Abstract
Heat shock proteins (HSPs) are ubiquitous polypeptides expressed in all living organisms that participate in several basic cellular processes, including protein folding, from which their denomination as molecular chaperones originated. There are several HSPs, including HSPA5, also known as 78-kDa glucose-regulated protein (GRP78) or binding immunoglobulin protein (BIP) that is an ER resident involved in the folding of polypeptides during their translocation into this compartment prior to the transition to the Golgi network. HSPA5 is detected on the surface of cells or secreted into the extracellular environment. Surface HSPA5 has been proposed to have various roles, such as receptor-mediated signal transduction, a co-receptor for soluble ligands, as well as a participant in tumor survival, proliferation, and resistance. Recently, surface HSPA5 has been reported to be a potential receptor of some viruses, including the novel SARS-CoV-2. In spite of these observations, the association of HSPA5 within the plasma membrane is still unclear. To gain information about this process, we studied the interaction of HSPA5 with liposomes made of different phospholipids. We found that HSPA5 has a high affinity for negatively charged phospholipids, such as palmitoyl-oleoyl phosphoserine (POPS) and cardiolipin (CL). The N-terminal and C-terminal domains of HSPA5 were independently capable of interacting with negatively charged phospholipids, but to a lesser extent than the full-length protein, suggesting that both domains are required for the maximum insertion into membranes. Interestingly, we found that the interaction of HSPA5 with negatively charged liposomes promotes an oligomerization process via intermolecular disulfide bonds in which the N-terminus end of the protein plays a critical role.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat shock proteins (HSPs) are ubiquitous polypeptides present in all living organisms, from bacteria to humans. They participate in several basic cellular processes, including protein folding, assembly of multiprotein complexes, as well as protein transport and degradation. In this regard, they have been coined molecular chaperones (Hartl and Hayer-Hartl 2002). Although many HSPs are present constitutively in cells, other members are expressed or overexpressed during stress conditions, such as increases in temperature or exposure to harmful chemicals. The expression of these proteins facilitates a rapid recovery from the stress preserving survival and prompting protection from subsequent insults (De Maio 1999). Unfortunately, the protective ability of these proteins may be a factor for the prolonged endurance of cancer cells that displayed abnormally high expression levels of HSPs (Calderwood 2018; Lang et al. 2019).
The HSP family is composed of several members of different molecular weights that were initially used for their classification (e.g., Hsp70s, Hsp90s) that has now been simplified with a new nomenclature (Kampinga et al. 2009). Each of these groups is also composed of various members. For example, the Hsp70 (HSPA) family is made up of four proteins with different subcellular localizations. Hsc70 or HSPA8 (constitutive) and Hsp70 or HSPA1 (stress-inducible) are both present in the cytosol/nucleus, mortalin or HSPA9 is localized in the mitochondrial matrix, and 78-kDa glucose-regulated protein (GRP78), binding immunoglobulin protein (BIP), or HSPA5 is present in the ER lumen (De Maio 1999). A common feature of Hsp70s (HSPAs) is that they are flexible proteins containing two regions, the nucleotide-binding and peptide-binding domains (NBD and PBD, respectively), connected by a short hydrophobic linker. NBD is within the N-terminal end containing ATPase activity, whereas PBD is within the C-terminal end and is involved in the binding of client polypeptides (Mayer et al. 2001; da Silva and Borges 2011). These two domains coordinate the folding process that is mediated by a bidirectional heterotrophic allosteric mechanism driving a conformational change from an open state in the presence of ATP that established the initial interaction with the client polypeptide to a closed state upon conversion of ATP into ADP retaining the target polypeptide. This process is modulated by co-chaperone molecules (Young 2010; da Silva and Borges 2011).
Although the folding process occurs inside the cell, extensive and compelling evidence has shown that HSP are also present outside the cells, apparently acting as signaling molecules participating in the communications between cells (De Maio 2011; De Maio 2014). The mechanism for the export of cytosolic HSP into the extracellular environment is still unclear, particularly because these proteins do not have the respective signal for the translocation into the classical secretory pathway. Thus, the challenge for cytosolic HSPs to reach the extracellular space is crossing the hydrophobic environment provided by the plasmalemma. Although the translocation across the plasma membrane appears as thermodynamically impermissible, there are broad reports showing the presence of HSP, particularly Hsp70 (HSPA1), embedded into the plasma membrane of transformed cells (Multhoff and Hightower 1996). Moreover, the insertion of HSP into lipid membranes has been corroborated by several studies using artificial lipid bilayers (Arispe and De Maio 2000; Vega et al. 2008; Armijo et al. 2014; Macazo and White 2014; De Maio et al. 2019). The interaction of HSP with the plasma membrane is likely the first step for their export mechanism (De Maio 2011). One possibility is that membrane HSPs are released in association with exosomes from viable cells (Gastpar et al. 2005; Vega et al. 2008). However, HSPs may also be discharged from dying necrotic cells (Basu and Srivastava, 2000; De Maio and Vazquez 2013).
Among all HSPs, the only one that could follow the classical secretory pathway is HSPA5 that is present in the ER lumen (Lee 1992; Pobre et al. 2019). HSPA5 has the same structural characteristics of all HSPAs with two domains, NBD and PBD. In addition, HSPA5 presents a leading sequence at the N-terminus end that is responsible for its translocation into the ER lumen and an ER retention signal (KDEL) at the carboxyl-terminal end. HSPA5 participates in the translocation of newly synthesized polypeptides across the ER membrane (Casanova-Morales et al. 2018; Pobre et al. 2019), in protein folding and quality control (Haas and Wabl 1983), in targeting misfolded proteins for degradation (Gardner et al. 2013; Oikonomou and Hendershot 2020), and in the unfolded protein response (Luo and Lee 2013; Wang and Kaufman 2016).
HSPA5 was reported present on the plasma membrane (Suzuki et al. 1991; Delpino and Castelli 2002; Zhang et al. 2013), in which it acts as a signal-transducing receptor or co-receptor for soluble ligands (Zhang et al. 2010). Also, cell-surface HSPA5 appears to be involved in tumor survival, proliferation, and resistance (Pfaffenbach and Lee 2011). Interestingly, cell-surface HSPA5 is a potential binding site for various viruses, including Borna disease (Honda et al. 2009), Coxsackie, dengue virus serotype 2, and Japanese encephalitis (Kottom et al. 2018). Most recently, HSPA5 could be a receptor for the corona viral spike protein (Chu et al. 2018), including the novel SARS-CoV-2 (Ibrahim et al. 2020), responsible for the COVID-19 pandemic. Although the presence of HSPA5 within the plasma membrane is controversial since this protein lacks a consensus transmembrane domain, it has been proposed that several domains are indeed embedded into the membrane (Tsai et al. 2015), particularly the C-terminus end (Tseng et al. 2019). In order to establish a possible insertion mechanism, we investigated the interaction of HSPA5 with liposomes. We found that HSPA5 has great selectivity for negatively charged phospholipids, similar to HSPA1 (Arispe et al. 2004; Armijo et al. 2014). Moreover, HSPA5 N-terminal and C-terminal domains could independently interact with phospholipid membranes, but to a lesser extent than the full-length protein, suggesting a synergetic mechanism of both domains in binding to membranes. Finally, we observed that HSPA5 undergoes an oligomerization process upon membrane insertion mediated by intermolecular disulfide bond formation.
Methods
Sequence and alignment HSPA proteins and their expression/isolation
The amino acid sequences of HSPA5 (UniProtKB, P11021) and HSPA1 (UniProtKB, P0DMV8) were evaluated by the Clustal Omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/) and used for global alignment. Synthetic coding DNA for full-length human HSPA5 without the leading signal peptide for ER translocation (HSPA5-FL: AA 25 to 654), N-terminal end (HSPA5-N; AA 25 to 426), C-terminal end (HSPA5-C; AA 432 to 654), HSPA5 without KDEL retention motif (HSPA5-KDEL; AA 25 to 634), full-length HSPA1 (HSPA1-FL; AA 1 to 641), N-terminal (HSPA1-N; AA 1 to 377), and C-terminal (HSPA1-C; AA 383 to 641) were synthesized by Epoch Life Science Inc. in frame with the pET28a vector containing a His-tag epitope at the N-terminus end. Escherichia coli BL21 (DE3) cells were transformed with the constructs and grown at 37 °C in LB medium in the presence of kanamycin (50 μg/mL). Protein expression was induced (A600 nm ~ 0.6) by the addition of IPTG (0.4 mM) at 30 °C. After 4 h, the cells were harvested by centrifugation at 2600×g for 30 min. The cell pellet was lysed, and protein purification was performed as described by Dores-Silva et al. (2015). Briefly, proteins were isolated by a single chromatographic step on a Ni2+-immobilized affinity column, step-washed with 20 mM phosphate buffer pH 7.5 containing 100 mM NaCl. The protein was eluted in the same buffer with the addition of imidazole (500 mM). Proteins were dialyzed against 50 mM Tris-HCl buffer (pH 7.5). The purity of recombinant proteins was evaluated by lithium dodecyl sulfate-polyacrylamide gel electrophoresis (LDS-PAGE). The protein concentration was determined spectrophotometrically using the extinction coefficient calculated for each protein under native conditions.
Liposome preparation and incorporation of proteins
Liposomes were prepared using the extrusion method, as previously described (Lopez et al. 2016). Briefly, palmitoyl-oleoyl phosphocholine (POPC), palmitoyl-oleoyl phosphoserine (POPS), palmitoyl-oleoyl phosphoethanolamine (POPE), and cardiolipin, 1′,3′-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-glycerol (CL) (Avanti Polar Lipids) were dissolved in CHCl3 (10 mg/mL) and dried under a nitrogen gas stream. The dried lipid (400 μg) was resuspended in 50 mM Tris-HCl buffer (pH 7.5), vortexed 6 times for 30 s with intervals of 5 min. The suspension was extruded using a 100 nm membrane filter by 15 passages. The size of the liposomes (100 nm) was confirmed by Nano Tracking Analysis (NTA) using the Nanosight NS300 instrument. Liposomes (400 μg) were incubated with the respective proteins in 50 mM Tris-HCl buffer (pH 7.5) for 30 min at 25 °C with continuous agitation. The mixture was centrifuged at 100,000×g for 1 h at 4 °C. The pellet was resuspended (300 μL) in 100 mM Na2CO3 buffer (pH 11.5) and centrifuged at 100,000×g for 1 h at 4 °C. The pellet was solubilized in sample buffer in the presence or absence of 10 mM of β-mercaptoethanol (βM), and the proteins were resolved by LDS-PAGE and visualized by staining with Coomassie Brilliant Blue R-250 (Thermo Fisher Scientific, Waltham, MA).
Spectroscopy studies
The secondary structure of HSPA5-FL was assessed by circular dichroism in the absence or presence of POPS or CL liposomes. Circular dichroism measurements were performed using a J-815 spectropolarimeter (Jasco Inc.) coupled to the Peltier system PFD 425S. HSPA5-FL in Tris-HCl 50 mmol L−1 (pH 7.5) buffer at final concentrations between 5 and 10 μM was analyzed in 1 mm path-length cuvette at 25 °C. The tests were conducted before and after HSPA5-FL incorporation into the liposomes at a final concentration of 0.6 mg/mL. All data were normalized to the mean residue ellipticity ([Θ]) (Dores-Silva et al. 2015).
Isothermal titration calorimetry
The interaction of HSPA5-FL with POPS and CL liposomes was assessed by isothermal titration calorimetry (ITC) using an iTC200 microcalorimeter (GE Healthcare Life Sciences). Experiments were conducted by titrating seventeen aliquots of 2 μL of liposomes (POPS or CL) at 3 mM into 203.8 μL of 10–15 μM of HSPA5-FL, at 25 °C. The experimental isotherm curves were analyzed to yield the association constant (KA), apparent enthalpy change (ΔHapp), and stoichiometric coefficient (n), as previously described (Batista et al. 2015; Borges and Ramos 2006; Dores-Silva et al. 2015). The dissociation constant (KD) was obtained as the inverse of KA. The apparent Gibbs energy change (ΔGapp) was calculated using the relation [∆G]app = − RTlnKA, and the apparent entropy change [ΔS]app was determined by the equation: [∆G]app = [∆H]app – T[∆S]app.
Recombinant HSPA5 resistance to protease digestion
Recombinant HSPA5-FL or HSPA1-FL (5 μg) incorporated into POPS or CL liposomes (400 μg) was incubated with proteinase K (5 μg/mL) for 1 h at 25 °C in 50 mM Tris-HCl buffer pH 7.5, and the HSPA liposomes were centrifuged at 100,000×g for 1 h at 4 °C. Pellets were resuspended in LDS sample buffer in the presence of 10 mM of βM, resolved by LDS-PAGE, and visualized using Coomassie Brilliant Blue R-250 staining.
Aggregation assay
The aggregation of HSPA5-FL or HSPA1-FL and subdomains (20 μg) incorporated into liposomes (POPS or CL, 400 μg) was monitored by absorbance at 340 nm every 10 s for approximately 1 h while maintaining the temperature constant at 25 °C (Kiraly et al. 2020). HSPA5-FL and HSPA1-FL in the absence of liposomes were subjected to the same procedure, as controls. The assay was performed in quadruplicate.
Results
Comparison between HSPA5 and HSPA1: identity, production, and purification
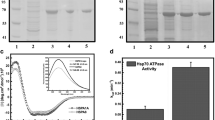
A comparison of the alignment of full-length HSPA5 and HSPA1 showed that they share about 64% identity in their primary sequence, with the N-terminal end displaying approximately 68% identity and the C-terminal end about 58% (Fig. 1a). HSPA5 has an ER translocation leading signal at the N-terminus end (Fig. 1a, shadow underlying box) and an ER retention signal (KDEL) at the C-terminus end (Fig. 1a, hexagon). HSPA5 contains two cysteine residues, one within the N-terminus end and the second within the C-terminal end (Fig. 1a, black boxes). In contrast, HSPA1 has five cysteine groups, three on the N-terminal end, and two on the C-terminal end (Fig. 1a, black boxes). The cysteine at the N-terminal end of HSPA5 is the only one in common with HSPA1 (Fig. 1a, black box with an arrow). We generated several HSPA5 constructs to investigate their interaction with lipid membranes, including full-length (HSPA5-FL), a deletion of the ER retention signal (HSPA5-KDEL), and the N- or C-terminus end domains (HSPA5-N and HSPA5-C, respectively). In addition, we generated full-length HSPA1 (HSPA1-FL) and both N- or C-terminus end domains (HSPA1-N and HSPA1-C, respectively). The proteins were produced in E. coli, purified by Ni2+-affinity chromatography with high purity (more than 92%), and visualized by LDS-PAGE (Fig. 1b and c, respectively).
HSPA5 and HSPA1 share a high identity, and the proteins were isolated with high purity. a The global alignment between HSPA5 and HSPA1 indicates 64% of identity, which is considered high. The N-terminal of these proteins presents approximately 68% (flat gray rectangle), and the C-terminal presents 58% of identity (dark gray flat rectangle). Cysteine groups are indicated by black boxes, two for HSPA5 and five for HSPA1. The ER translocation signal peptide is displayed by the shadow area, and the ER retention signal (KDEL) is indicated by a hexagon. b HSPA5-FL, HSPA5-KDEL, HSPA5-N, and HSPA5-C. c HSPA1-FL, HSPA1-N, and HSPA1-C. Proteins were purified as described in Methods. All steps for the production and isolation process were visualized by LDS-PAGE and Coomassie blue staining. All proteins were obtained with more than 92% purity.
Full-length and both N- or C-terminus end domains of HSPA5 and HSPA1 interact preferentially with negatively charged phospholipids
We evaluated the ability of HSPA5 to associate with membranes containing a variety of phospholipids. Unilamellar liposomes were made of POPC, POPS, POPE, or CL phospholipids, which are common components of cellular membranes. These liposomes were incubated with HSPA5-FL, HSPA5-KDEL, HSPA5-N, or HSPA5-C for 30 min at 25 °C. Then, liposomes were separated from the soluble protein by high-speed centrifugation (100,000×g), and the resulting liposome pellet was analyzed by LDS-PAGE as previously described (Armijo et al. 2014). HSPA5-FL showed a similar level of incorporation into liposomes made of POPS or CL, which are both negatively charged phospholipids (60% ± 6 and 54% ± 5, respectively). In contrast, the protein was not inserted into liposomes containing POPC or POPE (Fig. 2a). This finding is consistent with previously reported observations indicating preferential association of HSPA1 for negatively charged phospholipids (Arispe et al. 2004; Armijo et al. 2014). Removal of the ER retention signal (HSPA5-KDEL) did not affect the incorporation level into negatively charged liposomes in comparison with the full-length protein (Fig. 2a, Table 1). Both HSPA5-N and HSPA5-C also incorporated preferentially into negatively charged phospholipids, but the level of insertion was reduced as opposed to full-length protein (Fig. 2a, Table 1). These observations suggest that the full-length protein is required for maximum incorporation into the lipid bilayer, but each N- and C-terminal end domain has an intrinsic capacity to interact with liposomes. We also tested whether the N-terminal and C-terminal ends of HSPA1 were capable of getting incorporated into liposomes. The incorporation of these protein fragments into negatively charged phospholipid liposomes was also reduced with respect to full-length protein (Fig. 2b, Table 1).
HSPA5 and HSPA1 and full-length and both N- and C-terminus end domains interact preferentially with negatively charged liposomes. a HSPA5-FL, HSPA5-KDEL, HSPA5-N, and HSPA5-C and b HSPA1-FL, HSPA1-N, and HSPA1-C (4 μg) were incubated with liposomes made of POPC, POPE, POPS, and CL (400 μg) in 50 mM Tris-HCl buffer (pH 7.5) for 30 min at 25 °C with continuous agitation. The mixture was centrifuged at 100,000×g for 1 h at 4 °C. The pellet was resuspended (300 μL) in 100 mM Na2CO3 buffer pH 11.5 and centrifuged at 100,000×g for 1 h at 4 °C. Proteoliposomes were solubilized in sample buffer containing 10 mM βM, and the proteins were resolved by LDS-PAGE and visualized by staining with Coomassie Brilliant Blue R-250. HSPA5-FL, HSPA5, and HSPA1-FL showed high affinity for the negatively charged liposomes (POPS and CL). The same behavior was observed for the N-terminus and C-terminus end domain of both proteins. Percentages of incorporation into liposomes are presented in Table 1.
HSPA5 interaction with POPS and CL liposomes is mainly driven by entropic changes
The interaction of HSPA5-FL with POPS or CL liposomes was evaluated by ITC, and the isothermogram profile showed an exothermic reaction (Fig. 3a and b). The results indicated that interactions between HSPA5-FL and liposomes were entropically and enthalpically driven, with a higher entropic contribution. The thermodynamic signatures for HSPA5 interaction with POPS and CL liposomes are displayed in Fig. 3c. The thermodynamic parameters obtained from these analyses are summarized in Table 2. The interaction of both HSPA5 with POPS or CL displayed a ΔGapp about − 6000 cal mol, a very low enthalpy ΔHapp (− 300 ± 50 cal mol for HSPA5-POPS liposomes and − 400 ± 70 cal mol for HSPA5-CL liposomes), and high values for – TΔSapp (− 5800 ± 600 cal mol deg for HSPA5-POPS liposomes and − 6100 ± 500 cal mol−1 deg−1 for HSPA5-CL liposomes) indicating that the entropy variation has a critical role on the HSPA5-liposomes interaction. The KD for HSPA5 association with liposomes made with POPS or CL was 3.5 ± 0.8 μmol/L and 1.6 ± 0.5 μmol/L, respectively. We also assessed the interaction between HSPA5-N, HSPA5-C, HSPA1-N, and HSPA1-C with POPS and CL liposomes. No significant changes in energy variation were observed within the interaction with POPS or CL liposomes, at least in the tested conditions. The secondary structure of HSPA5, before and after insertion into POPS or CL liposomes, was evaluated by far-UV circular dichroism. No differences in secondary structures were observed before or after incorporation into POPS or CL liposomes indicating that the protein within a hydrophobic environment did not change significantly within the secondary structure (Fig. 4).
HSPA5 interaction with POPS and CL liposomes is mainly driven by entropic changes. The thermodynamics parameters were obtained using an iTC200 microcalorimeter. Seventeen 2-μL aliquots of POPS or CL liposomes at 3 mM were injected into 203.8 μL of 10–15 μmol/L recombinant HSPA5-FL, at 25 °C. All solutions were prepared in 50 mM Tris-HCl (pH 7.4) buffer. The experimental isotherm curves were analyzed to obtain the KA and ΔHapp. a HSPA5-FL interaction with POPS liposomes. b HSPA5 interaction with CL liposomes. Protein insertion into liposomes was entropically and enthalpically driven, with a higher entropic contribution. c Thermodynamic signatures for HSPA5 interaction with POPS and CL shown that the interaction of HSPA5-FL with POPS or CL liposomes has a discrete enthalpy contribution, and it is pretty much entropically driven.
HSPA5 secondary structure did not change after insertion into liposomes. Circular dichroism performed for HSPA5-FL (5–10 μM) before and after incorporation in POPS or CL liposomes (3 mM) in 50 mM Tris-HCl buffer pH 7.5. A J-815 spectropolarimeter coupled to the Peltier system, at 25 °C, was used in these experiments. The secondary structures of HSPA5-FL APO and after incorporated in POPS or CL liposomes presented no differences, suggesting that incorporation of protein into liposomes had no effect in the secondary structure.
HSPA5 and HSPA1 appear to interact differently with POPS or CL liposomes
In order to get an idea of the topology of HSPA5-FL and HSPA1-FL within liposomes, these two proteins, after insertion into the liposomes, were incubated with proteinase K for 30 min at 25 °C. Then, the liposomes were pelleted by high-speed centrifugation and analyzed by SDS-PAGE. HSPA5-FL and HSPA1-FL within POPS liposomes were highly susceptible to proteinase K degradation (Fig. 5a and b, respectively), with the recovery of discrete low molecular weight peptides (about 18 kDa) that were still associated within the liposomes (see bracket at the bottom of the gel). This observation suggests that a good portion of these proteins are exposed on the surface of the liposome.
HSPA5 and HSPA1 interact differently with POPS or CL liposomes. HSPA5-FL or HSPA1-FL (4 μg) were incubated with POPS or CL liposomes (400 μg) in 50 mM Tris-HCl buffer (pH 7.5) for 30 min at 25 °C with continuous agitation. The mixture was centrifuged at 100,000×g for 1 h at 4 °C. The pellet was resuspended (300 μL) in 100 mM Na2CO3 buffer pH 11.5 and centrifuged again at 100,000×g for 1 h at 4 °C. The pellet was resuspended in 50 mM Tris-HCl buffer pH 7.4 and incubated with proteinase K (5 μg/mL) for 30 min at 25 °C and centrifuged for 1 h at 100,000×g. The pellet was solubilized in sample buffer containing 10 mM βM, and the proteins were resolved by LDS-PAGE and visualized by staining with Coomassie Brilliant Blue R-250. a HSPA5 in POPS liposomes. b HSPA1-FL in POPS liposomes. c HSPA5-FL in CL liposomes. d HSPA1-FL in CL liposomes. Brackets indicate low molecular weight peptides retained within the liposomes after protease digestion.
In contrast, HSPA5-FL and HSPA1-FL inserted into CL liposomes were very resistant to proteinase K digestion (Fig. 5c and d, respectively), suggesting that these proteins are completely embedded into the lipid bilayer or within the liposome lumen. Another explanation for the resistance of HSPA5-FL and HSPA1-FL within CL liposomes to proteinase K digestion is that the proteins within these liposomes are sheltered, avoiding the interaction with the protease, which could be due to liposome aggregation. This possibility was evaluated by monitoring changes in absorbance at 340 nm (A340nm) at a constant temperature (25 °C) for 60 min. Indeed, the aggregation of HSPA5-FL or HSPA1-FL CL liposomes was evident (Fig. 6a). This observation was confirmed by direct visualization of the aggregates on 96-well plates on which the protein solution became cloudy suggesting the formation of particular forms or aggregates (Fig. 6b). In contrast, the proteins did not show signs of aggregation in the absence of liposomes as detected by the lack of changes in absorption (Fig. 6a) or direct visualization in which the solution remains clear (Fig. 6b). Furthermore, HSPA5-FL or HSPA1-FL within POPS liposomes or in the absence of liposomes did not show any aggregation process (Fig. 6c and d). These observations suggest that the resistance to proteinase K digestion is likely due to the aggregation process.
CL liposomes aggregated in the presence of Hsp70s. The aggregation of POPS or CL liposomes with or without incorporation of HSPA5-FL (a, b) or HSPA1-FL (c, d) in 50 mM Tris-HCl 50 buffer (pH 7.5) was monitored by the changes in absorbance at 340 nm every 10 s for 1 h at 25 °C. a and c aggregation pattern. b and d visualization of aggregates in 96-well plates.
HSPA5 and HSPA1 oligomerize upon insertion into POPS or CL liposomes via a disulfide bond within the N-terminal end domain
Prior studies have shown that several members of the HSPA family form dimers and oligomers in solution (Guidon and Hightower 1986; Benaroudj et al. 1996; Gao et al. 1996; Aprile et al. 2013), which was modulated by the presence of nucleotides (Kim et al. 1992; Benaroudj et al. 1996) or by temperature (Angelidis et al. 1999; Kiraly et al. 2020). Moreover, it was reported that HSPA1 formed high molecular weight oligomers within liposomes made of POPS with different types of saturation as determined by Western blotting after LDS-PAGE in the presence of reducing agents (Armijo et al. 2014). Thus, we investigate whether or not HSPA5-FL could also form oligomers after insertion into POPS or CL liposomes. We detected HSPA5-FL oligomers after insertion into POPS or CL liposomes only if the material was subjected to LDS-PAGE in the absence of βM, but not if the reducing agent was used (Fig. 7a and c, see bracket). Dimers were also detected when the HSPA5-N was incorporated into POPS or CL liposomes in the absence of βM (Fig. 7e and g), whereas these dimers were not detected if the C-terminus end of this protein was used even in the absence of βM (HSPA5-C data no shown). Similar observations were made for HSPA1-FL and HSPA1-N (Fig. 7b, d, f, and h).
Aggregation profile of HSPA5 and HSPA1 and the respective N-terminal end domain after incorporation into liposomes. HSPA5-FL and HSPA5-N or HSPA1-FL and HSPA1-N and HSPA1-N were incorporated into POPS (a, b, e, f) or CL (c ,d ,g ,h) liposomes in 50 mM Tris-HCl buffer (pH 7.5) for 30 min at 25 °C. The mixture was centrifuged at 100,000×g for 1 h at 4 °C. The pellet was resuspended (300 μL) in 100 mM Na2CO3 buffer (pH 11.5) and centrifuged at 100,000×g for 1 h at 4 °C. The pellet was solubilized in sample buffer containing or not 10 mM βM, and the proteins were resolved by LDS-PAGE and visualized by staining with Coomassie Brilliant Blue R-250. The presence of dimers and oligomers is indicated.
Discussion
The biological role of HSPs has bypassed their initial interest as intracellular molecular chaperones after their detection outside the cells. Extracellular HSPs appear to play a signaling function mediating the communication between cells or perhaps as stress sentinel beacons (De Maio 2011). A major question that has emerged is how these cytosolic HSPs, which lack a secretory signal, could escape the intracellular environment and reach the outside milieu. HSPs are not alone in this regard since many other cytosolic proteins follow the same pathway moving into the extracellular environment by an unclear mechanism, namely, the nonclassical secretory pathway (Nickel and Seedorf 2008). Although many mechanisms have been proposed to explain this alternative export pathway, the crossing of the plasma membrane is a necessary requirement for the process. However, the conventional wisdom states that the crossing of the plasma membrane by a protein without a bona fide hydrophobic domain is thermodynamically forbidden. In spite of these assumptions, HSPs, particularly Hsp70 (HSPA1), is present embedded into the plasma membrane of transformed cells (Multhoff et al. 1995; Multhoff and Hightower 1996). The disbelief in these observations was challenged by further studies demonstrating that several HSPAs were capable of inserting into lipid bilayers opening ion conductance pathways (Arispe and De Maio 2000; Vega et al. 2008; Macazo and White 2014). Even with this evidence, the mechanism for membrane insertion has remained elusive, particularly since these proteins lack a consensus hydrophobic domain that could explain membrane interaction.
In contrast with other HSPAs, HSPA5 (Grp78 or BIP) is present in the secretory pathway that opens the possibility for its traffic outside the cells. Indeed, HSPA5 was reported secreted into the extracellular milieu (Delpino and Castelli 2002; Zhang et al. 2013). Even more interesting, this protein was also detected on the surface of cancer cells (Delpino and Castelli 2002; Zhang et al. 2010, 2013). These observations have raised several intriguing questions. Firstly, how is HSPA5 exported outside the ER in spite of presenting a retaining signal (KDEL)? HSPA5 was found to escape this subcellular compartment after ER stress (Zhang et al. 2013). Secondly, how does the protein get inserted into the plasma membrane? Prior studies have suggested that HSPA5 displays several potential domains, particularly within the C-terminus end (Tseng et al. 2019), that could explain the insertion into the plasma membrane (Zhang et al. 2013; Tsai et al. 2015). In the current study, we directly addressed how HSPA5 could interact with lipid membranes by analyzing their association with liposomes. We found that HSPA5 also has the same specificity of other HSPAs for negatively charged phospholipids, such as phosphatidylserine and palmitoyloleoyl phosphatidylglycerol (Arispe et al. 2004; Schilling et al. 2009; Armijo et al. 2014). Moreover, both N- and C-terminus end domains of the protein could independently get inserted into negatively charged liposomes, but with less efficiency than the full-length protein, suggesting a synergy between the domains in binding to the liposomes. This observation supports the idea that multiple domains of HSPA5 are associated with the lipid membrane. However, the full-length protein is required for maximum insertion.
The insertion of HSPA5 into liposomes was mediated by entropic changes, probably due to changes in water molecules surrounding the protein, without affecting the secondary structure of the protein. However, the most interesting observation is that HSPA5, as well as HSPA1 undergoing an oligomerization process upon membrane insertion, is mediated by the formation of disulfide bonds. HSPA5 contains two cysteine groups, one at the beginning of the N-terminus end and the second at the C-terminus end (Fig. 1a). Although there is no evidence that these cysteine groups could form intramolecular bridges, we think that they produce intermolecular bonds coordinating the assembly of oligomeric complexes. This multi-polypeptide complex appears to assemble after insertion into the lipid bilayer since it was observed only in the presence of phospholipids (Fig. 6). Therefore, the lipid bilayer may provide an oxidative environment capable of forming these disulfide bridges. Moreover, we found that the N-terminus end domain was capable of forming dimers, whereas the C-terminus end was not. Thus, the cysteine within the N-terminus end of the proteins may be in the right conformation to form intermolecular complexes. A puzzling observation is that multiple oligomers were only observed in the case of the full-length protein, whereas only dimers were noticed when the N-terminus domain was inserted into the liposomes. A possible explanation for these results is that intermolecular disulfide bonds could be formed between the N-terminus end and the C-terminus domain of an adjacent polypeptide. Tandem repeats of HSPA5 in an antiparallel conformation may be assembled within the lipid bilayer (Fig. 8). These observations were also seen for HSPA1 that share the same cysteine group on the N-terminus end as HSPA5 (Fig. 1a). Indeed, cytosolic Hsp70 (HSPA1) formed antiparallel dimers (Morgner et al. 2015). Other studies have also detected the presence of HSPA1 oligomerization forms upon insertion into POPS liposomes (Armijo et al. 2014). Interestingly, DnaK, which contains only one cysteine group, formed dimers but not oligomers (Lopez et al. 2016). However, the best evidence that HSPA could form oligomers within membranes was provided by early reports showing that HSPA8 and HSPA1 form ion channels after insertion into artificial lipid bilayers (Arispe and De Maio 2000; Vega et al. 2008). The possibility for the assembly of HSPA multimeric complexes was confirmed by elegant studies using atomic force microscopy showing the formation of protein clusters upon insertion of HSPA1 into dipalmitoyl phosphatidylserine bilayers (Lamprecht et al. 2018).
A proteolysis approach was used to learn about the topology of HSPAs within the membrane of liposomes. HSPA1 and HSPA5 within POPS liposomes were very sensitive to proteinase K digestion, suggesting that a great portion of the protein is located outside the liposome. Several small regions of HSPA1 are indeed outside the liposomes and spread along the entire length of the protein (Lopez et al. 2016), and a 27-kDa peptide was found to be resistant to chymotrypsin digestion indicating that it was inserted into the liposome (Armijo et al. 2014). These observations may also be consistent with an antiparallel oligomerization complex, as described above, in which regions within both the N- and C-terminus end could be exposed on the surface of the liposome. In contrast, both HSPA1 and HSPA5 inserted into CL liposomes were resistant to proteinase K digestion. This observation is apparently due to reduced access for this protease since HSPA1/HSPA5 within CL liposomes forms large aggregates at 25 °C that likely hindered accessibility for digestion.
Although our results showed a great deal of selectivity of HSPA5 for negatively charged lipids, it is still unclear how the protein interacts with the natural membranes in which this protein resides. The lumen of the ER is not rich in phosphatidylserine moieties (Yeung et al. 2008) that may explain why the protein has not been detected in the membrane of this subcellular compartment. Similarly, phosphatidylserine is constrained to the inner side of the plasma membrane as a result of an active energy-dependent process to maintain the asymmetry of the plasma membrane (Leventis and Grinstein 2010). However, phosphatidylserine is capable of flipping out the plasma membrane by a process enhanced during cell activation (Elliott et al. 2005; Dillon et al. 2000) or pathological processes (Zwaal et al. 2005). Thus, HSPA5 may encounter phosphatidylserine microdomains within the surface of the plasma membrane that facilitates its insertion, particularly in cancer cells. Although several functions have been ascribed to cell-surface HSPA5, the biological significance of this phenomenon needs further exploration. HSPA5 also showed affinity for CL that is a major component (18%) of the inner mitochondrial membrane (Horvath and Daum 2013). The presence of HSPA5 has been detected within the mitochondrial matrix associated with the inner mitochondrial membrane upon activation of the unfolded protein response (Sun et al. 2006). The presence and possible interaction of HSPA5 with the CL-rich inner membrane may be part of a mechanism to maintain the proper import of proteins into the mitochondria, perhaps in cooperation with HSPA9 (mtHsp70), that is constitutively present in this compartment (De Maio 1999; Daugaard et al. 2007).
References
Angelidis CE, Lazaridis I, Pagoulatos GN (1999) Aggregation of hsp70 and hsc70 in vivo is distinct and temperature-dependent and their chaperone function is directly related to non-aggregated forms. Eur J Biochem 259:505–512
Aprile FA, Dhulesia A, Stengel F, Roodveldt C, Benesch JL, Tortora P, Robinson CV et al (2013) Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS One 8:e67961
Arispe N, De Maio A (2000) ATP and ADP modulate a cation channel formed by Hsc70 I acidic phospholipid membranes. J Biol Chem 275:30839–30843
Arispe N, Doh M, Simakova O, Kurganov B, De Maio A (2004) Hsc70 and Hsp70 interact with phosphatidylserine on the surface of PC12 cells resulting in a decrease in viability. FASEB J 18:1636–1645
Armijo G, Okerblom J, Cauvi DM, Lopez V, Schlamadinger DE, Kim J, Arispe N, de Maio A (2014) Interaction of heat shock protein 70 with membranes depends on the lipid environment. Cell Stress Chaperones 19:877–886
Basu S, Srivastava PK (2000) Heat shock proteins: the fountainhead of innate and adaptive immune responses. Cell Stress Chaperones 5:443–451
Batista FA, Gava LM, Pinheiro GM, Ramos CH, Borges JC (2015) From conformation to interaction: techniques to explore the Hsp70/Hsp90 network. Curr Protein Pept Sci 16:735–753
Benaroudj N, Triniolles F, Ladjimi MM (1996) Effect of nucleotides, peptides and unfolded proteins on the self-association of the molecular chaperone HSC70. J Biol Chem 271:18471–18476
Borges JC, Ramos CH (2006) Spectroscopic and thermodynamic measurements of nucleotide-induced changes in the human 70-kDa heat shock cognate protein. Arch Biochem Biophys 452:46–54
Calderwood SK (2018) Heat shock proteins and cancer: intracellular chaperones or extracellular signalling ligands? Philos Trans R Soc Lond B Biol Sci 373(1738). pii: 20160524
Casanova-Morales N, Quiroga-Roger D, Alfaro-Valdés HM, Alavi Z, Lagos-Espinoza MIA, Zocchi G, Wilson CAM (2018) Mechanical properties of BiP protein determined by nano-rheology. Protein Sci 27:1418–1426
Chu H, Chan CM, Zhang X, Wang Y, Yuan S, Zhou J, Au-Yeung RK et al (2018) Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem 293:11709–11726
da Silva KP, Borges JC (2011) The molecular chaperone Hsp70 family members function by a bidirectional heterotrophic allosteric mechanism. Protein Pept Lett 18:132–142
Daugaard M, Rohde M, Jäättelä M (2007) The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett 581:3702–3710
De Maio A (1999) Heat shock proteins: facts, thoughts, and dreams. Shock 11:1–12
De Maio A (2011) Extracellular heat shock proteins, cellular export vesicles, and the stress observation system: a form of communication during injury, infection, and cell damage. It is never known how far a controversial finding will go! Dedicated to Ferruccio Ritossa. Cell Stress Chaperones 16:235–249
De Maio A (2014) Extracellular Hsp70: export and function. Curr Protein Pept Sci 15:225–231
De Maio A, Vazquez D (2013) Extracellular heat shock proteins: a new location, and a new function. Shock 40:239–246
De Maio A, Cauvi DM, Capone R, Bello I, Egberts WV, Arispe N, Boelens W (2019) The small heat shock proteins, HSPB1 and HSPB5, interact differently with lipid membranes. Cell Stress Chaperones 14:947–956
Delpino A, Castelli M (2002) The 78 kDa glucose-regulated protein (GRP78/BIP) is expressed on the cell membrane, is released into cell culture medium and is also present in human peripheral circulation. Biosci Rep 22:407–420
Dillon SR, Mancini M, Rosen A, Schlissel MS (2000) Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J Immunol 164:1322–1332
Dores-Silva PR, Barbosa LR, Ramos CH, Borges JC (2015) Human mitochondrial Hsp70 (mortalin): shedding light on ATPase activity, interaction with adenosine nucleotides, solution structure and domain organization. PLoS One 10:e0117170
Elliott JI, Surprenant A, Marelli-Berg FM, Cooper JC, Cassady-Cain RL, Wooding C, Linton K, Alexander DR, Higgins CF (2005) Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat Cell Biol 7:808–816
Gao B, Eisenberg E, Greene L (1996) Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate. J Biol Chem 271:16792–16797
Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P (2013) Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol 5:a013169
Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, Multhoff G (2005) Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res 65:5238–5247
Guidon PT, Hightower LE (1986) Purification and initial characterization of the 71-kilodalton rat heat-shock protein and its cognate as fatty acid binding proteins. Biochemistry 128:257–266
Haas IG, Wabl M (1983) Immunoglobulin heavy chain binding protein. Nature 306:387–389
Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858
Honda T, Horie M, Daito T, Ikuta K, Tomonaga K (2009) Molecular chaperone BiP interacts with Borna disease virus glycoprotein at the cell surface. J Virol 83:12622–12625
Horvath SE, Daum G (2013) Lipids of mitochondria. Prog. Lipid Res 52:590–614
Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA (2020) COVID-19 spike-host cell receptor GRP78 binding site prediction. J Inf Secur 80:554–562
Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14:105–111
Kim D, Lee YJ, Corry PM (1992) Constitutive HSP70 oligomerization and its dependence on ATP binding. J Cell Physiol 153:353–361
Kiraly VTR, Dores-Silva PR, Serrão VHB, Cauvi DM, De Maio A, Borges JC (2020) Thermal aggregates of human mortalin and Hsp70-1A behave as supramolecular assemblies. Int J Biol Macromol 146:320–331
Kottom TJ, Hebrink DM, Limper AH (2018) Binding of Pneumocystis carinii to the lung epithelial cell receptor HSPA5 (GRP78). J Med Microbiol 67:1772–1777
Lamprecht C, Gehrmann M, Madl J, Römer W, Multhoff G, Ebner A (2018) Molecular AFM imaging of Hsp70-1A association with dipalmitoylphosphatidylserine reveals membrane blebbing in the presence of cholesterol. Cell Stress Chaperones 23:673–683
Lang BJ, Guerrero-Gimenez ME, Prince TL, Ackerman A, Bonorino C, Calderwood SK (2019) Heat shock proteins are essential components in transformation and tumor progression: cancer cell intrinsic pathways and beyond. Int J Mol Sci 20:pii: E4507
Lee AS (1992) Mammalian stress response: induction of the glucose-related protein family. Curr Opin Cell Biol 4:267–273
Leventis PA, Grinstein S (2010) The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 39:407–427
Lopez V, Cauvi DM, Arispe N, De Maio A (2016) Bacterial Hsp70 (DnaK) and mammalian Hsp70 interact differently with lipid membranes. Cell Stress Chaperones 21:609–616
Luo B, Lee AS (2013) The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 32:805–818
Macazo FC, White RJ (2014) Monitoring charge flux to quantify unusual ligand-induced ion channel activity for use in biological nanopore-based sensors. Anal Chem 86:5519–5525
Mayer MP, Brehmer D, Gässler CS, Bukau B (2001) Hsp70 chaperone machines. Adv Protein Chem 59:1–44
Morgner N, Schmidt C, Beilsten-Edmands V, Ebong IO, Patel NA, Clerico EM, Kirschke E, Daturpalli S, Jackson SE, Agard D, Robinson CV (2015) Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep 11:759–769
Multhoff G, Hightower LE (1996) Cell surface expression of heat shock proteins and the immune response. Cell Stress Chaperones 1:167–176
Multhoff G, Botzler C, Wiesnet M, Müller E, Meier T, Wilmanns W, Issels RD (1995) A stress-inducible 72-kDa heat-shock protein (HSP72) is expressed on the surface of human tumor cells, but not on normal cells. Blood 85:2124–2131
Nickel W, Seedorf M (2008) Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu Rev Cell Dev Biol 24:287–308
Oikonomou C, Hendershot LM (2020) Disposing of misfolded ER proteins: a troubled substrate’s way out of the ER. Mol Cell Endocrinol 500:110630
Pfaffenbach KT, Lee AS (2011) The critical role of GRP78 in physiologic and pathologic stress. Curr Opin Cell Biol 23:150–156
Pobre KFR, Poet GJ, Hendershot LM (2019) The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: getting by with a little help from Erdj friends. J Biol Chem 294:2098–2108
Schilling D, Gehrmann M, Steinem C, De Maio A, Pockley AG, Abend M, Molls M et al (2009) Binding of heat shock protein 70 to extracellular phosphatidylserine promotes killing of normoxic and hypoxic tumor cells. FASEB J 23:2467–2477
Sun FC, Wei S, Li CW, Chang YS, Chao CC, Lai YK (2006) Localization of GRP78 to mitochondria under the unfolded protein response. Biochem J 396:31–39
Suzuki CK, Bonifacino JS, Lin AY, Davis MM, Klausner RD (1991) Regulating the retention of t-cell receptor alpha chain variants within the endoplasmic reticulum: Ca(2+)-dependent association with BiP. J Cell Biol 114:189–205
Tsai YL, Zhang Y, Tseng CC, Stanciauskas R, Pinaud F, Lee AS (2015) Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J Biol Chem 290:8049–8064
Tseng CC, Zhang P, Lee AS (2019) The COOH-terminal proline-rich region of GRP78 is a key regulator of its cell surface expression and viability of tamoxifen-resistant breast. Cancer Cells 21:837–848
Vega VL, Rodriguez-Silva M, Frey T, Gehrmann M, Diaz JC, Steinem C, Multhoff G et al (2008) Hsp70 translocates into the plasma membrane after stress and is released into the extracellular environment in a membrane-associated form that activates macrophages. J Immunol 180:4299–4307
Wang M, Kaufman RJ (2016) Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529:326–335
Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S (2008) Membrane phosphatidylserine regulates surface charge and protein localization. Science 319:210–213
Young JC (2010) Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol 88:291–300
Zhang Y, Liu R, Ni M, Gill P, Lee AS (2010) Chaperone and unfolded protein response regulator GRP78/BIP. J Biol Chem 285:15065–15075
Zhang Y, Tseng CC, Tsai YL, Fu X, Schiff R, Lee A (2013) Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complex with PI3K and enhances PI(3,4,5)P3 production. PLoS One 8:e80071
Zwaal RF, Comfurius P, Bevers EM (2005) Surface exposure of phosphatidylserine in pathological cells. Cell Mol Life Sci 62:971–988
Funding
This work was supported by the National Institutes of Health (NIH) grants R01 GM098455-04 and R01 GM114473-01 and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) process: 2012/50161-8, 2014/16646-0, 2016/22477-1, 2017/07335-9, and 2017/26131-5.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM1
(TIF 1896 kb)
Rights and permissions
About this article
Cite this article
Dores-Silva, P.R., Cauvi, D.M., Coto, A.L.S. et al. Interaction of HSPA5 (Grp78, BIP) with negatively charged phospholipid membranes via oligomerization involving the N-terminal end domain. Cell Stress and Chaperones 25, 979–991 (2020). https://doi.org/10.1007/s12192-020-01134-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-020-01134-9