Abstract
Introduction
Dysregulated neutrophil function plays a significant role in the pathology of infections, cancer, cardiovascular diseases, and autoimmune disorders. Neutrophil activity is influenced by various cell populations, including macrophages, which are crucial regulators. However, the exact role of human macrophages in controlling neutrophil function remains unclear due to a scarcity of studies utilizing human cells in physiologically relevant models.
Methods
We adapted our “Infection-on-a-Chip” microfluidic device to incorporate macrophages within the collagen extracellular matrix, allowing for the study of interactions between human neutrophils and macrophages in a context that mimics in vivo conditions. The integration of THP-1 macrophages was optimized and their effect on the endothelial lumen was characterized, focusing on permeability and structural integrity. The device was then employed to examine the influence of macrophages on neutrophil response to infection with the bacterial pathogen Pseudomonas aeruginosa.
Results
Integration of THP-1 macrophages into the microfluidic device was successfully optimized, showing no increase in endothelial permeability or structural damage. The presence of macrophages was found to significantly reduce neutrophil transendothelial migration in response to Pseudomonas aeruginosa infection.
Conclusions
Our findings highlight the regulatory role of macrophages in modulating neutrophil responses, suggesting potential therapeutic targets to control neutrophil function in various diseases. The modified microfluidic platform offers a valuable tool for mechanistic studies into macrophage-neutrophil interactions in disease contexts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Significance Statement
-
The study of human macrophage-neutrophil interactions has been minimally explored in vitro and remains unclear due to limitations in existing experimental platforms.
-
We developed an Infection-on-a-Chip device through which we have shown that the presence of a human macrophage cell line reduced primary human neutrophil transendothelial migration in response to Pseudomonas aeruginosa infection.
-
Our research sheds light on the interaction between human neutrophils and macrophages, providing a platform to explore immune cell interactions in physiologically relevant conditions for potential clinical applications. Future studies should investigate how macrophages influence neutrophils, whether through soluble factors or physical interactions.
Introduction
The immune response is a complex yet coordinated process that involves the interaction of many cell populations. While current research has often focused on adaptive immune cells and their therapeutic potential [1], there is growing recognition for the importance of the innate immune system in human health and disease [2, 3]. Specifically, the importance of macrophage-neutrophil interactions in regulating the initiation and resolution of the immune response has been a topic of significant interest. Macrophages and neutrophils are innate immune cells whose primary role is to combat pathogens, but they also play a crucial role as first responders to sterile injury and environmental toxin exposures [4,5,6]. Innate immune cell dysfunction has been identified as a contributing factor to the pathology of diverse human diseases, including cancer, cardiovascular disease, autoimmune disorders, and fibrosis [7]. Yet, how neutrophils and macrophages regulate each other's functions in healthy tissue and how these interactions are altered in disease remain unclear.
Neutrophils, constituting 50–70% of all leukocytes in humans, are short-lived effector cells of the innate immune system. Upon pathogen exposure, they rapidly mobilize to sites of infection, where their antimicrobial capabilities are essential for pathogen elimination. However, in the context of tissue injury, the potent actions of neutrophils, including NETosis, reactive oxygen species production, cytokine release, and degranulation, can paradoxically lead to tissue damage [8]. The detrimental effect of neutrophil overactivation is evident in various medical conditions such as rheumatoid arthritis, acute respiratory distress syndrome, and cancer [9]. A potential strategy for treating these diseases lies in controlling neutrophil recruitment and function; however, success in this endeavor requires a comprehensive understanding of how neutrophils interpret and respond to the diverse signals and interactions within their environment.
Macrophages play a crucial role in both combating infections and promoting the resolution of inflammation. Upon infection, they initiate the secretion of pro-inflammatory mediators like tumor necrosis factor-alpha (TNF-α), interleukin-1 beta (IL-1β), along with nitric oxide, which activates other cells of the innate immune system and bolsters their antimicrobial defenses to effectively eliminate invading pathogens [10].
Neutrophils and macrophages express distinct membrane proteins, including G-protein-coupled receptors, chemokine receptors, and adhesion receptors that enable cell–cell communication and influence cell function [11]. Traditionally, macrophages were thought to interact with neutrophils by releasing cytokines to recruit them to sites of infection, then by clearing apoptotic neutrophils following immune resolution. However, recent studies employing advanced microscopy techniques have potentially discovered a more complex role for neutrophil-macrophage interactions in the innate immune response. Initial investigations in zebrafish found that macrophages directly facilitate neutrophil reverse migration through physical touch [12]. However, contrasting observations in mice indicate soluble signals are instead responsible for this phenomenon [13, 14]. These contradictory findings complicate our comprehension of these dynamic interactions, prompting contemplation on how macrophages and neutrophils interact in humans. While these studies done in mice and zebrafish are informative, translating these findings to the human context is crucial for a comprehensive understanding and successful application in clinical settings. Therefore, it is imperative to unravel the complexities of macrophage-neutrophil interactions in humans to clarify how these interactions regulate the immune response.
A major challenge in understanding cell–cell interactions between primary human cells is the limitations of existing experimental platforms. While in vitro systems enable the investigation of primary human neutrophil responses to individual signals or cellular interactions [15], they fall short in replicating the intricate milieu of signals, multicellular interactions, and the three-dimensional architectural complexity inherent in the in vivo setting [16]. Therefore, human macrophage-neutrophil interactions have been minimally explored in vitro beyond exposure to conditioned media [17].
To address these limitations, our research group has pioneered the use of an innovative Infection-on-a-Chip device, previously employed to study neutrophil interactions with endothelial cells and pathogens including gram-negative bacteria, gram-positive bacteria, and fungi [18,19,20,21,22]. The Infection-on-a-Chip device replicates essential elements of a three-dimensional, in vivo inflammatory environment, including an endothelial blood vessel, primary human immune cells, extracellular matrix proteins, and a source of live pathogens. In this study, we modify this platform to create a unique and robust in vitro model to unravel the dynamics of neutrophil-macrophage crosstalk. Using the modified device, we reveal a reduction in primary human neutrophil transendothelial migration (TEM) in the presence of macrophages during an inflammatory response. These findings enhance our understanding of macrophage-mediated regulation of the neutrophil response and present a novel experimental platform for investigating these critical cell–cell interactions.
Results
THP-1 Cells Embedded in Collagen Gels Have Higher Viability When Differentiated In Situ
To determine how macrophages influence neutrophil transendothelial migration, we needed to integrate macrophages into our Infection-on-a-Chip device and confirm their differentiation and viability. We first determined if THP-1 macrophages could maintain viability when embedded in collagen gels following differentiation with phorbol 12-myristate 13-acetate (PMA). To test this, THP-1 monocytes were differentiated into macrophages in a well plate for 2 days. They were then lifted and suspended in a collagen solution which was polymerized in a 48-well plate (Day 0). Cells were maintained in the well plate with fresh media for 3 days to determine cell viability following the embedding process (Fig. 1A).
THP-1 cells embedded in collagen gels have higher viability when differentiated in situ. A Schematic of the THP-1 monocyte differentiation process with PMA in well plates, followed by the embedding of macrophages in 1.5 mg/mL collagen on Day 3. B Representative images of THP-1 cells seeded in 1.5 mg/mL collagen on Day 3. Cells stained with calcein AM (live) and propidium iodide (dead). Scale bar, 250 µm. C Percent THP-1 viability after THP-1 cells were embedded in collagen quantified using live/dead fluorescent staining on Day 0 to Day 3. Data was quantified from 3 wells per condition across 3 independent experiments. D Schematic of the THP-1 differentiation process with monocytes added to collagen on Day 0 and directly differentiated in situ. E Representative images of THP-1 differentiated in 1.5 mg/mL collagen on Day 3. THP-1 stained with calcein AM (live) and propidium iodide (dead). Scale bar, 250 µm. F Percent THP-1 viability following THP-1 differentiation in situ quantified using live/dead fluorescent staining. Data was quantified from 3 wells per condition across 6 independent experiments. C, F Each bar represents the mean plus standard error of the mean (SEM). Asterisks represent the significance between conditions at each point. *P, 0.05; ns not significant
To quantify cell viability, THP-1 cells were imaged in the presence of the live/dead stains calcein AM (live) and propidium iodide (dead) on Days 0–3 (Fig. 1B). The initial viability on Day 0 was 88% but average viability decreased to 43% by Day 4, indicating that both the embedding process and subsequent incubation period decreased THP-1 cell survival (Fig. 1C).
While our initial viability results indicated macrophages could survive in collagen gels, to investigate the effect of macrophages on neutrophil transendothelial migration without confounding cell death, we required higher THP-1 viability. Therefore, we needed an improved differentiation protocol. Based on our initial viability data, we hypothesized the cell lifting and embedding process caused unwanted cell death and that directly differentiating the THP-1s in the collagen would lead to improved viability. To test our hypothesis, we first seeded THP-1 monocytes in the collagen gels and then differentiated them into macrophages for 2 days (Fig. 1D). We again used a live/dead stain to quantify cell viability over the 2-days differentiation and for 1 day following differentiation. THP-1 viability remained very high (~89%) throughout both the 2-days differentiation period and the 24-h incubation period (Fig. 1E, F). This method of differentiating monocytes into macrophages within collagen gels was used for the duration of the project due to the high level of cell viability achieved.
THP-1 Macrophages have High Levels of Differentiation and Viability in Collagen Gels
We next needed to quantify cell viability and differentiation in collagen gels from Day 0 to Day 4 to match the timeline of our future experiments using the Infection-on-a-Chip device. To accomplish this, we first seeded and differentiated THP-1 cells in collagen gels using PMA. We then isolated the cells on Day 2, immediately following differentiation, and Day 4, following the 2-days incubation period. We used flow cytometry to simultaneously measure THP-1 viability and differentiation.
Viability assessment was conducted by labeling THP-1 cells with ZombieViolet live/dead stain following removal from the collagen gels. THP-1 cells exhibited high viability levels, and there was no significant difference in viability between Day 2 and Day 4. There was also no significant difference in viability between PMA-exposed cells in collagen and THP-1 cells plated in well plates without PMA (Fig. 2A).
THP-1 macrophages have high levels of differentiation and viability in collagen gels of varying density. THP-1 monocytes were differentiated with PMA in 1.5 mg/mL and 4 mg/mL collagen for 2 days (Days 1–2), then treated with cell media for an additional 2 days (Days 3–4). In each graph, differentiated cells are compared to control cells plated in well plates without PMA (“NO PMA”). A Percent THP-1 viability for control cells and PMA-treated cells embedded in 1.5 mg/mL collagen on Day 2 and Day 4. B Percent THP-1 monocyte (CD11blo) differentiation into macrophages (CD11bhi) for control cells and PMA-treated cells plated in 1.5 mg/mL collagen on Day 2 and Day 4. C Percent THP-1 cell viability on Day 4 for control cells and PMA-treated cells plated in 1.5 and 4 mg/mL collagen. D Percent THP-1 monocyte (CD11blo) differentiation into macrophages (CD11bhi) on Day 4 for control cells and PMA-treated cells plated in 1.5 mg/mL and 4 mg/mL collagen. Each bar represents the estimated marginal mean plus standard error of the mean (SEM) for each condition. Data was quantified from 6 lumens across 3 independent experiments. Asterisks represent significance between conditions at each point *P, 0.05; **P, 0.01; ***P, 0.001; ns not significant
CD11b was used as an indicator for THP-1 monocyte-to-macrophage differentiation as previously described [23], where an increase in CD11b expression indicates macrophage differentiation.
CD11b expression was assessed using flow cytometry with a fluorescent anti-CD11b antibody and compared to a control population seeded in well plates. Following 48 h of PMA treatment in collagen gels, approximately 89% of THP-1 cells differentiated into macrophages with a non-significant increase to 98% by Day 4 (Fig. 2B). Moreover, there was a significant increase in CD11b+ cells in these conditions compared to THP-1 cells seeded in well plates with no PMA treatment. This suggests that PMA promotes the differentiation of THP-1 cells in collagen.
THP-1 Viability and Differentiation are not Altered in Higher-Density Collagen
We carried out our initial viability and differentiation experiments in 1.5 mg/mL collagen; however, our Infection-on-a-Chip devices use 4 mg/mL collagen gels to mimic the ECM of an in vivo microenvironment. To validate that THP-1 cells maintain high viability and undergo successful differentiation in 4 mg/mL collagen gels, we compared viability and CD11b expression between THP-1 cells differentiated in 1.5 mg/mL and 4 mg/mL collagen gels using flow cytometry. Notably, there were no significant differences in viability (1.5 mg/mL = 95%, 4 mg/mL = 92%) (Fig. 2C) or differentiation levels (1.5 mg/mL = 98%, 4 mg/mL = 95%) on Day 4 between cells seeded in 1.5 mg/mL or 4 mg/mL collagen (Fig. 2D). Furthermore, there was no significant difference between THP-1 viability in collagen and the control cells in well plates without PMA (Fig. 2C). Additionally, the percentage of differentiated cells obtained with PMA in both collagen conditions was statistically higher than the control cells without PMA (Fig. 2D). Overall, increasing the collagen density from 1.5 to 4 mg/mL collagen does not impact THP-1 viability and differentiation, indicating THP-1 cells will remain alive and differentiated in the Infection-on-a-Chip devices for the full length of the experiment.
THP-1s Differentiate and Remain Viable in the Infection-on-a-Chip Device
While we saw high THP-1 viability and differentiation in collagen gels polymerized in a well plate, our Infection-on-a-Chip device contains human umbilical vein endothelial cells (HUVECs), has a different geometry, and uses different volumes than a well plate. Therefore, we next wanted to confirm that THP-1 cells would differentiate and remain viable in our device. To assess differentiation and viability, THP-1 cells were differentiated and then co-cultured with HUVECs in the device. THP-1 monocytes in 4 mg/mL collagen gels were first loaded into the device and differentiated with PMA for 48 h. On Day 2, following differentiation, endothelial cells were seeded in the lumen and HUVEC media was added to the lumen. The lumen monolayer formed over an additional 48 h until Day 4 (Fig. 3A). Fluorescent live/dead stains, calcein AM, and propidium iodide were added to devices on Day 2 and Day 4 to quantify viability. As calcein AM stains all living cells, we only imaged outside the lumen monolayer to ensure all the live cells counted were macrophages, not endothelial cells (Fig. 3B). There was no significant difference in viability between Day 2 (75%) and Day 4 (78 %) (Fig. 3C). To determine if THP-1 monocytes were able to differentiate into macrophages in the Infection-on-a-Chip devices, we analyzed differentiation levels using CD68, which has increased expression in macrophages. We found there was significantly more THP-1 differentiation in the devices treated with PMA for 2 days (63%) than those that were not treated with PMA (44%), as indicated by a higher percentage of CD68-positive cells (Fig. 3D, E). Notably, 2 days of exposure to PMA, as opposed to 4 days, resulted in a higher percentage of CD68-positive cells (Fig. S1). These findings substantiate the ability of THP-1 cells to differentiate and maintain viability within our Infection-on-a-Chip device.
THP-1 monocytes differentiate and remain viable in the Infection-on-a-Chip device. THP-1 cells were seeded in the device in 4 mg/mL collagen and differentiated for 48 h (Days 1–2). Endothelial cells were then seeded and viability and differentiation were evaluated following co-culture (Days 3–4). A Schematic of the Infection-on-a-Chip device model with the endothelial lumen in the middle of the device with the side ports on each side surrounded by the embedded macrophages in 4 mg/mL collagen. The cross-sectional area of the HUVEC lumen is shown by the gray box. B Representative fluorescence images on Day 2 and Day 4 for live (calcein AM, green) and dead (propidium iodide, red) cells in the collagen in the device. The white lines represent the edges of the lumen. Scale bars = 150 µm. C Percent viability of THP-1 cells embedded in 4 mg/mL collagen in the device on Day 2 and Day 4. Data was quantified from 6 devices per condition across 8 independent experiments. Each bar represents the mean plus standard error of the mean (SEM) for each condition. ns not significant. D Representative fluorescence images on Day 2 of all THP-1 (Phalloidin, red) and differentiated THP-1 (anti-CD68, blue) cells in the collagen in the device. (Top) THP-1 cells that were exposed to cell media for 48 h. (Bottom) THP-1 cells that were exposed to PMA for 48 h. Scale bar = 150 µm. E Percent of differentiated THP-1 cells on Day 2 in the device with and without PMA exposure through CD68 positive cell quantification. F Representative fluorescence images of differentiated THP-1 cells within the collagen matrix of the device after 48 h of PMA exposure. THP-1 cells exposed to PMA were stained with Phalloidin (red, top left), anti-CD68 (blue, top right), and anti-CD163 (green, bottom left). All stains are overlaid in the bottom right image. Scale bar = 150 µm. G Percent of the CD68+ THP-1 cell population that are CD163+ (M2-like) within the in the device after 2 and 4 days of PMA exposure. Data was quantified from 6 devices per condition across 3 independent experiments. Each bar represents the mean plus standard error of the mean (SEM) for each condition. Asterisks represent significance between conditions at each point *P, 0.05; **P, 0.01; ***P, 0.001; ns not significant.
To discern whether THP-1 macrophages in collagen exhibit an M1 or M2 phenotype, we assessed the expression of CD163, which is indicative of an M2 phenotype [24]. We determined the percentage of CD68 positive cells that were also CD163 positive at Day 2 (77%) and Day 4 (57%), and found the majority of THP-1 macrophages displayed an M2 phenotype 2 days of exposure to PMA when embedded in collagen (Fig. 3F, G).
HUVECs Generate a Patent Lumen and Retain Proper Barrier Function in the Presence of Macrophages
The formation of a patent endothelial lumen vessel with significant barrier function is critical for evaluating neutrophil transendothelial migration in the Infection-on-a-Chip device. To determine if the presence of macrophages altered the structural integrity of the endothelial lumen in our device, we conducted a series of analyses to visualize lumen formation and permeability.
We visualized lumen integrity by fabricating lumens in the absence (Fig. 4A–D) or presence (Fig. 4E–H) of THP-1 macrophages then staining for actin (phalloidin), cell nuclei (Hoescht), and tight junction formation (anti-VE-cadherin). The phalloidin stain revealed no disruptions or gaps in endothelial cell α-actin filaments in lumens with macrophages (4E) compared to those without (Fig. 4A). The Hoechst stain (nuclei) showed no noticeable differences in endothelial cell distribution in the absence (Fig. 4B) or presence (Fig. 4F) of macrophages. Finally, the anti-VE-cadherin stain showed no changes in tight junction formation between endothelial cells, without (Fig. 4C) or with macrophages (Fig. 4G). Overall, we found no observable differences when comparing endothelial lumens without (Fig. 4D) and with (Fig. 4H) macrophages in the collagen further than an increase in the endothelial monolayer sprouting toward the collagen matrix, indicating that the addition of macrophages did not alter the integrity of the lumen structure.
HUVECs generate a patent lumen and retain proper barrier function in the presence of macrophages. Representative images of lumens formed within the Infection-on-a-Chip device, both without (A–D) and with THP-1 macrophages (E–H). A–H Lumens were stained using Phalloidin (A, E) to highlight α-actin filaments, Hoechst (B, F) for nuclei visualization, and anti-VE-cadherin (C, G) to reveal tight junctions. Merged images (D, H) present a comprehensive view of HUVEC cells stained with Hoechst, Anti-VE-Cadherin, and Phalloidin. (Left) Top view of the HUVEC lumen within the device. (Right) Side view of the HUVEC lumen within the device. Scale bar, 100 mm. I FITC-dextran diffusion from lumens without THP-1 (left column) and with THP-1 macrophages embedded in the ECM (right column) and exposed to media (top row) or Pseudomonas aeruginosa (bottom row) at 0 (red), 5 (blue), 10 (yellow), and 15 (green) minutes. Pixel intensity across the images was measured and normalized to the intensity of the lumen center at t = 0 min, providing insights into the dynamic behavior of the endothelial barrier. The x-axis depicts the distance across a region centered around each lumen, spanning three-lumen widths in diameter (in arbitrary units).
Previous research has demonstrated that neutrophil interaction with endothelial cells through ICAM-1 binding to the ß2 integrin is critical for efficient extravasation [18]. Additionally, endothelial cells at high passage numbers (P12) have been reported to exhibit reduced ICAM-1 expression [25]. Therefore, to confirm ICAM-1 was expressed in response to Pseudomonas aeruginosa, we measured ICAM-1 levels in HUVECs at passages 4–7, as used in our experiments. Significant ICAM-1 expression was evident in all passage numbers (4–7) and no significant change in ICAM-1 expression across passages (Fig. S2).
We next wanted to determine whether the addition of macrophages changes the barrier function of our endothelial lumens in the presence of Pseudomonas aeruginosa. To visualize lumen permeability, we imaged the diffusion of FITC-dextran out of lumens in the absence or presence of THP-1 macrophages and the absence or presence of Pseudomonas aeruginosa over 15 min (Fig. 4I). We found that the addition of Pseudomonas aeruginosa increases endothelial lumen permeability, which matches observations in previous studies [18, 26] and further supports the applicability to in vivo results. Notably, we found the addition of macrophages to the device did not result in a change in the permeability of the endothelial barrier. Overall, these data suggest that the addition of THP-1 macrophages to the collagen does not alter the structure or integrity of the lumen vessel.
THP-1 Macrophages Decrease Neutrophil TEM
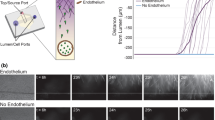
Given our data showing high macrophage viability and differentiation in the Infection-on-a-Chip device along with our observations that the endothelial lumen structure and permeability were not altered by the presence of macrophages, we were prepared to investigate how the presence of THP-1 macrophages influences neutrophil transendothelial migration during an infectious response. To visualize and quantify neutrophil transendothelial migration, endothelial lumens were seeded with primary human neutrophils in the absence (Fig. 5A), or presence (Fig. 5B) of THP-1 macrophages in the collagen. Pseudomonas aeruginosa was added to the top port of the device to simulate an infection and initiate neutrophil recruitment.
THP-1 macrophages reduce neutrophil transendothelial migration during an infectious response. A, B Representative images of neutrophils extravasating the endothelial monolayer at 2-h intervals towards Pseudomonas aeruginosa without (A) and with (B) THP-1 macrophages in the device. 20% of neutrophils stained with calcein AM. The gradient direction is shown in greyscale on the left. The white line indicates the edge of the endothelial lumen. Scale bar, 200 µm. C Quantification of neutrophil transendothelial migration at 2-h intervals without (red) and with (blue) THP-1 macrophages. Each bar represents the mean plus standard error of the mean (SEM). Data was quantified from 6 lumens per condition across 5 independent experiments. Asterisks represent significance between conditions at each point *P, 0.05; **P, 0.01; ***P, 0.001; ns not significant
Interestingly, we found that relatively fewer neutrophils extravasated into the macrophage-containing collagen compared to the cell-free collagen control (Fig. 5C). This difference reached significance after 6 h and remained significant through 10 h post-infection. The difference in neutrophil transendothelial migration increased over time, indicating a distinct impact of THP-1 macrophages on the neutrophil transendothelial migration dynamics within our Infection-on-a-Chip devices.
Discussion
The specific orchestration of interactions between macrophages and neutrophils is crucial for an efficient immune response. However, the precise mechanisms by which macrophages influence neutrophil recruitment and extravasation during human infection remain elusive. As a first step in addressing this knowledge gap, our study incorporated THP-1 macrophages into the collagen extracellular matrix of our Infection-on-a-Chip microfluidic device to study how human macrophages modulate neutrophil extravasation in response to infection. Our study confirms that THP-1 macrophages undergo differentiation, and M2 polarization, and maintain viability within the collagen matrix without disrupting the model endothelial blood vessel. Notably, we find reduced human primary neutrophil transendothelial migration in the presence of THP-1 macrophages during infection with Pseudomonas aeruginosa. Collectively, these insights provide initial evidence for macrophage-mediated regulation of the neutrophil response to infection and, most critically, present a novel experimental platform for systematic evaluation of these crucial cell–cell interactions. Future studies will now be able to utilize our platform to determine the precise mechanisms of human macrophage regulation of neutrophil function by examining the interaction between primary human neutrophils and macrophages in a physiologically relevant context.
An essential first step in investigating macrophage-neutrophil interactions was successfully integrating macrophages into the tissue compartment of our microfluidic device. While microfluidics have been used for studying cell–cell interactions in the context of tissue engineering, often employing synthetic hydrogels [27], we wanted to embed macrophages in physiologically relevant collagen I hydrogels to emulate the microenvironment in vivo.
Several studies have investigated the significance of collagen concentration on macrophage phenotype. Larsen et al. reported no discernible change in proliferation or viability in varied gel concentrations [28]. However, their research used RAW264.7 mouse macrophage-like cells that do not require differentiation in contrast to our study which employed THP-1, human macrophage-like cells that require a differentiation step. Despite these distinctions, we similarly observed no differences in viability or differentiation rates for THP-1 macrophages embedded in 1.5 versus 4 mg/mL collagen hydrogels, in agreement with these results. While our use of a human immortalized cell line may introduce variations in phenotype compared to primary macrophages, future studies will use primary macrophages to confirm our findings and offer additional insights into human macrophage behavior.
In other microfluidic platforms, the presence of macrophages has been shown to alter endothelial phenotypes; therefore, it was important for us to validate the integrity of the endothelial cell vessel in our Infection-on-a-Chip device in the presence of THP-1 macrophages. Previous studies have found that macrophages influence endothelial cell morphology and branched network formation leading to angiogenesis [29]. Although not quantified, we observed a qualitative increase in the sprouting of endothelial cells when macrophages were present, which aligns with these results. While macrophages may affect endothelial sprouting, the effect of this sprouting on neutrophil transendothelial migration has not been described in literature.
Moreover, we found the presence of macrophages did not lead to an increase in the permeability of the endothelial vessel. Endothelial cell permeability increases during inflammation in vivo and we have previously seen this increased permeability in our device in response to an inflamed environment [30]. Therefore, these results indicate that collagen-embedded macrophages do not create an inflamed environment before infection in our device. These results differ from previous reports, likely due to significant differences in experimental setup. Middelkamp et al. found increased fibrin deposition and clot formation in the presence of macrophages and, therefore, hypothesized the macrophages caused an inflamed endothelium; however, they saw no change in actin structure in vessels with or without macrophages, in line with our lumen characterization results [31]. Notably, unlike the Middelkamp study, where the phenotype of the embedded macrophages was not determined, our investigation revealed a high percentage of CD163-positive macrophages, indicative of a predominantly M2 phenotype. In fact, we found a significant fraction of our macrophages were M2 prior to infection, further supporting evidence that macrophages are not creating a pro-inflammatory environment. Moreover, Middelkamp et al. and Yu et al. used macrophages that were differentiated in a plate and then OxLDL-treated or polarized before being introduced into the system, and both reported that macrophage polarization alters macrophages' impact on endothelial cells. In contrast to these studies, our THP-1 macrophages were differentiated once embedded in collagen in situ with PMA [31]. Taken together, both existing research and our results suggest that the presence of THP-1 macrophages, differentiated in our system, does not induce an inflammatory state in the device. Future studies in our laboratory will quantify cytokine secretion in the presence of endothelial cells and macrophages. Additionally, previous research has shown that macrophages can exhibit diverse phenotypic states, which depend on the type and progression of bacterial infection. In our study, macrophages predominantly exhibited M2 phenotypes within our device [32]. While this finding provides valuable insights into potential mechanisms for reducing neutrophil extravasation, it also represents a limitation, highlighting the need for future research to intentionally polarize macrophages and investigate the differences between M1 and M2 phenotypes in infection contexts.
The role of macrophage-neutrophil interactions in regulating neutrophil recruitment and resolution remains unclear, in part due to conflicting studies that report different signaling requirements and mechanisms. In 2014, Tauzin et al. reported that direct neutrophil-macrophage contact following tissue damage in zebrafish leads to neutrophil resolution through reverse migration [33]. In contrast, a study by Wang et al. found macrophages play no role in neutrophil resolution using a sterile injury model in mice [13]. Yet only 1 year later in 2018, Loynes et al., using a similar tissue wounding model in zebrafish as Tauzin et al., found that macrophages do promote neutrophil resolution, but identified the secretion of PGE2 to be critical with no requirement for direct contact [34]. While these studies collectively underscore the significance of macrophage-neutrophil interactions in governing neutrophil recruitment and resolution, the discrepancies in their findings, potentially stemming from differences in animal models or inflammatory stimuli, contribute to the ongoing uncertainty regarding the underlying mechanisms governing these interactions. Moreover, while reverse migration of primary human neutrophils has been observed in simple in vitro microfluidic models [35], an investigation of primary human neutrophil-macrophage interactions in a physiologically relevant environment had previously not been accomplished. Our study sought to address these gaps by analyzing the primary human neutrophil response in the presence of macrophages in a model infectious microenvironment. Notably, we found the presence of macrophages caused a reduction in neutrophil transendothelial migration to Pseudomonas aeruginosa infection over 6–10 h. Our findings align with the previous studies that find that macrophages play a significant role in the neutrophil response to inflammation and its resolution process [33, 34]. Furthermore, the predominantly M2 phenotype exhibited by the macrophages prior to the introduction of bacteria and neutrophils into the device may elucidate their role in attenuating the neutrophil response. This observation aligns with previous work by Su et al., demonstrating that M2 macrophages can mitigate the neutrophil response in sepsis by impeding neutrophil recruitment and neutrophil extracellular trap (NET) formation [36]. Further studies are needed to determine if the effects we see are due to soluble factors, physical touching, or both and whether the mechanisms are distinct from the pro-resolution pathways previously reported. Collectively, our results indicate an important role for macrophages in regulating the human neutrophil response to infection.
Our Infection-on-a-Chip device is designed to closely replicate physiological conditions, allowing for a targeted examination of the intricate physical and chemical mechanisms governing neutrophil-macrophage interactions during the innate immune response. This innovative platform bridges the gap between traditional in vitro systems and complex in vivo environments, unlocking novel therapeutic possibilities for clinical applications by unraveling the nuanced and dynamic interplay between neutrophils and macrophages in a controlled yet realistic environment. Future research will focus on characterizing key secretory factors influencing the inflammatory environment and investigating whether macrophages alter neutrophils through soluble factors or physical interactions.
Materials and Methods
THP-1 Cell Culture
THP-1 cells (ATCC TIB-202) were cultured in T-25 flasks using complete THP-1 media, which consists of RPMI medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Fisher), 1% penicillin-streptomycin (Gibco), and 0.05 mM 2-mercaptoethanol (Sigma-Aldrich). These cells were used from passages 10 to 17. Cells were passaged at 2 × 106 cells/mL and reseeded at a concentration of 2.5 × 105 cells/mL.
Human Umbilical Vein Endothelial Cell (HUVEC) Culture
Pooled Human Umbilical Vein Endothelial Cells (HUVEC, 50-305-964, Promocell Gmb HC12203) were cultured in T-75 flasks and maintained in Endothelial Growth Media (EGM-2, NC9525043, Lonza Walkersville CC3162) until they reached 80% confluence. These cells were used from passages 2 to 7. The media was changed every 2 days. Cells were detached by rinsing with Hank’s Balanced Salt Solution, then using Trypsin EDTA solution (0.05% Trypsin and 0.02% EDTA in phosphate-buffered saline (1X PBS) without Ca2+ or Mg2+) (50189662FP, ATCC) and Trypsin Neutralizing Solution (5% FBS in 1X PBS) (50189663FP, ATCC). They were then split and reseeded at a concentration of 375,000 cells per 15 mL.
Pseudomonas aeruginosa Strain K (PAK) Culture
LB agar was prepared following the manufacturer’s instructions to create LB plates. Pseudomonas aeruginosa was streaked onto LB plates and incubated at 37 °C for 16 h. Following incubation, the plates were stored in a refrigerator at 4 °C for up to 30 days. A single colony from the LB plates was grown overnight in 5 mL of LB broth using a bacterial shaker at 37 °C. Afterward, 1 mL of the cultured solution was diluted in 4 mL of fresh LB broth and further cultured in a bacterial shaker at 37 °C for 1.5 h. 1 mL of the bacterial culture was pelleted by centrifugation (17,000×g for 1 min) and resuspended in 100 μL of EGM-2. The optical density (OD) of the bacterial solution was measured at 600 nm, and it was then diluted in EGM-2 to achieve an OD of 5.
Infection-on-a-Chip Device Fabrication
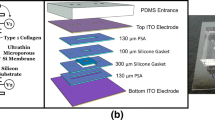
Microfluidic devices were fabricated as previously described [37]. Briefly, silicon wafers were patterned with SU-8-100 (Kayaku Advance Materials) through soft lithography to create top and bottom masters. Polydimethylsiloxane (PDMS) (Sylgard 184, Electron Microscopy Science) was added to the masters and polymerized for 4 h at 60 °C. The PDMS layers were aligned, and a PDMS rod (0.337 mm inner diameter) was introduced. The devices were bonded to a glass-bottom MatTek dish (MatTek Corporation) using oxygen plasma.
Hydrogel and Infection-on-a-Chip Device Setup
Following UV sterilization for 15 min, the microfluidic devices were exposed to a 1% polyethyleneimine (PEI) solution in deionized (DI) water (03880, Sigma Aldrich, St. Louis, Missouri) and incubated for 10 min. After this, a 0.1% glutaraldehyde (GA) solution in DI water (G6257, Sigma Aldrich) was pipetted into the device and incubated for 30 min. The devices were then washed three times with DI water. Type I Rat Tail Collagen (354249, Corning), neutralized to a desired pH of 7.2 at a concentration of 4 mg/mL, was introduced into the devices. The collagen solution polymerized around the PDMS rod in the central chamber. After polymerization, the PDMS rod was delicately removed, leaving the lumen structure in place. The lumens were subsequently seeded with HUVECs at a concentration of 2 × 104 cells/µL and incubated overnight on a rotator. HUVECs were allowed to grow and form a monolayer, with twice-daily EGM-2 media changes for 2 days.
THP-1 Differentiation and Cell Viability Experiments in Collagen
To assess cell viability in collagen via microscopy, 200 µL of collagen solution (Corning or Ibidi) was added to each well of an 8-well coverslip plate (Ibidi) and incubated for 30 min at 37 °C with 5% CO2. Afterward, 200 µL of complete THP-1 media was added to each well. Live/dead staining was conducted using a solution containing THP-1 cell media, 2 μg/mL propidium iodide (PI) (MilliporeSigma), and 500 ng/mL calcein AM (ThermoFisher).
To determine cell viability and differentiation inside the devices, the differentiation process started on Day 0. Devices were prepared, and collagen-embedded THP-1 monocytes were added at a concentration of 5 × 105 cells/mL. THP-1 monocytes were then exposed to complete THP-1 media supplemented with 50 ng/mL PMA. Media was changed through the side ports of the device every 24 h for 2 days.
For flow cytometry experiments to assess viability and differentiation, we prepared 1.5 mg/mL and 4 mg/mL collagen gels with 5 × 105 THP-1 monocytes/mL. 200 µL of the collagen gels were seeded into each well of a 48-well plate. After polymerization, 200 µL of complete THP-1 media with 50 ng/mL PMA was added to each well, and media changes occurred every 24 h for 2 days. For the non-differentiated control, THP-1 monocytes were added to well plates without collagen and maintained with complete THP-1 media. The media was changed every 24 h.
HUVEC Permeability Assay
HUVEC permeability assays were performed on Day 4, 2 days after HUVECs were seeded in the device as previously outlined. The EGM-2 media was added inside the lumen, aspirated, and substituted with a 125 ug/mL solution of 10 kDa fluorescein isothiocyanate (FITC)-Dextran (Sigma-Aldrich).
For lumens exposed to PAK, 3 μL of Pseudomonas aeruginosa at an optical density (OD) of 5 was added to the top port of the device and incubated at 37 °C, 5% CO2 for 1 h before adding FITC inside of the lumen.
HUVEC Staining Assay
To assess a proper barrier formation by the HUVECs, the HUVEC staining assay was conducted on Day 4, 2 days after HUVECs were seeded in the device as previously outlined. Initially, 1X PBS was added and aspirated through side ports of the lumen twice, and prewarmed 4% paraformaldehyde (PFA) was introduced inside the lumen and incubated for 1 h at 37 °C, 5% CO2. The lumen was then washed three times with 1X PBS. PBST (0.1% Tween 20 in 1X PBS) was then added to the side port of the lumen and incubated at room temperature for 10 min. A staining solution with 1:200 Hoechst (Thermo Fisher), 1:1000 Phalloidin (Abcam), and 5:600 Anti-VE-Cadherin (BD) in 1X PBS was prepared. The PBST was aspirated, and the staining solution was added to the inside of the lumens. The device was then incubated and protected from light at 4 °C overnight. On Day 5, the staining solution was aspirated, and the lumen was washed three times with 1X PBS.
To ensure the staining did not introduce interference or noise and to validate the integrity of the results, control experiments were performed. These controls included staining lumens in the presence of macrophages with solely Anti-VE-Cadherin and Hoechst, or with only Phalloidin and Hoechst.
To quantify ICAM-1 expression in HUVECs, we stained and imaged HUVECs in wells using 1:200 anti-ICAM1 (R&D Systems), 1:200 Hoechst (Thermo Fisher), and 1:1000 Phalloidin (Abcam) in 1X PBS, as previously described after a 2-h exposure to PAK at an optical density (OD) of 5.
THP-1 Infection-on-a-Chip Cell Viability Assay
THP-1 viability was assessed on Day 2 and Day 4, following the preparation of the devices and the differentiation of THP-1 as previously described. A solution containing 500 ng/mL of calcein AM (Invitrogen) and 2 μg/mL of PI (Sigma) was prepared and introduced into the device through the side ports on Day 2 or Day 4
THP-1 Infection-on-a-Chip Cell Differentiation Assay
THP-1 differentiation was assessed on Day 2, and Day 4, and THP-1 phenotyping was assessed on Day 0, Day 2, and Day 4 following the preparation of the devices and the differentiation of THP-1s as previously explained. Briefly, the upper PDMS layer of the device was carefully removed, THP-1 cells were fixed with 4% paraformaldehyde for 1 h, rinsed three times for 2 h with 1X PBS, and permeabilized using a buffer containing 0.20% Triton-X for 45 min at 37 °C with 5% CO2. Following permeabilization, the cells underwent three 30-min incubations at 37 °C, 5% CO2 with 1X PBS for thorough washing. Next, a dilution of 1:1000 of Phalloidin (Abcam) in 1X PBS was added to each device and incubated at 4 °C overnight. The next day, the cells underwent three 30-min incubations at room temperature with 1X PBS washes. Next, the cell surface was blocked and permeabilized using a solution of 5% BSA in 0.1% PBS-Tween 80 for 24 h at 37 °C with 5% CO2.
To assess THP-1 differentiation, the cells were treated with rabbit primary anti-CD68 antibodies (ab213363, Abcam) diluted 1:100 in a dilution buffer consisting of 1X PBS and 1% BSA and incubated for 48 h at 37 °C with 5% CO2. To assess THP-1 phenotype, the cells were treated with rabbit primary antibodies anti-CD68 (ab213363, Abcam) diluted 1:100 and anti-CD163 (130-099-969, Miltenyi) diluted 1:11 in a dilution buffer consisting of 1X PBS and 1% BSA and incubated for 48 h at 37 °C with 5% CO2.
After the primary antibody incubation, the cells were washed with 1X PBS in two rounds of 2-h incubations at 37 °C to remove any residual primary antibodies. Following this, a secondary antibody (goat anti-rabbit IgG Alexa Fluor 405) was applied at a dilution of 1:200 in 1X PBS with 1% BSA for 24 h at 37 °C with 5% CO2. Subsequently, the cells were washed twice with 1X PBS, and incubated for 2 h at 37 °C with 5% CO2 during each wash.
Neutrophil Isolation
All blood samples were collected following institutional review board-approved protocols in adherence to the principles of the Declaration of Helsinki at the University of Colorado-Boulder. Peripheral blood neutrophils were isolated from healthy donors using the MACSxpress Neutrophil Isolation Kit (Miltenyi Biotec), following the manufacturer's instructions. Informed consent was obtained from donors at the time of the blood draw, following institutional review board procedures.
Neutrophil TEM
Neutrophil transendothelial migration was assessed in devices both with and without THP-1 macrophages. Following neutrophil isolation, 20% of the neutrophil population was stained with 500 ng/mL calcein AM (ThermoFisher) for 10 min at room temperature, while the remaining 80% remained unstained. After staining, neutrophils were centrifuged at 200×g for 5 min. The stained and unstained neutrophils were then combined and resuspended at a concentration of 6 × 106 cells/mL. The neutrophils were then seeded into the lumen chamber. 3 μL of Pseudomonas aeruginosa at an optical density (OD) of 5 was added to the top port of the device. Confocal images were captured every 10 min for 10 h to visualize neutrophil transendothelial migration.
Flow Cytometry
Flow cytometry analysis was conducted using a BD FACS Celesta Flow Cytometer. For the acquisition of THP-1 cells plated in wells, media was aspirated, Trypsin EDTA solution was added, and the cells were incubated for 20 min at 37 °C with 5% CO2. Upon detachment, 500 µL of complete THP-1 media was added to each well, followed by centrifugation at 200×g for 5 min. To obtain THP-1 cells embedded in collagen, 200 µL of 8 mg/mL collagenase I (ThermoFisher) was added to each well. After 30 min of incubation at 37 °C with 5% CO2, collagen was gently agitated using a pipette and incubated for an additional 10 min. Subsequently, 500 µL of complete THP-1 media was added to each well, and the cells were pipetted out and centrifuged at 200×g for 5 min. After obtaining cells in both conditions, the pellets were resuspended in a 1:200 dilution of ZombieViolet in flow cytometry buffer (2 µM EDTA + 0.5% BSA in 1X PBS), followed by a 15-min incubation at room temperature. After 15 min, 2 µL anti-CD11b antibody (130-110-614, Miltenyi) was introduced, and incubated for 10 min at 4 °C. Post-incubation, 1 mL of flow cytometry buffer was added to each tube, and tubes were centrifuged at 200×g for 5 min. The cells were resuspended in 500 µL of flow cytometry buffer and analyzed using the flow cytometer with the 405 nm laser (450/40 bandpass filter) for Zombie Violet and the 561 nm laser (780/60 bandpass filter) for the CD11b antibody. This entire process was executed on days 2 and 4, following the seeding of THP-1 cells in wells or embedding them in type I collagen, as detailed in previous sections.
Image Processing and Data Acquisition
Nikon A1R HD25 Laser Scanning Confocal Microscope built on the Nikon Ti2-E Inverted Microscope System with a Nikon 10x/0.45 (NA) objective was used for acquiring all images. Fluorescence images were analyzed through NIS Elements software and Fiji (ImageJ). For all the viability and differentiation calculations, 3 images were taken from each device for each condition to determine the average values.
Cell Viability in Well-Plates: To assess cell viability in the well plates without collagen, cells were imaged using the 488-channel (live cells, calcein AM, green) and the 561-channel (dead cells, PI, red) and counted using the cell counting feature in Nikon Elements software. This was achieved through Bright Spot Detection employing the Clustered Method, with specific parameters set at 21 µm, a contrast of 10 for calcein AM, and a contrast of 20 for PI. The obtained values were then normalized to the initial count of living cells from Day 0.
Cell Viability in the Infection-on-a-Chip Device: To assess cell viability in the devices, max intensity projections were generated from fluorescent z-stack images. Cells were imaged under the same conditions as the well plates. The obtained values were then normalized to the initial count of living cells from Day 0.
Human umbilical vein endothelial cells (HUVECs) were stained and imaged using a confocal microscope with the following laser settings: nuclei were visualized with Hoechst using a 405 nm laser (blue), α-actin was visualized with Phalloidin using a 561 nm laser (red), tight junctions were visualized with Anti-VE-Cadherin using a 488 nm laser (green), and ICAM-1 was visualized with Anti-ICAM using a 488 nm laser (green). To quantify ICAM-1 staining, the mean pixel intensity within three different 1200 × 860 µm regions of interest (ROIs) was calculated and corrected by subtracting the mean pixel intensity of a 300 × 80 µm background segment within each of the three different well regions. To ensure comparability between samples, we assessed the number of nuclei within the same 1200 × 860 µm ROIs. The pixel intensity of ICAM-1 staining was then normalized by dividing the mean pixel intensity by the number of nuclei counted in each respective ROI. This normalization ensured that variations in cell density did not confound the intensity measurements.
Endothelial Cells Monolayer Permeability Assessment: To evaluate the permeability of endothelial cell monolayers, pixel intensities of FITC were measured at single z-planes using ImageJ. Images were obtained using the 488 nm laser (FITC, calcein AM, green). This analysis was conducted at the 5-, 10-, and 15-min time points.
Cell Differentiation in 4 mg/mL Collagen and the Infection-on-a-Chip Device: The differentiation percentage was determined by quantifying cells stained with Phalloidin and anti-CD68 within the field of view. The phenotyping percentage was determined by quantifying cells stained with Phalloidin, anti-CD68, and anti-CD163 within the field of view. Cells were imaged using the 405 nm (CD68, anti-CD68, blue), the 561 nm (α-actin, Phalloidin, red), and 488 nm-channel (CD163, anti-CD163, green) lasers and counted using the cell counting feature in Nikon Elements software. This was achieved through Bright Spot Detection employing the Clustered Method, with specific parameters set at 16 µm, a contrast of 10 for Phalloidin, a contrast of 20 for anti-CD68, and a contrast of 20 for anti-CD163. The percentage of differentiated THP-1s was calculated as the number of CD68+ THP-1 cells divided by the total number of cells (stained with phalloidin) and multiplied by 100. The percentage of CD163+ THP-1s was calculated as the number of CD163+ THP-1 cells divided by the number of CD68+ cells.
TEM Data Acquisition: Transendothelial migration was analyzed as previously described [21]. Briefly, the 488 nm (live cells, calcein AM, green) laser was used to obtain images, and max intensity projections in the z-axis were generated using Nikon Elements software. These videos were then transferred to FIJI (ImageJ) for further analysis. Neutrophils were analyzed in the center of the device to eliminate edge effects. The analysis of extravasated neutrophils involved creating a 690 μm × 345 μm rectangular region of interest at the top edge of the lumen in the device's center. The number of neutrophils within this region was counted at each time point and normalized to the initial number of neutrophils in the lumen, to account for variability in cell loading. The initial number of neutrophils was determined by counting within a 345 μm × 122.5 μm rectangular region drawn within the center of the lumen at the first time point.
Flow Cytometer Data Acquisitions: Flow cytometric data collection employed a BD FACS Celesta Flow Cytometer, and subsequent analysis utilized FlowJo software.
Data Presentation and Statistical Analysis
The study's data are pooled from three or more independent experimental replicates. Statistical analysis was conducted using RStudio (version 2022.07.1 + 554). Transendothelial migration experiment parameters, specifically normalized extravasated neutrophils, underwent comparison between conditions using analysis of variance (ANOVA). Results are summarized in terms of estimated marginal means and standard errors. Pairwise comparisons were performed with Tukey’s adjustment and P values are denoted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001, indicating the level of significance.
Data availability
All data are available upon reasonable request from the corresponding author.
Abbreviations
- HUVEC:
-
Human umbilical vein endothelial cells
- THP-1:
-
Human acute monocytic leukemia cell line
- PMA:
-
Phorbol 12-myristate 13-acetate
- PAK:
-
Pseudomonas aeruginosa Strain K
References
Iwai, Y., S. Terawaki, and T. Honjo. PD-1 blockade inhibits hematogenous spread of poorly immunogenic tumor cells by enhanced recruitment of effector T cells. Int. Immunol. 17(2):133–144, 2005.
Coffelt, S. B., M. D. Wellenstein, and K. E. De Visser. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 16(7):431–446, 2016.
Nielsen, S. R., and M. C. Schmid. Macrophages as key drivers of cancer progression and metastasis. Mediat. Inflamm. 2017:9624760, 2017.
Marsland, B. J., et al. Nippostrongylus brasiliensis infection leads to the development of emphysema associated with the induction of alternatively activated macrophages. Eur. J. Immunol. 38(2):479–488, 2008.
Neumann, K., B. Schiller, and G. Tiegs. NLRP3 inflammasome and IL-33: Novel players in sterile liver inflammation. Int. J. Mol. Sci. 19(9):2732, 2018.
Shen, H., D. Kreisel, and D. R. Goldstein. Processes of Sterile Inflammation. J. Immunol. 191(6):2857–2863, 2013.
Bouchery, T., and N. Harris. Neutrophil-macrophage cooperation and its impact on tissue repair. Immunology and cell biology. 97(3):289–298, 2019.
Mcdonald, B., et al. Intravascular danger signals guide neutrophils to sites of sterile. Inflammation. 330(6002):362–366, 2010.
Soehnlein, O., and L. Lindbom. Phagocyte partnership during the onset and resolution of inflammation. Nat. Rev. Immunol. 10(6):427–439, 2010.
Wynn, T. A., A. Chawla, and J. W. Pollard. Macrophage biology in development. Homeostasis Dis. 496(7446):445–455, 2013.
Magnarin, M., et al. Human neutrophils specifically interact with human monocyte-derived macrophages monolayers. Inflammation. 24(1):89–98, 2000.
Mathias, J. R., et al. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J. Leukocyte Biol. 80(6):1281–1288, 2006.
Wang, J., et al. Visualizing the function and fate of neutrophils in sterile injury and repair. Science. 358(6359):111–116, 2017.
Woodfin, A., et al. The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nat. Immunol. 12(8):761–769, 2011.
Yoo, S. K., et al. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature. 480(7375):109–112, 2011.
Barros-Becker, F., et al. Live imaging reveals distinct modes of neutrophil and macrophage migration within interstitial tissues. J. Cell Sci. 130(22):3801–3808, 2017.
Manda-Handzlik, A., et al. Secretomes of M1 and M2 macrophages decrease the release of neutrophil extracellular traps. Sci. Rep. 13(1):15633, 2023.
Hind, L. E., et al. Interaction with an endothelial lumen increases neutrophil lifetime and motility in response to P aeruginosa. Blood. 132(17):1818–1828, 2018.
Hind, L. E., et al. Immune cell paracrine signaling drives the neutrophil response to A. fumigatus in an infection-on-a-chip model. Cell. Mol. Bioeng. 14(2):133–145, 2021.
Ingram, P. N., L. E. Hind, J. A. Jiminez-Torres, A. Huttenlocher, and D. J. Beebe. An accessible organotypic microvessel model using iPSC-derived endothelium. Adv. Healthc. Mater. 7(2):1700497, 2018. https://doi.org/10.1002/adhm.201700497.
Richardson, I. M., et al. Diverse bacteria elicit distinct neutrophil responses in a physiologically relevant model of infection. iScience.27(1):108627, 2024.
Richardson, I. M., C. J. Calo, and L. E. Hind. Microphysiological systems for studying cellular crosstalk during the neutrophil response to infection. Front. Immunol.12:661537, 2021.
Lu, R., and P. M. Pitha. Monocyte differentiation to macrophage requires interferon regulatory factor 7. J. Biol. Chem. 276(48):45491–45496, 2001.
Komohara, Y., J. Hirahara, et al. AM-3K, an anti-macrophage antibody, recognizes CD163, a molecule associated with an anti-inflammatory macrophage phenotype. J. Histochem. Cytochem. 54(7):763–771, 2006.
Khaidakov, M., X. Wang, and J. L. Mehta. Potential involvement of LOX-1 in functional consequences of endothelial senescence. PLoS ONE.6(6):e20964, 2011.
Ganter, M., J. Roux, G. Su, et al. Role of small GTPases and αvβ5 integrin in Pseudomonas aeruginosa-induced increase in lung endothelial permeability. Am. J. Respir. Cell Mol. Biol. 40(1):108–118, 2009.
Pang, L., J. Ding, X. X. Liu, Z. Kou, L. Guo, X. Xu, and S. K. Fan. Microfluidics-based single-cell research for intercellular interaction. Front. Cell Dev. Boil.9:680307, 2021.
Larsen, A. M. H., et al. Collagen density modulates the immunosuppressive functions of macrophages. J. Immunol. 205(5):1461–1472, 2020.
Yu, J., et al. Reconfigurable open microfluidics for studying the spatiotemporal dynamics of paracrine signalling. Nat. Biomed. Eng. 3(10):830–841, 2019.
Hind, L. E., E. B. Lurier, M. Dembo, et al. Effect of M1–M2 polarization on the motility and traction stresses of primary human macrophages. Cel. Mol. Bioeng. 9:455–465, 2016.
Middelkamp, H., H. Weener, T. Gensheimer, et al. Embedded macrophages induce intravascular coagulation in 3D blood vessel-on-chip. Biomed. Microdevices. 26(1):2, 2024.
Atri, C., F. Z. Guerfali, D. Laouini, et al. Role of human macrophage polarization in inflammation during infectious diseases. Int. J. Mol. Sci. 19(6):1801, 2018.
Tauzin, S., et al. Redox and Src family kinase signaling control leukocyte wound attraction and neutrophil reverse migration. J. Cell Biol. 207(5):589–598, 2014.
Loynes, C. A., et al. PGE 2 production at sites of tissue injury promotes an anti-inflammatory neutrophil phenotype and determines the outcome of inflammation resolution in vivo. Sci. Adv. 4(9):eaar8320, 2018.
Hamza, B., et al. Retrotaxis of human neutrophils during mechanical confinement inside microfluidic channels. Integr. Biol. 6(2):175–183, 2014.
Su, Y., J. Gao, P. Kaur, and Z. Wang. Neutrophils and macrophages as targets for development of nanotherapeutics in inflammatory diseases. Pharmaceutics. 12(12):1222, 2020.
Jiménez-Torres, J. A., et al. LumeNEXT: a practical method to pattern luminal structures in ECM gels. Adv. Healthc. Mater. 5(2):198–204, 2016.
Acknowledgments
Flow cytometry was conducted in the Flow Cytometry Shared Core Facility at CU Boulder with instruments supported by NIH grant S10ODO21601. Water Confocal Microscope images were conducted in the BioFrontiers Institute Advanced Light Microscopy Core at CU Boulder (RRID: SCR_018302).
Funding
This work was supported by grants from the National Institutes of Health (R35 GM1 46737A) and by the 2023-24 National Institutes of Health Molecular Biophysics Training Scholarship.
Author information
Authors and Affiliations
Contributions
AIR, HKW, and LEH designed the research and wrote the manuscript. AIR, HKW, and MS performed the experiments. AIR, HKW, TK, MS, and LEH analyzed and interpreted the data.
Corresponding author
Ethics declarations
Conflict of interest
The authors AIR, HKW, TK, MS, and LEH have no conflicts of interest to declare.
Ethical Approval
This study does not involve animal subjects. All blood samples were collected following institutional review board-approved protocols in adherence to the principles of the Declaration of Helsinki at the University of Colorado-Boulder.
Additional information
Associate Editor Michael R. King oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ignes-Romeu, A., Weppner, H.K., Kaur, T. et al. THP-1 Macrophages Limit Neutrophil Transendothelial Migration in a Model Infection. Cel. Mol. Bioeng. (2024). https://doi.org/10.1007/s12195-024-00813-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12195-024-00813-2