell adhesion with the application of physiological fluid shear stress,38,43, possibly because of a reduced number of focal adhesions compared to non-porous materials.6 While we have recently solved this issue without a loss of permeability by adding a low density of 3 µm pores in a background of abundant nanopores,43 in the current paper we overcame the challenge by applying a layer of collagen I (2 mg mL−1) and fibronectin (0.17 mg mL−1) to the SiO2 membrane surface prior to seeding cells. Note that in the absence of the nanomembrane, collagen will sag with the application of flow, thus while the membrane is not the primary substrate for cells in our device, it is essential for the construction of a stable system.
Device Fabrication
The μSiM-MVM consists of two main compartments: luminal (‘top’ or ‘apical’) and abluminal (‘bottom’ or ‘basal’) chambers separated by the nanomembrane. Channel and spacer layers were comprised of silicone gasket materials (Trelleborg Sealing Solutions; Trelleborg, Sweden) cut with a conventional craft-cutting tool (Silhouette Cameo; Lindon, Utah) and assembled in a layer-by-layer fashion as described elsewhere.7,25,37,43 The dimension of the top channel (230 µm in height) produces a physiological shear stress of 10 dynes cm−2 at a flow rate of 1.19 mL min−1.
There were two key departures from our prior device assemblies. First was the use of indium tin oxide (ITO) coated-glass to serve as electrodes and the floor and ceiling layers of our fluidic device. ITO sheets (PF-65IN) were commercially purchased from Delta Technologies (Colorado, USA) and consist of 200 nm of In2O3/Au/Ag film thermally deposited on ~ 100 μm thick polyethylene terephthalate (PET) substrate with a manufacturer reported resistivity of 10 Ω cm and an optical transmittance > 78%. The planar electrode design is the ideal configuration for MPS as it produces parallel electrical field lines in an MPS device, minimizing high baseline impedances that arise from requiring current to pass through microfluidic access ports.26 Since only two film-substrates are used to create four electrodes, each film was scored using the craft-cutter to physically separate the ITO films on each side of the insulate, thus creating two separate electrodes on a single planar sheet of ITO. Silver wires of 250 µm diameter (A-M Systems, Carlsborg, WA) were bonded to ITO films using silver epoxy (8331S, MG Systems, Surrey, B.C., Canada) and cured at 70 °C overnight to achieve high conductance wire bonding (Supplemental Fig. S1).
The second departure from our previous devices was the use of pressure sensitive adhesive (PSA) layers for device assembly (468MP; 3 M, Minnesota, USA) in addition to our conventional use of UV-ozone bonding for layer assembly.7,25,37,43 The use of PSA allowed us to overcome the difficulty of bonding UV-activated silicone to ITO. The use of PSA also allowed us to assemble the bottom half of the device after the addition of collagen I to the ‘trench’57 on the underside of the silicon chip. The PSA was patterned and cut using the same craft-cutter used for silicone patterning. Ultimately, the central parts of the device consisting of the membrane chip, spacer and gasket layers were bonded with UV-ozone, both the top and bottom channels were patterned from PSA, and the device was enclosed with ITO layers (Fig. 1a). As with our prior devices, a polydimethylsiloxane (PDMS) block was used to facilitate access to both top and bottom fluidic channels. PSA was again used to facilitate bonding between the ITO and the PDMS block.
Microvascular Mimetic (µSiM-MVM) platform: design and assembly. (a) 2D schematic showing apical (luminal) and basal (abluminal) compartments separated by a silicon nanomembrane, which also acts as a scaffold for incorporating type 1 collagen gel as an extracellular matrix (ECM) mimic. Top and bottom channels are accessible using independent inlet/outlet ports. Indium tin oxide (ITO) electrodes are assembled to enable electrical resistance measurement. (b) Exploded 3D view of the microfluidic system illustrating the silicone gaskets, pressure sensitive adhesive (PSA) and electrodes. (c) Assembled device without electrodes (for visual clarity) highlighting the miniaturized nature and the transparency of the platform.
Cell Culture Protocols
Pooled HUVECs and MCDB-131 Complete Medium were obtained from VEC Technologies (Rensselaer, NY). Prior to device assembly, the silicon nanomembranes were coated by adding rat tail collagen I into the top sealing layer (Enzo Life Sciences, Farmingdale, NY) at a density of 2 mg mL−1 and adjusting the pH to 7.4 to promote polymerization. After gel formation, the membrane was further coated with 0.17 mg mL−1 of human plasma fibronectin (BD Biosciences, Mississauga, ON) for 1 h at room temperature. In cases where the trench was also filled with collagen I, a ~ 2 µL drop was added to the trench prior to addition of the bottom channel and ITO layer. After the coating process, the devices were fully assembled and filled with MCDB-131 media for 1–2 h prior to cell seeding. Confluent monolayers of HUVECs were trypsinized from tissue culture flasks and seeded inside the devices (top chamber) at the density of ∼ 40,000 cells cm−2. Cells were grown under static conditions for ∼ 24 h (37 °C, 5% CO2) and then subjected to shear stress [10 dynes cm−2] for an additional 24 h before any experiments. HUVECs were used prior to passage 7. While the apical media was subjected to continuous perfusion, the basal media was changed regularly to avoid evaporation and replenish nutrients. The entire flow circuit including the peristaltic pump, media/nutrient reservoir, fluidic capacitor and the μSiM-MVM device was placed inside the incubator to maintain a proper gaseous environment and temperature conditions suitable for EC growth (Supplemental Fig. S2). The flow system supporting the μSiM-MVM was recently described elsewhere.43 When stated, 5–50 U mL−1 of thrombin (Sigma, St. Louis, MO) was added to the ECs for 20 min to induce changes in permeability.
Neutrophil Assays
Human blood was collected from peripheral veins of healthy individuals in heparin-coated tubes. Neutrophils were immediately isolated from whole blood using a 1-step polymorph solution (Accurate Chemical, Westbury, NY) as previously described.7,43 The isolation procedure took less than 2 h, and neutrophils were used within the next 3–4 h.
For transmigration assays, neutrophils were suspended in MCDB-131 medium (to acclimate to the EC media) at a density of 107 mL−1 and 100 µL of the neutrophil rich solution was infused into the apical chamber (106 neutrophils). Transmigration was induced with 10 nM fMLP (Sigma Aldrich, St. Louis, MO) suspended in MCDB-131. The use of fMLP was determined based on its role in directing neutrophils in localized bacterial infection, as well as its lack of influence on EC barrier integrity. This solution was used to replace the media in the basal chamber (50 μL). Before the introduction of fMLP, a phase image of the apical chamber was obtained for counting the initial number of neutrophils. For transmigration observations, the chambers were either kept inside the incubator, or placed on the heated stage on a phase microscope. Neutrophil migration-blocking was achieved by adding a low volume of concentrated anti-CD29 (β1) monoclonal antibody P5D2 (Biolegend, San Diego, CA) in PBS to the solution of neutrophils in MCDB-131 immediately prior to their addition to the apical chamber. The final concentration of anti-CD29 was 1 µg per 100 µL (i.e., 1 µg per 106 neutrophils).
In neutrophil transmigration experiments with interlaced phase and fluorescence, HUVECs were pre-labeled with a solution of Alexa Fluor 488 conjugated anti-vascular endothelial (VE)-Cadherin monoclonal antibody (Santa Cruz Biotechnology; Dallas, TX). Alternating phase and fluorescence images were collected with a fully incubated microscope (Etaluma LS560; Carlsbad, CA) at 6 frames per min (fpm) over 30 min.
TEER and Impedance Measurements
The TEER measurements were achieved using an EVOM Voltohmmeter (WPI Inc., Sarasota, FL). The silver wires were connected to the EVOM Voltohmmeter using custom-designed breadboard circuits. The circuits and the devices were placed in the incubator, while the EVOM Voltohmmeter was placed outside using connecting cables. TEER measurements were performed at 12.5 Hz using a low amplitude square-wave AC current (10 µA), and the corresponding potential drop is internally recorded to yield the final resistance in ohms as displayed on the Voltohmmeter. The EVOM Voltohmmeter also has an analog output and was connected to an Agilent Digital Multimeter 34401a (Keysight, Santa Rosa, CA) using coaxial cables. The multimeter was controlled using the manufacturer’s software installed on a Windows PC. Thus, ultimately, the TEER data acquisition was fully automated and the temporal evolution of electrical resistance was recorded digitally on a computer for further analysis and interpretation.
While TEER measurement requires tetrapolar output, impedance measurement requires only two electrodes—one on either side of the membrane. For impedance measurements, the output was connected to custom-designed breadboard circuit, which was further connected to VersaSTAT Potentiostat Galvanostat (Princeton Applied Research, Oak Ridge, TN) to conduct frequency sweep. The VersaSTAT instrument was controlled using the manufacturer’s software installed on the Windows PC. The frequency sweep was performed from 10 Hz to 106 Hz with a sampling frequency of 10 points per decade with Data Quality (DQ) set to 3 (number of measurements averaged) for measurements with transient phenomenon, or 50 points per decade and DQ set to 5 for time-insensitive measurements. While the higher DQ improves the robustness of the data acquisition, it also increases the total time of the sweep, and can reduce the ability to capture time-sensitive changes. The impedance data at 12.5 and 1000 Hz was analyzed in MATLAB using custom-written programs for further analysis (Supplemental Fig. S3).
Electron Microscopy
Endothelial monolayers grown in the µSiM-MVM were fixed with 25 µL of media containing 2.5% glutaraldehyde for 10 min. Silicon membranes were then carefully extracted from the device and transferred to 50% ethanol in a six well plate. The samples were dehydrated in sequential ethanol steps (10% increase per step) to 100% ethanol. To prepare for electron microscopy, the samples were then dried using a Tousimis SamdriR-PVT-3D (Tousimis, Rockville, MD, USA) critical point dryer and then sputter-coated with 10 nm of gold in a Denton PVD (Denton Vacuum, Moorestown, NJ, USA) sputter coater. Samples were imaged in a Zeiss Auriga (Carl Zeiss Microscopy GmbH, Jena, Germany) field emission scanning electron microscope with a 10 kV accelerating voltage.
In Situ Measurements of Transmembrane Diffusion
FITC-dextran (10 kDa) was added to MCDB-131 complete media at a concentration of 1 mg mL−1. Neutrophils were suspended in FITC-dextran media with or without anti-CD29 antibody as previously described. The neutrophil solution was perfused into the apical channel of EC-seeded µSiM devices and devices were transferred to a heat-stage atop an epifluorescence microscope. Fluorescent images were collected every minute for 20 min in the basal channel, adjacent to, but not directly beneath the nanoporous membrane window (50 µm from the nanoporous membrane edge). Images were imported to a custom designed MATLAB script, where fluorescence intensity (grayscale) was averaged parallel to the membrane edge, and plotted over time.
Quantitative Framework for In Situ Molecular Permeability
In a follow-up effort, we sought to develop a quantitative framework for interpreting in situ molecular diffusion measurements in terms of monolayer permeability. This work employed open µSiM devices without any accompanying electronics as described in Masters et al.,34 and human pulmonary aorta endothelial cells (HPAECs) instead of HUVECs because the permeability of HPAECs were well-characterized by our team in conventional Transwell™ devices. HPAECs (Cell Applications, Inc.; San Diego, CA) were cultured as previously described.18 HPAECs were seeded in open top µSiM devices and grown to confluency over 48 h. HPAECs were used prior to passage 6. Once the monolayer was established, apical media was removed and replaced with 50 µL of fresh media. Devices were transferred to an Olympus IX70 microscope stage (Tokyo, Japan) and an Olympus LUCPlanFLN 40X objective was focused to the periphery of the long edge of the free-standing porous nanomembrane. Once in focus, 50 μL of a 400 μg mL−1 solution of FITC-dextran was added to the apical well and fluorescent time-lapse imaging was initiated. Images were collected by an ORCA-ER digital camera (Hamamatsu, Japan) once a minute for 10 min with an exposure of 100 ms to minimize effects of photobleaching. Following the completion of the time-lapse, a 200 μg mL−1 solution for FITC-dextran was carefully added to the basal channel of the μSiM and a final image was collected. Resulting images were imported into a custom-made MATLAB script to determine the fluorescence intensity 50 μm from the freestanding membrane edge.
While the analytical solution (detailed in the supplemental material) of the experimental system is well described, it does not account for many of the geometric features of our μSiM. To overcome this potential room for error, finite element modeling software, COMSOL, was used to computationally recreate our experiment. A 2-D cross-sectional geometry of our device was built to exact dimensions in COMSOL. A 10 μm thick box was placed directly above the ‘membrane’ to act as a model cell monolayer. A 200 μg mL−1 solution was assigned to the ‘trench’ portion of the device (inverted trapezoid) to act as the input FITC-dextran solution. The diffusive species was assigned a free diffusion coefficient of 7.44 × 10−11 m2 s−1 to reflect the properties of a 40 kDa FITC-dextran solution in 37°C media (based on a supplier provided Stoke’s radius of 45 Angstroms). The diffusion coefficient in the monolayer region Dm was adjusted based on the monolayer permeability PE according to:
where L is the thickness of the monolayer region (10 μm).
To evaluate transport across the monolayer, a point was placed at the top edge of the bottom channel, 50 μm from the edge of the ‘membrane’ to act as the microscope focal point. The change in concentration through that point was recorded over 10 min as in our experimental setup. A parametric sweep of varying monolayer permeabilities was performed to build a database of potential solutions ranging from PE of 1 × 10−7 to 1 × 10−4 cm s−1. These solutions were fitted to experimental data to obtain permeability coefficients.
Transwell™ Measurements of Molecular Permeability
HPAECs were seeded in Costar Transwell™ inserts (3 µm pores, 12 mm inserts) and grown to confluency over 48 h. After reaching confluency, apical media was replaced with a 200 μg mL−1 solution of 40 kDa FITC-dextran in cell culture media and cells were incubated at 37 °C, 5% CO2 for 2 h. Basal media was then collected, and FITC-dextran concentration was measured in a fluorescence plate reader. Permeability of the EC monolayer was calculated using Eq. (2)3,42:
where P represents the permeability coefficient, JF is the flux of FITC-dextran across the EC and membrane, A is the membrane area, and Ci is the concentration of FITC-dextran initially added to the apical well. With a system permeability coefficient calculated, the EC permeability was isolated using Eq. (3):
where PE is the permeability coefficient relating to the EC monolayer, P is the system permeability as calculated in Eq. (2), and PM is the permeability of a cell-free membrane.
Results
The µSiM-MVM: An EC Barrier Imaging Platform with Integrated Flow and Electronics
The µSiM-MVM concept and device assembly is show in Fig. 1. Assembly details are described in the methods. The µSiM-MVM device is an advancement on our prior µSiM devices which were with either simple transmembrane culture chambers12,34 or closed flow cells.37,43 Our goal here was to integrate electronic measurements of monolayer integrity with our flow cell device and to introduce a backside collagen gel as abluminal extracellular matrix (ECM) space. Because the creation of a trench on the backside of the silicon chip is a natural outcome of the fabrication process51, we use this space to create the abluminal compartment without occluding the microfluidic channel on the basal side of the device. We used SiO2 membranes patterned with 3 µm pores35 at a density of 8 × 105 cm−2 (~ 5% porosity). As we’ve shown previously, cells do not adhere well to these membranes under flow even with ECM coatings.37 This is possibly due to the fact that pores create large spans without focal adhesions, thus reducing the overall focal adhesion density6 and compromising their ability to resist shear forces. To overcome this, we also added a collagen I gel layer atop the membrane, which, along with a topical fibronectin coating, enabled the cells to remain attached under physiological levels of shear. The introduction of the collagen gel layers required that we use pressure sensitive adhesives as gasket layers to enable µSiM device assembly after the addition of the gel.
The final advancement over our prior studies was the incorporation of ITO as transparent electrodes. We include these as the apical and basal terminal layers that enclosed our device. We used them both in a tetrapolar configuration for TEER measurements and in a dual electrode configuration for impedance measurements during the more time sensitive studies involving neutrophil transmigration. In both cases we configured the electrodes to be directly above the membrane to minimize non-parallel field lines which can cause high background resistance in microfluidic designs.20,26
Enhanced EC Barrier and Alignment with Physiological Shear
Enhanced barrier function is a well-established response to flow conditioning of ECs with physiological levels of shear.11,44 Additionally, the elongation and alignment of ECs in the direction of flow with the application of physiological shear is one of the oldest observations in cell culture flow chambers.15 We considered these to be fundamental requirements for the successful operation of the μSiM-MVM. We also reasoned that the shear-induced elevation of EC barrier function would be the ideal starting point for detecting a potentially small impact of neutrophil transmigration on permeability.
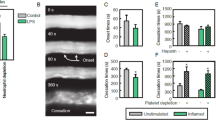
To examine the ability of the μSiM-MVM to support shear conditioning of ECs, we first established the evolution of TEER values when ECs were grown under static conditions for 48 h. We found that a clear plateau was established after about 24 h (Fig. 2a). In a subset of these studies we introduced 10 dynes cm−2 of physiological shear stress, a value which is well above the threshold for alignment15 and barrier enhancement44 and is consistent with the shear values of post-capillary venules where neutrophil diapedesis occurs.41 Cells remained adherent under flow, confirming that the addition of a collagen gel and fibronectin coating were effective for enabling EC adhesion to SiO2 with 3 µm pores under flow.37 Within 10 h of the onset of flow, EC monolayers achieved a new TEER plateau value more than 50% higher than that achieved with static growth (Fig. 2a). This elevated barrier function could be quickly disrupted (< 20 min) with thrombin. In the same experiments we confirmed that physiological shear-induced elongation and alignment of ECs (Fig. 2b). To our knowledge, this is the first time an MPS vascular model has been used to monitor the shear-induced alignment of live cells by phase microscopy while simultaneously monitoring TEER. The result highlights the value of using optically5 and electrically50 transparent silicon nanomembranes for the construction of MPS.
TEER and EC morphology under shear. (a) TEER measurement was performed over 48–52 h post cell seeding in the µSiM-MVM. TEER rises during first 24 h (red curve), after which it remains stable in the absence of shear (blue curve) or progresses to an elevated level in the presence of shear (brown curve). The addition of thrombin immediately causes the TEER to decay below 50% of its maximum value in less than half an hour (black dashed line). Error bars represent standard error of mean with at least n = 5. (b) Endothelial cells grown under static condition for 24 h demonstrate cobblestone morphology as observed in the phase image on the top left, and the corresponding image analysis in MATLAB reveals the isotropic orientation of the cell population. Endothelial cells exposed to shear stress of 10 dynes cm−2 for 24 h exhibit uniformly aligned morphology with a majority of the cells parallel to the flow direction (Y axis).
Neutrophil Transmigration in the μSiM-MVM Platform
To examine the impact of neutrophil transmigration on EC barrier function, we first needed to ensure the ability of neutrophils to migrate through the μSiM-MVM in response to a chemoattractant applied to the basal compartment. This question was particularly relevant given the addition of collagen gels above and below the membrane. It was not clear if these gels would hinder neutrophil transmigration, fMLP diffusion, or both. To test this, we added freshly isolated neutrophils (107 mL−1) to the apical chamber with: (1) ECs atop an apical gel with a basal gel added to the trench, and (2) ECs atop an apical collagen gel only. Neutrophils were then stimulated by the basal addition of 10 nM fMLP. After 30 min neutrophils were found in similar numbers on floor of the basal chamber (17.54 ± 4.29% in absence of basal gel and, 19.89 ± 5.52% in the presence of basal gel (n = 3, mean ± standard error); Fig. 3a). Since the addition of the second 300 µm thick collagen gel did not have any apparent impact on the rate of migration, we concluded that the gels did not represent a significant hinderance to either the diffusion of fMLP or neutrophil migration. The endothelial layer, on the other hand, clearly provided a barrier to transmembrane migration because when these experiments were conducted in the absence of this layer (and the top gel required for stability under shear) the number of transmigrated neutrophils increased substantially (Fig. 3a).
Neutrophil transmigration through a collagen matrix. (a) Even in the presence of endothelial cells and supplemented collagen I gel, neutrophils transmigrate to the abluminal channel. The presence of basal gel (below the nanomembrane) does not significantly reduce the amount of migrated cell population as observed from the representative images of cells on the cover-slip 3 h after addition (17.54 ± 4.29% in absence of basal gel, and 19.89 ± 5.52% in the presence of basal gel, n = 3 each; the percentage of cells calculated by dividing the number of cells in the basal chamber 3 h later to the number of cells in the top chamber at the onset of migration). In the absence of endothelial cells, neutrophils (hPMN) migrate fastest due to potential absence of any steric inhibition. Scale bar = 100 μm. (b) Scanning electron microscopy (SEM) analysis shows the ability of hPMNs to transmigrate through the collagen gel, eventually residing on the underlying nanomembrane.
In a related experiment, we developed a technique to peel back the endothelial and collagen layer from the microporous membrane after fixing, extracting and critically point drying the membrane chip (Fig. 3b). This allowed us to confirm the presence of neutrophils at layer interfaces by scanning electron microscopy (SEM). In a single SEM image, neutrophils could be found above the endothelial layer, emerging from the collagen layer, and on the microporous membrane where they were co-localized with pores. Remarkably, the endothelial monolayer remained intact with the shear-induced alignment still evident in the preparation (Fig. 3b).
As we previously demonstrated with nanoporous37 and dual scale nanomembranes,43 transmigration across endothelium can be imaged in live cell microscopy on microporous SiO2 membranes (Fig. 4). In phase contrast microscopy, leukocytes transformed from phase bright atop the endothelial layer to phase dark as they migrate beneath the layer as flattened cells (Fig. 4a). In the current experiments we also noted phase bright gaps developing between ECs as neutrophils egressed. In follow-up experiments, we labeled VE-cadherin with fluorescent antibodies and interlaced fluorescence and phase-contrast images in time-lapse microscopy to clearly see the impact of neutrophils on endothelial junctions (Fig. 4b). The temporary, localized disruption of endothelial junctions is consistent with prior observations by others,48 and supports our hypothesis that some disruption of barrier function should be detectable with an elevated number of concurrent transmigration events and a sufficiently sensitive measurement configuration.
Optical clarity of μSiM devices permit real-time observations of EC junctions. (a) Phase time lapse images of neutrophils (yellow arrow) transmigrating through a HUVEC monolayer reveal potential separations in EC junctions (blue arrow). Scale bar = 10 μm. (b) Live immunolabeling of HUVEC junctional proteins (VE-cadherin; green) further highlights the disruption of the EC barrier during a neutrophil transmigration event. Scale bar = 20 μm.
Regulating Neutrophil Subendothelial Migration
To investigate the impact of neutrophil transmigration on barrier function, we sought a function blocking antibody specific to the transmigration step. Because of the role of β2 integrins in both endothelial adhesion and transendothelial migration,31 we selected instead to target β1 (CD29) integrins. The β1 integrins appear constitutively at a low level but are upregulated in response to β2 engagement and neutrophil activation56 and have a key role for migration on and through the EC basement membrane.4 Unlike β2 or β3 integrins, β1 integrins appear to have no role in collagen-based migration32 or initial EC adhesion.4 Thus, while both β2 and β1 integrins contribute to the transmigration across the barrier, only the β1 integrins appear to have a role specific to the perivascular stage. Additionally, the previously observed remedial effects of β1 inhibition in a murine model of sepsis make this a particularly interesting target for future inflammatory therapeutics.30,45
To confirm the effectiveness of β1 blocking, we repeated our studies by phase and electron microscopy in the presence of the blocking antibody. In these studies, neutrophils were pre-incubated with control media (Fig. 5a) or an excess of anti-CD29 blocking antibody (Fig. 5b) before adding the solution to the apical chamber of the µSiM-MVM with a fMLP challenge from the basal chamber. We reasoned that the excess antibody should also help block any newly expressed β1 integrins that appear upon activation. By taking the ratio of the number of neutrophils found on the floor of the basal chamber after 3 h to the total number of neutrophils in the luminal surface of the ECs at time zero, we quantified the percentage of neutrophils that fully transmigrated the device in response to a basal 10 nM fMLP challenge. We found reduction in the percentage of transmigrated neutrophils with the β1 blocking regimen (Fig. 5c). Consistent with this result, we did not find any anti-CD29 treated neutrophils associated with either the membrane or the collagen gel in delaminated samples viewed by SEM (Fig. 5d).
Neutrophils migrate uninhibited in the interstitial space in the absence of β1 integrin blocking (a) but remain trapped in the subendothelial space in the presence of the blocking antibody against β1 integrins (b). (c) Percent migration from the apical to basal compartments under the influence of basally added fMLP over the duration of 3 h reduces significantly in response to β1 integrin blocking. Error bars represent standard error of mean; n = 3 for each condition. (d) The disruption of 3D migration of β1 integrin-blocked neutrophils was again revealed under SEM. The yellow inset shows the zoomed-in portion of the membrane indicating the absence of neutrophils, which have egressed beyond the collagen gel. The entire membrane-area was thoroughly scanned to identify any neutrophils present on the porous membrane.
Impact of Neutrophil Transmigration on Barrier Function
Having established a method to hinder neutrophil transmigration in the presence of an fMLP gradient, we then sought to test our hypothesis that transmigration would result in changes in EC monolayer permeability that could be detectable in the μSiM-MVM platform. As before, we grew HUVECs under physiological flow to enhance barrier properties. We found that our low frequency TEER measurements were prone to temporal fluctuations on minute time scales. These fluctuations would obscure the monolayer response, so we used low noise impedance spectroscopy at 1000 Hz to monitor changes over the time course of neutrophil transmigration. As seen in Fig. 6a, the introduction of neutrophils to the µSiM-MVM device caused a reduction in impedance values over the 20 min window we observed transmigration, and this decline was partially rectified with our β1 blocking antibody treatment. In a second set of experiments, we examined the diffusion of 10 kD FITC-dextran from the apical to basal chamber during fMLP-induced neutrophil transmigration. Again, we saw a clear effect of the blocking antibody on the rate of fluorescence transfer between the two chambers (Fig. 6b). Thus, by two different in situ measures of permeability—impedance and small molecule diffusion—we saw that limiting neutrophil transmigration via β1 blocking also limited increases in EC permeability.
Endothelial cell monolayer disruption due to neutrophil transmigration: (a) Differential decay in the net impedance during neutrophil migration with (red curve) and without (blue curve) the blocking of β1 integrins on the neutrophil surface. Impedance values for each individual scan was normalized to its initial value before averaging, to yield a common starting point for comparison. (For each test condition, n = 3) Error bars represent standard error of the mean. Final time points were statistically compared by t test, *p < 0.05. (b) The changes in the diffusion of FITC-dextran was monitored during the extravasation of untreated neutrophils (blue curve) or β1 integrin blocked neutrophils (red curve). The increase in the fluorescent signal was greater for the untreated neutrophils than for antibody-treated neutrophils, confirming the permeability-regulating effects of antibody treatment. (n = 5 for blue curve, and n = 2 for red curve) Error bars represent standard error of the mean. Final time points were statistically compared by t test, *p < 0.05.
In Situ Small Molecule Permeability
Interpretation of small molecule permeability across different platforms requires an analytical framework that captures the contribution of the model barrier without influence from the particular system geometry. Inspired by our ability to see subtle changes in small molecule diffusion through a monolayer in the µSiM-MVM, we undertook the task of developing such a framework for a platform similar to the one utilized here. As we have done in the analysis of transport through nanomembranes,7,26,49 we developed both analytical (supplement) and computational models with the understanding that the analytical model is simpler but only a computational model can capture the complex geometry of our nanomembrane chip formats. Both models featured a cell monolayer with a small molecule permeability (cm s−1) parameter atop a membrane with negligible resistance compared to the monolayer and system permeabilities.28,49
Focusing on the computational model because of its greater potential for accuracy, we ran a series of small molecule diffusion simulations with ‘sampling’ done along the bottom of the chip 50 µm away from the monolayer and membrane. Barrier permeabilities between 10−4 and 10−7 cm s−1 were run to create a ‘look-up’ database that could be used to interpret experiments (Supplemental Fig. S4).
We conducted conventional Transwell™ permeability measurements on confluent HPAECs using 40 kDa FITC-dextran to determine a confluent monolayer permeability of 1.06 × 10−5 ± 1.57 × 10−6 cm s−1 (mean ± standard error). In parallel, we conducted in situ permeability measurements on HPAECs in open-well Transwell™-mimetic µSiM devices37 (Fig. 7a). In both assays we added 40 kDa FITC-dextran as a bolus to the apical well. In the µSiM experiment, the evolution of the fluorescent tracer was recorded using a ×40 objective (Fig. 7b). Fluorescence intensity values across each image were recorded in a custom designed MATLAB script. The average fluorescence intensity at the chip surface and 50 μm from the membrane edge was then normalized to the initial intensity (Ix/Io; where Io is equal to the MATLAB-determined intensity when the working solution of FITC-dextran is added to the basal channel) and plotted over time (Fig. 7c). These data were matched to COMSOL solutions with the least root mean squared error between the numerical results and the data. The corresponding permeability values are shown in Fig. 7d. The lack of a statistically significant difference between the Transwell™ and µSiM permeability values provides confidence that a µSiM-based in situ assay can be used to provide conventional monolayer permeability values.
Quantitative analysis of in situ small molecule permeability. (a) Open-top μSiM devices were seeded with HPAECs and grown to confluency. (b) The diffusive properties of 40 kDa FITC-dextran through the monolayer were recorded in the basal compartment using an epifluorescence scope and ×40 objective. (c) Changes in normalized fluorescence intensity 50 μm from the membrane edge were compared to expected results based on conventional Transwell™ permeability values and a COMSOL simulation of the experimental configuration. Root mean square error (RMSE) represents the deviation of the experimental results from the expected. “Cell free” represents media only devices, while “Native” represents HPAEC seeded devices. (d) Additionally, conventional permeability values were determined and compared to values recorded in Transwell™ systems. A paired t-test showed a lack of statistical significance between the two experimental configurations (n = 3). Error bars represent standard error of mean.
Discussion
While we have long established the benefits of using silicon-based nanomembranes in place of track-etched membranes for cell culture applications,1,5,35,40 we have only recently applied these membranes to the creation of dual chamber devices (the µSiM platform) to mimic tissue barriers.34,37 Of particular interest to us is the creation of vascular mimetics in which ECs are grown on the nanomembrane substrate where they interface with the cellular and plasma components of blood on the apical (luminal) side of the device, and extravascular elements on the basal (abluminal) side. We have already shown that the optical clarity of nanomembranes enables high quality live cell imaging of leukocyte transmigration across endothelial barriers.37,43
In the current paper we provide some missing validation of the microvascular mimetic platform (µSiM-MVM) by showing that ECs align and establish tighter barriers in response to shear conditioning (Fig. 2), and we explore the integration of permeability assays (TEER; impedance; and fluorescent dextran transport) into the device platform. We also establish techniques for adding collagen layers to the apical and basal side of the membrane, where it provides for both advanced adhesion under flow and an extravascular mimetic for leukocyte migration in 3D. Finally, we apply the µSiM-MVM to an elusive question: Does leukocyte transmigration alone increase small molecule permeability? In both electrical and fluorescence-based assays, we find clear evidence that it does so (Fig. 6).
Transendothelial migration of leukocytes occurs via a complex series of binding events, which often depends on leukocyte type/subtype, inherent endothelial characteristics, the endothelium’s activation state, and which inflammatory stimuli are involved.39 As knowledge in the field has developed, the lingering debate of coupled vs. uncoupled transmigration and permeability has seemingly been resolved.16 As the transmigrating leukocyte passes between endothelial cells, the lateral border recycling compartment (LBRC) allows for migration without the need for endothelial cell retraction. As the LBRC actively excludes VE-cadherin, fluorescence imaging will show paracellular gaps if staining for this junctional molecule33—an observation we have reproduced with live imaging in the µSiM-MVM (Fig. 4b). The actual gaps between cells, however, may be much smaller. Additionally, several studies have demonstrated that VE-cadherin quickly returns and reseals the cell–cell junctions following diapedesis.52 Of note, there have been increases in permeability associated with leukocyte binding to the endothelium, as discovered by Gautum et al. and further permeability changes due to generation of reactive oxygen species (ROS), but none of these studies have found permeability associated with the actual transmigration event.19 Even more notable, on a large scale, changes in permeability in numerous inflammatory studies appear separated both spatially and temporally from leukocyte transmigration.2,47 These studies, however, operate at large scales, and may miss small movements of particles across the monolayer which could only be detected in an MPS device with in situ assays. Here we show that inhibiting fMLP-driven transmigration of neutrophils via β1 integrin blocking partially restored EC barrier function after the introduction of neutrophils, leading to our contention that there is are small, localized permeability changes associated with the transmigration event itself.
Whether small increases in EC permeability during leukocyte transmigration are physiologically important is not clear, but the point stands that bringing assays closer to cells in MPS devices can reveal subtle responses that macroscopic, including in vivo measurements, may miss. It is for this reason that the integration of assays into devices is the next frontier in the evolution of MPS concepts.27,36 While the integration of electrical3,8,20,55 and fluorescence-based permeability assays into MPS has been done by others before,54 our inclusion of these assays in the µSiM platform enables their combination with live cell microscopy of leukocyte dynamics at the endothelial interface. Furthermore, the decreased experimentation time required to obtain permeability values in the µSiM vs. the Transwell™ (10 min vs. 2 h) and minimal cell requirements may promote the adoption of this assay in future studies.
Abbreviations
- μSiM-MVM:
-
Microfluidic silicon membrane-enabled microvascular mimetic
- TEER:
-
Transendothelial electrical resistance
- HUVECs:
-
Human umbilical vein endothelial cells
- fMLP:
-
N-Formylmethionyl-leucyl-phenylalanine
- ECs:
-
Endothelial cells
- MPS:
-
Microphysiological system
- ITO:
-
Indium tin oxide
- PSA:
-
Pressure sensitive adhesive
- VE-cadherin:
-
Vascular endothelial cadherin
- HPAECs:
-
Human pulmonary aorta endothelial cells
- ECM:
-
Extracellular matrix
- ROS:
-
Reactive oxygen species
- SEM:
-
Scanning electron microscopy
References
Agrawal, A. A., B. J. Nehilla, K. V. Reisig, T. R. Gaborski, D. Z. Fang, C. C. Striemer, P. M. Fauchet, and J. L. McGrath. Porous nanocrystalline silicon membranes as highly permeable and molecularly thin substrates for cell culture. Biomaterials 31:5408–5417, 2010.
Baluk, P., P. Bolton, A. Hirata, G. Thurston, and D. M. McDonald. Endothelial gaps and adherent leukocytes in allergen-induced early- and late-phase plasma leakage in rat airways. Am. J. Pathol. 152:1463–1476, 1998.
Booth, R., and H. Kim. Characterization of a microfluidic in vitro model of the blood-brain barrier (mubbb). Lab. Chip 12:1784–1792, 2012.
Burton, V. J., L. M. Butler, H. M. McGettrick, P. C. Stone, H. C. Jeffery, C. O. Savage, G. E. Rainger, and G. B. Nash. Delay of migrating leukocytes by the basement membrane deposited by endothelial cells in long-term culture. Exp. Cell Res. 317:276–292, 2011.
Carter, R. N., S. M. Casillo, A. R. Mazzocchi, J. S. DesOrmeaux, J. A. Roussie, and T. R. Gaborski. Ultrathin transparent membranes for cellular barrier and co-culture models. Biofabrication. 9:015019, 2017.
Casillo, S. M., A. P. Peredo, S. J. Perry, H. H. Chung, and T. R. Gaborski. Membrane pore spacing can modulate endothelial cell-substrate and cell-cell interactions. ACS Biomater. Sci. Eng. 3:243–248, 2017.
Chung, H. H., C. K. Chan, T. S. Khire, G. A. Marsh, A. Clark, Jr, R. E. Waugh, and J. L. McGrath. Highly permeable silicon membranes for shear free chemotaxis and rapid cell labeling. Lab. Chip 14:2456–2468, 2014.
Chung, H. H., M. Mireles, B. J. Kwarta, and T. R. Gaborski. Use of porous membranes in tissue barrier and co-culture models. Lab. Chip 18:1671–1689, 2018.
Cohnheim, J. Entzündung und eiterung. Arch. Pathol. Anat. 40:1–79, 1867.
Colbert, J. F., and E. P. Schmidt. Endothelial and microcirculatory function and dysfunction in sepsis. Clin. Chest Med. 37:263–275, 2016.
Cucullo, L., M. Hossain, W. Tierney, and D. Janigro. A new dynamic in vitro modular capillaries-venules modular system: cerebrovascular physiology in a box. BMC Neurosci. 14:18, 2013.
de Mesy Bentley, K. L., R. Trombetta, K. Nishitani, S. N. Bello-Irizarry, M. Ninomiya, L. Zhang, H. L. Chung, J. L. McGrath, J. L. Daiss, H. A. Awad, S. L. Kates, and E. M. Schwarz. Evidence of staphylococcus aureus deformation, proliferation, and migration in canaliculi of live cortical bone in murine models of osteomyelitis. J. Bone Miner. Res. 32:985–990, 2017.
Del Maschio, A., A. Zanetti, M. Corada, Y. Rival, L. Ruco, M. G. Lampugnani, and E. Dejana. Polymorphonuclear leukocyte adhesion triggers the disorganization of endothelial cell-to-cell adherens junctions. J. Cell Biol. 135:497–510, 1996.
DesOrmeaux, J. P., J. D. Winans, S. E. Wayson, T. R. Gaborski, T. S. Khire, C. C. Striemer, and J. L. McGrath. Nanoporous silicon nitride membranes fabricated from porous nanocrystalline silicon templates. Nanoscale. 6:10798–10805, 2014.
Dewey, Jr, C. F., S. R. Bussolari, M. A. Gimbrone, Jr, and P. F. Davies. The dynamic response of vascular endothelial cells to fluid shear stress. J. Biomech. Eng. 103:177–185, 1981.
Filippi, M.-D. Neutrophil transendothelial migration: updates and new perspectives. Blood 133:2149, 2019.
Gautam, N., P. Hedqvist, and L. Lindbom. Kinetics of leukocyte-induced changes in endothelial barrier function. Br. J. Pharmacol. 125:1109–1114, 1998.
Goitre, L., P. V. DiStefano, A. Moglia, N. Nobiletti, E. Baldini, L. Trabalzini, J. Keubel, E. Trapani, V. V. Shuvaev, V. R. Muzykantov, I. H. Sarelius, S. F. Retta, and A. J. Glading. Up-regulation of nadph oxidase-mediated redox signaling contributes to the loss of barrier function in krit1 deficient endothelium. Sci Rep. 7:8296, 2017.
He, P. Leucocyte/endothelium interactions and microvessel permeability: coupled or uncoupled? Cardiovasc. Res. 87:281–290, 2010.
Henry, O. Y. F., R. Villenave, M. J. Cronce, W. D. Leineweber, M. A. Benz, and D. E. Ingber. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (teer) measurements of human epithelial barrier function. Lab. Chip 17:2264–2271, 2017.
Huh, D., D. C. Leslie, B. D. Matthews, J. P. Fraser, S. Jurek, G. A. Hamilton, K. S. Thorneloe, M. A. McAlexander, and D. E. Ingber. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Trans. Med. 4:159ra147, 2012.
Ishimatsu, R., J. Kim, P. Jing, C. C. Striemer, D. Z. Fang, P. M. Fauchet, J. L. McGrath, and S. Amemiya. Ion-selective permeability of an ultrathin nanoporous silicon membrane as probed by scanning electrochemical microscopy using micropipet-supported ities tips. Anal. Chem. 82:7127–7134, 2010.
Jarcho, S. Cohnheim on inflammation 2. Am. J. Cardiol. 29:246–247, 1972.
Jarcho, S. Cohnheim on inflammation. I. Am. J. Cardiol. 29:247–249, 1972.
Johnson, D. G., T. S. Khire, Y. L. Lyubarskaya, K. J. Smith, J. P. Desormeaux, J. G. Taylor, T. R. Gaborski, A. A. Shestopalov, C. C. Striemer, and J. L. McGrath. Ultrathin silicon membranes for wearable dialysis. Adv. Chronic Kidney Dis. 20:508–515, 2013.
Khire, T. S., B. J. Nehilla, J. Getpreecharsawas, M. E. Gracheva, R. E. Waugh, and J. L. McGrath. Finite element modeling to analyze teer values across silicon nanomembranes. Biomed. Microdev. 20:11, 2018.
Kieninger, J., A. Weltin, H. Flamm, and G. A. Urban. Microsensor systems for cell metabolism—from 2d culture to organ-on-chip. Lab. Chip 18:1274–1291, 2018.
Kim, E., H. Xiong, C. C. Striemer, D. Z. Fang, P. M. Fauchet, J. L. McGrath, and S. Amemiya. A structure–permeability relationship of ultrathin nanoporous silicon membrane: a comparison with the nuclear envelope. J. Am. Chem. Soc. 130:4230–4231, 2008.
Leick, M., V. Azcutia, G. Newton, and F. W. Luscinskas. Leukocyte recruitment in inflammation: basic concepts and new mechanistic insights based on new models and microscopic imaging technologies. Cell Tissue Res. 355:647–656, 2014.
Lerman, Y. V., and M. Kim. Neutrophil migration under normal and sepsis conditions. Cardiovasc. Hematol. Disord. Drug Targets 15:19–28, 2015.
Ley, K., C. Laudanna, M. I. Cybulsky, and S. Nourshargh. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7:678–689, 2007.
Luo, D., H. M. McGettrick, P. C. Stone, G. E. Rainger, and G. B. Nash. The roles of integrins in function of human neutrophils after their migration through endothelium into interstitial matrix. PLoS ONE 10:e0118593, 2015.
Mamdouh, Z., A. Mikhailov, and W. A. Muller. Transcellular migration of leukocytes is mediated by the endothelial lateral border recycling compartment. J. Exp. Med. 206:2795, 2009.
Masters, E. A., A. T. Salminen, S. Begolo, E. N. Luke, S. C. Barrett, C. T. Overby, A. L. Gill, K. L. de Mesy Bentley, H. A. Awad, S. R. Gill, E. M. Schwarz, and J. L. McGrath. An in vitro platform for elucidating the molecular genetics of s. Aureus invasion of the osteocyte lacuno-canalicular network during chronic osteomyelitis. Nanomedicine 2019. https://doi.org/10.1016/j.nano.2019.102039.
Mazzocchi, A. R., A. J. Man, J.-P. S. DesOrmeaux, and T. R. Gaborski. Porous membranes promote endothelial differentiation of adipose-derived stem cells and perivascular interactions. Cell. Mol. Bioeng. 7:369–378, 2014.
Modena, M. M., K. Chawla, P. M. Misun, and A. Hierlemann. Smart cell culture systems: integration of sensors and actuators into microphysiological systems. ACS Chem. Biol. 13:1767–1784, 2018.
Mossu, A., M. Rosito, T. Khire, H. L. Chung, H. Nishihara, I. Gruber, E. Luke, L. Dehouck, F. Sallusto, F. Gosselet, J. McGrath, and B. Engelhardt. A silicon nanomembrane platform for the visualization of immune cell trafficking acros the human blood-brain barier under flow. J. Cereb. Blood Flow Metab. 39:395–410, 2018.
Mossu, A., M. Rosito, T. Khire, H. Li-Chung, H. Nishihara, I. Gruber, E. Luke, L. Dehouck, F. Sallusto, F. Gosselet, J. L. McGrath, and B. Engelhardt. A silicon nanomembrane platform for the visualization of immune cell trafficking across the human blood-brain barrier under flow. J. Cereb. Blood Flow Metab. 39:395–410, 2019.
Muller, W. A. Transendothelial migration: unifying principles from the endothelial perspective. Immunol. Rev. 273:61–75, 2016.
Nehilla, B. J., N. Nataraj, T. R. Gaborski, and J. L. McGrath. Endothelial vacuolization induced by highly permeable silicon membranes. Acta Biomater. 10:4670–4677, 2014.
Papaioannou, T. G., E. N. Karatzis, M. Vavuranakis, J. P. Lekakis, and C. Stefanadis. Assessment of vascular wall shear stress and implications for atherosclerotic disease. Int. J. Cardiol. 113:12–18, 2006.
Pardridge, W. M., D. Triguero, J. Yang, and P. A. Cancilla. Comparison of in vitro and in vivo models of drug transcytosis through the blood-brain barrier. J. Pharmacol. Exp. Ther. 253:884–891, 1990.
Salminen, A. T., J. Zhang, G. R. Madejski, T. S. Khire, R. E. Waugh, J. L. McGrath, and T. R. Gaborski. Ultrathin dual-scale nano- and microporous membranes for vascular transmigration models. Small 15:e1804111, 2019.
Santaguida, S., D. Janigro, M. Hossain, E. Oby, E. Rapp, and L. Cucullo. Side by side comparison between dynamic versus static models of blood-brain barrier in vitro: a permeability study. Brain Res. 1109:1–13, 2006.
Sarangi, P. P., Y.-M. Hyun, Y. V. Lerman, A. P. Pietropaoli, and M. Kim. Role of β1 integrin in tissue homing of neutrophils during sepsis. Shock (Augusta, GA). 38:281–287, 2012.
Sarelius, I. H., and A. J. Glading. Control of vascular permeability by adhesion molecules. Tissue Barriers. 3:e985954, 2015.
Schnoor, M., F. P. L. Lai, A. Zarbock, R. Kläver, C. Polaschegg, D. Schulte, H. A. Weich, J. M. Oelkers, K. Rottner, and D. Vestweber. Cortactin deficiency is associated with reduced neutrophil recruitment but increased vascular permeability in vivo. J. Exp. Med. 208:1721, 2011.
Shaw, S. K., P. S. Bamba, B. N. Perkins, and F. W. Luscinskas. Real-time imaging of vascular endothelial-cadherin during leukocyte transmigration across endothelium. J Immunol. 167:2323–2330, 2001.
Snyder, J. L., A. Clark, D. Z. Fang, T. R. Gaborski, C. C. Striemer, P. M. Fauchet, and J. L. McGrath. An experimental and theoretical analysis of molecular separations by diffusion through ultrathin nanoporous membranes. J. Membr. Sci. 369:119–129, 2011.
Snyder, J. L., J. Getpreecharsawas, D. Z. Fang, T. R. Gaborski, C. C. Striemer, P. M. Fauchet, D. A. Borkholder, and J. L. McGrath. High-performance, low-voltage electroosmotic pumps with molecularly thin silicon nanomembranes. Proc. Natl. Acad. Sci. U.S.A. 110:18425–18430, 2013.
Striemer, C. C., T. R. Gaborski, J. L. McGrath, and P. M. Fauchet. Charge- and size-based separation of macromolecules using ultrathin silicon membranes. Nature 445:749–753, 2007.
Su, W.-H., H.-I. Chen, and C. J. Jen. Differential movements of ve-cadherin and pecam-1 during transmigration of polymorphonuclear leukocytes through human umbilical vein endothelium. Blood 100:3597, 2002.
Sumagin, R., J. M. Kuebel, and I. H. Sarelius. Leukocyte rolling and adhesion both contribute to regulation of microvascular permeability to albumin via ligation of Icam-1. Am. J. Physiol. Cell Physiol. 301:C804–C813, 2011.
Terrell-Hall, T. B., A. G. Ammer, J. I. Griffith, and P. R. Lockman. Permeability across a novel microfluidic blood-tumor barrier model. Fluids Barriers CNS. 14:3, 2017.
Walter, F. R., S. Valkai, A. Kincses, A. Petnehazi, T. Czeller, S. Veszelka, P. Ormos, M. A. Deli, and A. Der. A versatile lab-on-a-chip tool for modeling biological barriers. Sens. Actuators B Chem. 222:1209–1219, 2016.
Werr, J., E. E. Eriksson, P. Hedqvist, and L. Lindbom. Engagement of β2 integrins induces surface expression of β1 integrin receptors in human neutrophils. J. Leukoc. Biol. 68:553–560, 2000.
Winans, J., K. Smith, T. Gaborski, J. Roussie, and J. McGrath. Membrane capacity and fouling mechanisms for ultrathin nanomembranes in dead-end filtration. J Memb Sci. 499:282–289, 2016.
Acknowledgments
The authors gratefully acknowledge E. Luke and R. Breidenstein for their efforts in device fabrication. The research reported here was supported by the US Public Health Service through the NIH under Award Numbers R01 HL125265 (R.E.W. and J.L.M.), R35 GM119623 (T.R.G.), and R01 141131 (A.J.G.). A.T.S. is supported under NIH Award Number T32 HL066988.
Conflict of interest
Authors Tejas S. Khire, Alec T. Salminen, Harsha Swamy, Kilean S. Lucas, Molly C. McCloskey, Raquel E. Ajalik, Henry H. Chung, Richard E. Waugh, and Angela J. Glading declare that they have no conflict of interest. Authors Thomas R. Gaborski and James L. McGrath are co-founders of SiMPore, an early-stage company commercializing ultrathin silicon-based technologies.
Research Involving Human and Animal Studies
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients for being included in the study. No animal studies were performed for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Michael R. King oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khire, T.S., Salminen, A.T., Swamy, H. et al. Microvascular Mimetics for the Study of Leukocyte–Endothelial Interactions. Cel. Mol. Bioeng. 13, 125–139 (2020). https://doi.org/10.1007/s12195-020-00611-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-020-00611-6