Abstract
C3HC4-type zinc finger proteins are known to play important roles in various plant processes including regulation of growth and development, signaling networks, responses to abiotic stresses etc. The current study identifies and explores the involvement of TaZnF in plant stress response, mainly heat stress. TaZnF belongs to C4HC3-type zinc finger transcription factor. Phylogenetic analysis of TaZnF revealed strong sequence similarity to Brachypodium distachyon, a model system for crop species. Gene expression studies have revealed its role under diverse stress conditions including heat and cold conditions. The transcript level of TaZnF was found to be highest in seed and starts at the post anthesis period 3-5DAA, a more sensitive stage resulting in a negative influence on the yield of crop species. TaZnF possesses transcriptional activity. Overexpression of TaZnF in Arabidopsis thaliana conferred improved tolerance to both basal and high-temperature stress as observed from various assays examining their growth and development. The transgenics were recovered and showed early flowering compared to wild-type. They had larger primary roots, more lateral branching, bigger, and more numerous leaves, resulting in heavier fresh weight. Enhanced growth and early recovery resulted in bigger plants with more yield. Additionally, the overexpression Arabidopsis transgenics also showed considerable tolerance to cold and oxidative stress. These observations suggest that TaZnF acts as a positive regulator of thermal stress and thus can be of great significance in understanding and improving temperature stress tolerance in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is the second largest crop grown annually on more than 220 million hectares of cropland worldwide. As a crop that prefers relatively cool temperatures, wheat is sown in most parts of the world in late autumn or early winter and harvested before early summer. Of the various abiotic stress conditions, heat stress is the major environmental stress that is found to affect wheat productivity. According to reports wheat growing areas face continual heat stress while terminal heat stress is found to affect 40% of irrigated crop areas in over 50 countries (Fischer and Byerlee 1991; Reynolds et al. 2001). High temperatures above 30 °C are reported to affect the grain weight by reducing grain filling via suppression of photosynthesis and starch synthesis in the endosperm.
Therefore, it is important to understand and unravel the molecular mechanisms of stress-responsive genes in plants. These stress conditions lead to an array of molecular, biochemical, physiological, and morphological responses in plants so that they can cope with these adverse conditions. The adaptations at the molecular level are modulated by a large number of transcription factors (TF) mediating the stress response. These TF are known to be major and essential regulators of various plant cellular and physiological responses, especially the environmental stimuli. These include several families like zinc finger, NAC, CUCs (cup-shaped cotyledon) AP2, WRKY, and some bZIP families which have been found to play a major role in the stress response pathway.
Zinc-binding proteins form one of the largest and best studied transcription factor families in eukaryotes, displaying variable secondary structures and enormous functional diversity. ZFP families have been divided into nine types according to their structural and functional difference, i.e., C2H2, C8, C6, C3HC4, C2HC, C2HC5, C4, C4HC3, and CCCH (C and H represent cysteine and histidine, respectively). This zinc finger binds a zinc ion in order to stabilize the three-dimensional structure through its cysteine and histidine residues (Takatsuji 1998). Previous studies of RING zinc finger proteins from various plants show that they are linked to a range of environmental stress processes, for example, drought and other abiotic stresses as well as disease resistance.
C3HC4-type RING finger proteins constitute the largest family in the plant kingdom and play important roles in many of the physiological processes of plant life. The RING domain of RING finger proteins was first identified as a DNA-binding motif in the transcription factor TFIIIA from Xenopus laevis (Brown et al. 1985; Miller et al. 1985). In addition to DNA, RING domains are reported to bind RNA, protein, and lipid substrates. The RING motif is small, consisting of four pairs of ligands that bind two ions. On the basis of the fifth co-ordination site (C/H), the RING family has been further categorized into seven types with different conserved motifs present: RING-H2, RING-HC, RING-v, RING-D, RING-S/T, RING-G, and RING-C2. Functions attributed to the RING domain itself include protein-protein interactions and ubiquitination (Kosarev et al. 2002; Stone 2005; Lim et al. 2010). Most RING finger proteins belong to E3 ubiquitin ligases that mediate the transfer of ubiquitin to target proteins and play important roles in diverse aspects of cellular regulation in plants, e.g., OsCOP1, a C3HC4-type RING finger proteins functions as an E3 ubiquitin ligase, which target photomorphogenesis-promoting transcription factors for ubiquitylation and degradation (Arnim and Deng 1993). Other C3HC4-type RING finger genes have also been identified and studied in rice, e.g., OsCOIN1 (OsRHC13), OsCOP1 (OsRHC11), OsXB3.1 (OsRHC24), and OsRHC1. These C3HC4 have also been studied in another well-known model plant, Arabidopsis (Jung et al. 2013).
The C3HC4 zinc finger has also been well studied in Arabidopsis and indicates their role in various environmental stress processes, e.g., drought and other abiotic stresses like salt, cold and heat stress, as well as disease resistance (Wang et al. 2006; Cheung et al. 2007; Kam et al. 2007; Zhang et al. 2007; Islam et al. 2009; Yang et al. 2014). Besides their role in various abiotic stresses, they also function in development and signaling processes linked to various stress processes like light perception, and peroxisome formation during root, and seed development (Pepper and Chory 1997; Xu and Li 2003; Chen and Ni 2006; Wang et al. 2006; Prestele et al. 2010).
In this study, we characterized the stress inducibilty of TaZnF and identified various other regulatory cues (spatial, temporal, and varietal specific cues) that may modulate its inducibilty. This study analyzes how changes in expression levels would affect the robustness of plants under stress conditions. TaZnF is C4HC3-type zinc finger, identified through the subtractive hybridization heat stress library of Triticum aestivum (CPAN1676). We found that the transcript is induced in response to heat and cold treatments. Moreover, overexpression of TaZnF in Arabidopsis shows enhanced tolerance to heat, cold, and oxidative stress. Together, these results suggest a positive role for TaZnF in mediating plant stress responses.
Materials and methods
Plant material, growth conditions, and stress treatments
Four genotypes of bread wheat (Triticum aestivum L.) cv.PBW343, HD2329, K7903, C306, and Arabidopsis ecotype Col0 were used throughout the present study. For seedling based studies, wheat plants were grown in plastic trays in a growth chamber maintained at 20 ± 1 °C; 16-h light and 8-h dark photoperiods in a daily cycle (Conviron, Canada) and Arabidopsis plants were grown on half-strength MS medium in Petri plates at 22 ± 1 °C and 16-h light and 8-h dark photoperiods in a daily cycle. For other stages of plant development, plants were grown in potted soil and soilrite mix for wheat and Arabidopsis, respectively. For stress treatments, plants were transferred to growth chambers set at specified temperatures and heat stress was provided for different time periods. After heat stress treatment, the control and stressed tissues were sampled, flash frozen in liquid nitrogen, and stored at −80 °C until RNA isolation. Remaining seedlings were returned to the growth chamber for recovery. For developing grains, potted wheat plants grown in the departmental garden were transferred for 2 h to the growth chambers maintained at 37 °C at 3, 5, 7, 10, and 20 days after anthesis. Sampling of tissues like lemma, palea, awn, glume, and developing grains was done from spikes of control and stressed plants at 37 °C and was stored at −80 °C until RNA isolation. For stress treatment, 10-day-old seedlings of PBW343 was subjected to heat stress (37 and 42 °C for 2 h), cold (4 °C for 2 h), salt (150 mM NaCl for 2 h), and dehydration (2% mannitol for 2 h). For a heat stress experiment, 1-week-old wild-type and transgenics were given heat stress treatment, i.e., 42 °C for 2 h and then kept to recover for further analysis. For oxidative stress response, 21-day-old seedlings of both wild and transgenics were subjected to methyl vilogen treatment, i.e., 50 μM for 4 h. For cold stress, 1-week-old seedlings were kept at 4 °C for 24 h and left for recovery for 20 days for further analysis.
Cloning of wheat TaZnF for transactivation studies
For the measurement of the transactivation activity, full CDS of TaZnF was cloned in pGBKT7 vector (containing GAL4 DNA-binding domain) by restriction based cloning for which EcoRI and BamHI restriction sites in the 5′ and 3′ primers were and positive clones were transformed in yeast strain Y2H109. A Y1H assay was undertaken and a drop out assay was performed on both SD-W and SD-HW media. The transformed yeast cells were able to grow both on SD-W and SD-HW media. The LacZ activity was observed in a medium containing 200 mg/L X-gal.
Phylogenetic and bioinformatic analysis
Identification of other plant TaZnF orthologs was achieved via bioinformatics and phylogenetic analysis. Full-length orthologous protein sequences were identified by searching the phytozome online database. To authenticate, the full-length protein sequence of the selected topmost (one or two) hit results from phytozome database were rechecked in NCBI database. Finally, these confirmed proteins from different plant species were used for full-length protein alignment in clustal × software. Manual curation in the alignment was performed and a final phylogenetic tree was constructed by the neighbor joining method with a bootstrap value of 1000.
Transcription profiling by RT-qPCR
Total RNA from different Arabidopsis plants was isolated using the RNeasy plant mini kit (Qiagen, Germany) according to the manufacturer’s instructions, including on-column DNaseI treatment to remove genomic DNA contamination. Two micrograms of the total RNA was used as template to synthesize cDNA employing the high-capacity cDNA archive kit (Applied Biosystems, USA) and mixed with 200 nM of each primer and SYBR green PCR master mix (Applied Biosystems) for real-time PCR analysis, using the ABI Prism 7000 sequence detection system and software (PE Applied Biosystems) according to the manufacturer’s protocol. The specificity of the reactions was verified by melting curve analysis. At least two independent RNA isolations were used for cDNA synthesis, and each cDNA sample was subjected to real-time PCR analysis. Actin was used as an internal control.
Construction of binary vectors and plant transformation
For overexpression studies, a 756 bp fragment (cDNA) containing the TaZnF open reading frame was PCR amplified from pGEM-T Easy:TaZnF plasmid with Topo- TaZnF -F and TaZnF R primers (Table S1). The amplicon was inserted into pENTR/D-TOPO vector (Invitrogen) for construction of entry clone and sequenced. This cassette was then mobilized into the destination vector pMDC32 by LR-clonase reaction to generate pMDC32::TaZnF vector. Arabidopsis thaliana Col0 were transformed by the floral dip method (Clough and Bent 1998). For analysis of different transgenic lines under various abiotic stresses, T3 homozygous seeds were used and the results presented represent at least three independent experiments. The data presented represent at least ten replicates and values are represented with average means of these experiments with standard error bars.
Chlorophyll fluorescence measurements
PSII activity was measured according to Krause and Weis (Krause and Weis 1991). Measurements of modulated chlorophyll fluorescence emission from the upper surface of the leaf were done using a pulse amplitude modulation fluorometer (Junior-PAM chlorophyll fluorometer, H.Walz, Germany). Leaves were dark-adapted for 20 min before measuring the induction of fluorescence. Measurements of various PSII functions such as maximum photosynthetic efficiency (Fv/Fm), effective photosynthetic efficiency (YII), and electron transport rate (ETR) were recorded in rosette leaves at specified time points in at least ten plants per line viz. WT, and transgenics.
Measurements of total chlorophylls
Chlorophylls estimation was done by the non-maceration method according to (Hiscox and Israelstam 1979). Leaf samples (0.05 g) were taken from both control and salt stress treated plants in three different replicates and were incubated in 5 ml of DMSO at 65 °C for 4 h in dark. Absorbance was recorded at 645 nm, 663 nm in a (Beckman DUTM 640B, Beckman Instruments Inc., USA) spectrophotometer, and chlorophyll contents were calculated according to the following formula.
where,
- V:
-
volume of DMSO in mL
- X:
-
path length, 1 cm
- W:
-
fresh weight in grams.
Measurement of membrane stability
Cell membrane stability was measured according to (Bajji et al. 2002). For this, seedlings were submerged in 5 ml of distilled water in test tubes and kept at 30 °C for 30 min and then electrical conductivity (C1) was measured. Then, these seedlings were autoclaved for 15 min and C2 was measured. Cell injury was calculated using the equation:
Proline estimation
Proline levels were measured according to Bates et al. (Bates et al. 1973). For this, 100 mg of tissue was ground in 1 ml of 3% sulphosalicyclic acid. It was centrifuged at 15000 rpm for 15 min. The supernatant obtained was divided and 400 μl of glacial acetic and 400 μl of ninhydrin were added and incubated at 100 °C for 1 h. After incubation, reaction was stopped by keeping the sample in ice and 800 μl of toluene was added to the sample and was vortexed vigorously. The supernatant was transferred to new MCT and absorbance was recorded at 520 nm in a UV-Visible spectrophotometer (U-2810 Spectrophotometer, Hitachi, Japan). Proline content was calculated using the equation:
ROS estimation
To check the amount of ROS (reactive oxygen species) produced in Arabidopsis transgenics and WT plants, staining with DAB (3,3-diaminobenzidine) (Shraudner et al. 1998) and NBT (nitro blue tetrazolium chloride) was done (Jabs et al. 1996). For this, 14-day-old seedlings (control and heat stress) of Arabidopsis WT and overexpression transgenics were subjected to heat stress (37 °C for 2 h) and oxidative stress was mimicked by methyl vilogen treatment (50 μM for 4 h). The plants were then stained by incubating in NBT (2 mM NBT powder, 20 mM phosphate buffer) and DAB (100 mM phosphate buffer, 0.05% v/v Tween 20, 200 mM Na2PO4, and pH 3.0) overnight. On the next day, the seedlings were washed with MQ water and subjected to removal of chlorophyll by dipping them in bleaching solution (ethanol, acetic acid, and glycerol in a ratio of 3:1:1) for 2 h. The plants were then visualized under a bright light microscope and pictures were taken for comparison of the visible places of action of ROS in transgenics and the WT Arabidopsis plants.
Results and discussions
Sequence analysis and phylogenetic analysis
The TaZnF zinc finger protein belongs to C4HC3-type zinc finger family. It contains one RING domain at 41–81 amino acid position, RING-v (40–82), two low complexity regions at position 18–38, and 194–215 and one transmembrane region at 231–250 a.a. based on SMART domain analysis (Fig. 1b). Comparative analysis of TaZnF protein sequence with other known zinc finger protein sequences reveals the presence of conserved RING motif (Fig. 1c). Phylogenetic analysis study using full-length TaZnF protein sequence deduced from the CDS was carried over using Clustal X as software by the neighborhood-joining method keeping 1000 bootstrap values. Overall, it was observed that TaZnF protein falls in a separate monocot clade, while its orthologues from dicot species form a distinct clade of their own. TaZnFP showed highest similarity to Brachypodium distachyon, family protein (Fig. 2).
Domain organization of TaZnF and comparison with its orthologue proteins in other plant species. a Nuclear localization signal prediction of TaZnF by NucPred tool. b Schematic representation of various domains in TaZnF protein by SMART tool. Pink color represent low complexity region, brown color represent RING domain and blue color the trans-membrane region. c Full length protein alignment of TaZnF with its orthologue from monocot and dicot species by Clustalx2
Phylogenetic analysis of the relationships between TaZnF and its C3HC4-type RING finger genes in other species. The multiple alignment was done using ClustalW2 and the dendrogram was built using MEGA4.0 software with the neighbor-joining and pair-wise deletion options as a consensus of 1000 bootstrap replicates
Transcription profiling of TaZnF under heat and other abiotic stresses
To understand the function of TaZnF, transcription level of TaZnF was analyzed in 10-day-old PBW343 subjected to heat stress (basal level, i.e., 37 °C and high temperature, i.e., at 42 °C) for 2 h. The RT-PCR analysis showed that the transcript level was induced under heat stress treatment; however, a slight decrease in the transcript level was observed at 42 °C in comparison to 37 °C of heat stress (Fig. 3a). Thus, in the present study through transcription profile studies, we have found that this TaZnF is heat inducible and similar results have been reported for CaRZFP (Zeba et al. 2009), Arabidopsis ZAT12 (Davletova 2005), and OsRZFP34 (Hsu et al. 2014).
Transcription profiling of TaZnF transcription factor under various abiotic stress condition and in various tissues. Change in transcript abundance of TaZnF in a 10-day-old seedlings of PBW343 under heat (37 and 42 °C for 2 h), cold (4 °C for 2 h), NaCl (150 mM for 2 h), and mannitol (2% for 2 h). b Shoot and various tissues from mature plants of PBW343 under heat stress at 37 °C. The transcription level at control, i.e., 0 h of treatment was normalized as 1.0 and the result shown are the means ± SDs of at least three independent experiments
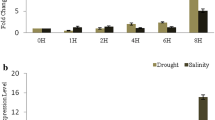
Transcription profiling revealed that TaZnF was also induced under various abiotic stresses in seedlings. The transcript level was almost 30-fold higher under stimulated drought stress and 10-fold higher under salt stress compared to that under control conditions (Fig. 3a). In addition to its role in heat stress, TaZnF is found to be up-regulated in other abiotic stress like cold, drought and salt with the highest level of transcript under salt stress. This result corroborates other reports where Arabidopsis ZAT12 is known to function in osmotic and salt stress (Davletova 2005) and BrRZFP1 is found to be induced under salt, dehydration, and cold stress (Jung et al. 2013). Similarly, TaZnFP belonging to CCCH-type was also found to be induced under drought, NaCl, cold, and ABA (Min et al. 2013). Another report by Xu et al. 2014, showed that TaRZ1, TaRZ2, and TaRZ3 were induced under abiotic stress conditions like salt, drought, and cold stress. RZ belongs to the GRP family, has a CCHC-type zinc finger motif which is present between the N-terminal RRM domain and the C-terminal glycine-rich region. Thus, these studies together indicate that TaZnF, a C4HC3 type zinc finger might function and play a crucial role under both heat and other abiotic stresses.
In addition to its role in abiotic stresses, the zinc finger domain has been identified in many disease resistance genes and has been reported to participate in disease resistance in plants. Similarly, Gupta et al. 2012 analyzed the protein sequences of 70 R genes from several crops and found that 26 proteins had zinc finger domains along NBS-LRR domain with leucine rice repeats. Another zinc finger protein, i.e., TaRZ1 was found to play role in defense response. The Arabidopsis transgenic expressing TaRZ1 showed enhanced resistance to Pseudomonas syringae (Xu et al. 2015). Thus, these studies together indicate the miscellaneous role of zinc finger protein in both biotic and abiotic responses.
Subsequent analysis was done to ascertain the involvement of any temporal or spatial cues on the behavior of TaZnF under heat stress conditions. For this purpose, transcription analysis was extended to various reproductive tissues (anther, ovary, lemma, palea, glumes, and awn) and in seeds at various development stages (3DAA, 5DAA, 7DAA, 10DAA, and 20DAA). Real-time PCR analysis showed broad expression of TaZnF across tissues with high-transcript level in palea and awn among the reproductive tissues and in seed at 3DAA and 5DAA (Fig. 3b). Such differential pattern has been reported for TaZFHD1, a zinc finger-homeodoamin from wheat previously by Abu-Romman (2014). It was found to express differentially during spike development. It showed preferential expression in spike half emerged, completely emerged, at half anthesis stage and also in emerging awns. Thus, these results suggest the role of zinc finger in wheat inflorescence and seed development. Moreover, it is reported that in many temperate cereal crops, both grain number and grain weight is affected by heat stress, with a decline in yield proportional to the increase in temperature during flowering and grain filling stage (Porter and Semenov 2005). Moreover, high-temperature stress during reproductive development is reported to affect the grain size and quality in wheat by decreasing the grain weight, mass, and also the sugar content of kernel (Shah and Paulsen 2003). Therefore, we can conclude that high levels of TaZnF transcript in awn and in seeds at 3 and 5 days of anthesis may have some role in enhancing tolerance under high-temperature stress and would thereby further contribute to enhancement in grain weight or yield in wheat.
Next, we were interested in understanding the role of TaZnF in recovery post heat stress. For this, 10-day-old seedlings after heat stress treatment were kept in growth room conditions for recovery for 2 to 4 h. It was observed that during recovery the transcript levels gradually increased with an increase in duration of recovery to almost 6–8-fold level (Fig. 4a). Together, these studies suggest the possible roles of TaZnF in both heat stress and recovery. The above experiments clearly demonstrate that TaZnFs are under dynamic regulation and respond to spatial, temporal, and environmental cues in a unique manner.
Transcription profiling of TaZnF transcription factor during high-temperature stress and recovery in various genotypes of Triticum aestivum. Change in transcript abundance of TaZnF in 10-day-old seedlings of various genotypes subjected to high-temperature stress at 37 and 42 °C for 2 h and followed by 2 and 4 h of recovery, a PBW343, b HD2329, c K7903, and d C306. The transcription level of TaZnF at control, i.e., 0 h of treatment was normalized as 1.0 and the results shown are the means ± SDs of at least three independent experiments
Varietal specific regulation of TaZnF under heat stress
Further studies were conducted to exploit the biodiversity of wheat cultivars to understand better the dynamics of the stress response. In order to compare the nature of regulation in different cultivars of wheat, a comparative transcript profile was undertaken from four different genotypes of wheat Triticum aestivum. To conduct this study, two heat sensitive varieties PBW343 and HD2329 and two heat tolerant varieties C306 and K7903 were selected. Ten-day-old seedlings of theses genotypes were subjected to heat stress at 37 °C and 42 °C for 2 h followed by recovery for 2 and 4 h, respectively. From the real-time analysis, it was observed that transcript level is upregulated at 37 °C, and under recovery conditions the transcript level gradually increases with increase in duration of recovery period in PBW343 (Fig. 4a). A similar transcription profile is observed at high-temperature stress treatment, thereby suggesting the role of TaZnF under both heat stress and recovery period. Moreover, the similar transcription pattern at both temperatures suggests no temperature specific regulation. However, in genotype HD2329, there is a slight increase in transcription levels followed by a decrease to that of the controls with an increase in the duration of recovery at both stress temperatures (Fig. 4b). In K7903 at 37 °C, there is a slight increase in the transcription level followed by a decrease during the recovery period. In contrast to 37 °C, the transcription level increases significantly at 42 °C followed by decrease with increase in duration of recovery (Fig. 4c). However in cultivar C306, the transcript is induced significantly at 37 °C followed by a decline in the transcription level. Interestingly, the transcription level rises significantly with an increase in the duration of recovery. A similar transcription profile is observed at 42 °C (Fig. 4d). It is clear from the above analysis that heat stress induces specific and unique profiles in different varieties under different stress conditions, further defining its possible role in maintaining the robustness of the plant under stress conditions.
Transactivational activity assay
TaZnF is a transcription factor and smart domain analysis revealed the presence of RING domain, a marked feature of C3CH4 type zinc finger transcription factor. A yeast assay system was employed to determine the transcriptional potential of TaZnF. The transformed yeast cells were able to grow both on SD-W and SD-HW media, suggesting that TaZnF was able to activate the transcription of the histidine reporter gene and confirming the transcription activation potential (Fig. 5a–b). This result co-related with quantitative β galactosidase activity assay (colony filter lift assay) using 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) as a substrate where the colonies turn blue due to the production of B-galctosidase. Thus, it was hypothesized that TaZnF has a functional motif which acts as a transactivator.
Overexpression of TaZnF gene increases basal and high thermotolerance of transgenics
To study the physiological functions of TaZnF, Arabidopsis transgenics that constitutively overexpress TaZnF under the control of CaMV35S promoter were raised. The transgenics lines overexpressing TaZnF showed high transcription levels of TaZnF in comparison to wild-type (Fig. 6c). ZnFs are well reported to respond to various abiotic and biotic stress conditions, therefore initial studies were undertaken to examine the effect under heat stress conditions. For this, 1-week-old transgenics were subjected to heat stress at 42 °C for 2 h and then returned to their original growth conditions for recovery. After 15 days of recovery period, transgenics plants revived earlier and showed faster and more robust growth as compared to wild-types (Fig. 6a). They displayed an increase in root length, leaf number, leaf size, rosette diameter, and accumulated a greater biomass than wild-type (Table 1). This increase in biomass was seen to be associated with enhanced photosynthetic efficiency as leaf photosynthesis is highly sensitive to high temperature stress (Berry and Bjorkman 1980). Among all photosynthetic functions, photosystem II (PSII) is the most heat sensitive (Berry and Bjorkman 1980; Havaux 1992). Heat stress also induces the dissociation of the peripheral antenna complex of PSII from its core complex (Gounaris et al. 1984), a loss of the oxygen evolving complex activity of PSII (Enami et al. 1994; Yamane et al. 1998), as well as an inhibition of electron transfer from OA to Qa at the acceptor side (Blum 1988). It has been reported that heat stress in excess above 40 °C affects antenna complexes, and over 30 °C, there is a decrease in overall PSII photochemistry (Briantais et al. 1996). Therefore, the effect of heat stress was investigated by measuring the maximum photosynthetic efficiency (Fv/Fm) of PSII. Fv/Fm of transgenics was found to be higher under heat stress as compared to wild-type (Fig. 7a). Similarly, effective photosynthetic efficiency (YII) and electron transport rate (ETR) were also studied and transgenics showed better YII and ETR response than wild-type under similar conditions (Fig. 7b–c).
TaZnF overexpressing plants showing increased tolerance to high-temperature stress. One-week-old seedlings of both wild-type and transgenics were given heat stress at 42 °C for 2 h and left for recovery at culture room conditions. a Phenotype observed after 15 days of recovery. b One and half months after transferring the wild-type and transgenic lines to pots. c Transcription profile of TaZnF in wild-type and overexpression transgenic lines of Arabidopsis. The transcription level in wild type (WT) was normalized as 1.0 and the results shown are the means ± SDs of at least three independent experiments
Arabidopsis transgenics overexpressing TaZnF subjected to high-temperature stress. Effect on various photosynthetic parameters like a photosynthetic efficiency (Fv/Fm). b Effective photosynthetic efficiency (YII); c electron transport rate; d membrane stability index (MSI %); e chlorophyll content (see text for details)
Membranes are thought to be the site of primary physiological injury (Blum 1988) and an early event in plant response to heat stress (Georgieva and Georgieva 1999). The membrane damage results in leakage which further results in chlorophyll spilling and degradation in the stroma. Therefore, measurement of solute leakage from tissue can be used to estimate damage to membranes. The membrane stability of transgenics was almost 100 folds higher in comparison to wild-type plants under heat stress (Fig. 7d). The chlorophyll content was also analyzed as chlorophyll synthesis is known to be sensitive to heat stress and serves as a good indicator for heat stress (Gosavi et al. 2014). Chlorophyll content (i.e., Chl a, Chl b, total chlorophyll content) was found to be significantly higher; two- to threefold higher in transgenics than in wild-type after stress (Fig. 7e). The transgenics matured earlier, i.e., within 1½ month, had longer, and more numerous productive siliques. These also showed considerably higher seed yield as the number of siliques per plant and yield per plant were found to be higher (Fig. 8c–d). The silique length was also found to be greater than in wild-types as indicated in Fig. 8a. Thus, it can be inferred that TaZnF confers increased tolerance to high-temperature stress as measured by faster recovery and robust growth of transgenics and also by increased membrane stability, chlorophyll retention, higher photosynthetic efficiency etc.
Effect of heat stress on yield parameters. One-week-old seedlings of both wild-type and transgenics were given heat stress at 42 °C for 2 h and left to recover for almost 2½ month for analysis of various parameters related to yield like a length of siliques; b seed morphology; c number of siliques per plant; d yield per plant
Overexpression of TaZnF provides enhanced tolerance via amelioration of oxidative stress
It is well documented that under adverse conditions such as high temperature there is an increase in ROS concentration levels in plants (Liu and Huang 2000; Gür et al. 2010). Therefore, to further examine the involvement of oxidative stress, ROS accumulation was measured under both heat stress and oxidative stress treatments by NBT and DAB staining. To measure the amount of ROS accumulation under heat stress, 14-day-old plants of both transgenics and wild-type were subjected to heat stress at 37 °C for 2 h. Under heat stress conditions, superoxide anion as measured by NBT staining was found to be lower in transgenic lines in comparison to wild-type plants (Fig. 9a). Similarly, the quantity of ROS generated under oxidative stress was measured by DAB staining. For oxidative stress treatment, 21-day-old seedlings of both wild and transgenics were subjected to methyl vilogen treatment (50 μM for 4 h). Under oxidative stress, hydrogen peroxide levels were found to be significantly lower in transgenics as compared to the wild-types (Fig. 9b). Moreover, plants are known to produce antioxidant enzymes as part of an enzymatic scavenging system when ROS are produced as a response to heat stress (Sharkey 2005). Therefore, transcription level of various oxidative stress marker genes like APX 1, APX 3, APX 4, APX 6, CAT1, and CAT2 was also analyzed. The transcription profiling revealed higher levels of these genes in the transgenics in comparison to the wild-type under heat stress (Fig. 9c). This enhanced tolerance to oxidative stress is consistent with the transcription level of the oxidative response marker gene. There are similar reports by Davletova et al. (Davletova et al. 2005), where they have reported zinc finger protein Zat12 involvement in oxidative stress response in Arabidopsis. Transcriptional profiling of Zat12-overexpressing plants and wild-type plants subjected to H2O2 stress has shown that constitutive expression of Zat12 in Arabidopsis results in the increased expression of oxidative- and light stress response transcripts. Therefore, this could be one of the reasons that constitutive overexpression of TaZnF leads to enhanced plant growth with increased tolerance under heat stress by low accumulation of ROS.
TaZnF overexpressing Arabidopsis plants showed increased tolerance to oxidative stress. a Oxidative stress response of 14-day-old WT and TaZnF overexpression Arabidopsis seedlings examined by comparing the accumulation of O2 and H2O2 radicals under heat stress, i.e., at 37 °C for 2 h. b Oxidative stress treatment by subjecting leaf of 21-day-old seedlings of both wild and transgenics under by treatment by methyl viologen (50 μM) for 4 h by DAB staining. c Transcription profiling of various oxidative stress-related marker genes in transgenic lines of Arabidopsis plants overexpressing TaZnF. The transcription level in wild-type (WT) was normalized as 1.0 and the result shown are the means ± SDs of at least three independent experiments
TaZnF also provides cold tolerance to transgenic Arabidopsis
Transgenics were also evaluated to assess the role of TaZnF under cold stress. Seedlings were grown on half-strength MS medium and was given cold stress at 4 °C for 24 h and then kept under culture room conditions for recovery. The transgenics were found to show faster and more robust growth: two- to threefold shoot length, five to six times longer roots, leaf numbers, leaf size, rosette diameter, and accumulated greater biomass than wild-type (Fig. 10a–c and Table 2). Moreover, the transgenics flowered and attained maturity earlier, i.e., in 1½ month and produced higher numbers of siliques and thus showed considerably higher seed yield (Fig. 10a). They performed better under cold stress with respect to photosynthetic efficiency (Fv/Fm) of PSII (Fig. 10e), membrane stability (Fig. 10f), proline accumulation (Fig. 10g), and chlorophyll content (Fig. 10h). This further establishes the additional role of TaZnF in conferring tolerance to cold stress. Our studies corroborate those of Jung et al. (Jung et al. 2013) where they have reported that overexpression of BrRZFP1 confers high tolerance to cold stress in Nicotiana tabacum. Similarly, TaRZ2 was reported to confer increased tolerance to cold stress in Arabidopsis (Xu et al. 2014). Together, these studies suggest that TaZnF has multi-abiotic stress connection and indicates an important role for TaZnF in stress tolerance and crop improvement.
Overexpressing TaZnF plants show increased tolerance to cold stress. a Phenotype observed after cold stress given to 1-week-old seedlings for 24 h and left at recovery for 20 days; b measure of plant height, shoot length and root length; c estimation of fresh weight; d number of flowering plants; e photosynthetic efficiency (Fv/Fm); f membrane stability index (MSI); g proline content. h chlorophyll content measured after stress treatment
Transcription profiling of stress marker genes
To gain an insight into how the overexpression of TaZnF leads to enhanced tolerance to various stresses, transcription profiling for stresses such as cold, heat and other stress-related marker genes were undertaken in transgenics and wild-type. Marker genes like HSF, MYB2, COR47, and LEA4-5 were found to be upregulated in transgenics with maximum transcript level in C3.4 lines, followed by C3.7 and C3.3 compared to wild-type (Fig. 11). The increased transcription level of marker genes in transgenics under stress conditions could be one of the reasons for their increased tolerance of thermal stress conditions. Together with previous observations, these results suggest that TaZnF may be a pivotal factor in the regulation of stress response genes in plants and may thus play a major role in various thermal stress-related processes.
Conclusions
In conclusion, the C4HC3-type zinc finger TaZnF from wheat was cloned and characterized and its function was investigated in transgenic Arabidopsis plants. Overexpression of the TaZnF in Arabidopsis plants conferred increased tolerance to heat, oxidative, and cold stress. These results indicate that TaZnF transcription factor is an important determinant of stress response in plants and changes in its expression level in plants could increase tolerance to various abiotic stresses.
References
Abu-Romman S (2014) Molecular cloning and expression analysis of zinc finger-homeodomain transcription factor TaZFHD1 in wheat. S Afr J Bot 91:32–36
Bajji M, Kinet JM, Lutts S (2002) The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul 36:61–70
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Berry J, Bjorkman O (1980) Photosynthetic response and adaptation to temperature in higher plants. Annu Rev Plant Physiol 31:491–543
Blum A (1988) Plant breeding for stress environments. CRC Press, Inc., Boca Raton
Briantais JM, Dacosta J, Goulas Y, Ducruet JM, Moya I (1996) Heat stress induces in leaves an increase of the minimum level of chlorophyll fluorescence, Fo: a time-resolved analysis. Photosynth Res 48:189–196
Brown RS, Sander C, Argos P (1985) The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett 186:271–274
Chen M, Ni M (2006) RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J 46:823–833
Cheung MY, Zeng NY, Tong SW, Li FWY, Zhao KJ, Zhang Q, Sun SS-M, Lam HM (2007) Expression of a RING-HC protein from rice improves resistance to Pseudomonas syringae pv. tomato DC3000 in transgenic Arabidopsis thaliana. J Exp Bot 58:4147–4159
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Davletova S (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139:847–856
Enami I, Kitamura M, Tomo T, Isokawa Y, Ohta H, Katoh S (1994) Is the primary cause of thermal inactivation of oxygen evolution in spinach PS II membranes release of the extrinsic 33 kDa protein or of Mn? Biochim Biophys Acta BBA - Bioenerg 1186:52–58
Fischer RA, Byerlee DB (1991) Trends of wheat production in the warmer areas: major issues and economic considerations. In: Saunders DA (ed) Wheat for nontraditional, warm areas. CIMMYT, Mexico, pp 3–27
Georgieva K (1999) Some mechanisms of damage and acclimation of the photosynthetic apparatus due to high temperature. Bulg J Plant Physiol 25:89–99
Gosavi G, Jadhav A, Kale A, Gadakh S, Pawar B, Chimote V (2014) Effect of heat stress on proline, chlorophyll content, heat shock proteins and antioxidant enzyme activity in sorghum (Sorghum bicolor) at seedlings stage. Indian J Biotechnol 13:356–363
Gounaris K, Brain ARR, Quinn PJ, Williams WP (1984) Structural re-organisation of chloroplast thylakoid membranes in response to heat-stress. Biochim Biophys Acta 766:198–208
Gupta SK, Rai AK, Kanwar SS, Sharma TR (2012) Comparative analysis of zinc finger proteins involved in plant disease resistance. PLoS One 7:e42578
Gür A, Demirel U, Özden M, Kahraman A, Çopur O (2010) Diurnal gradual heat stress affects antioxidant enzymes, proline accumulation and some physiological components in cotton ( Gossypium hirsutum L.) Afr J Biotechnol 9:1008–1015
Havaux M (1992) Stress tolerance of photosystem II in vivo: antagonistic effects of water, heat, and photoinhibition stresses. Plant Physiol 100:424–432
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Hsu KH, Liu CC, Wu SJ, Kuo YY, Lu CA, Wu CR, Lian PJ, Hong CY, Ke YT, Huang JH et al (2014) Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol Biol 86:125–137
Islam MM, Tani C, Watanabe-Sugimoto M, Uraji M, Jahan MS, Masuda C, Nakamura Y, Mori IC, Murata Y (2009) Myrosinases, TGG1 and TGG2, redundantly function in ABA and MeJA signaling in Arabidopsis guard cells. Plant Cell Physiol 50:1171–1175
Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273:1853–1856
Jung YJ, Lee IH, Nou IS, Lee KD, Rashotte AM, Kang KK (2013) BrRZFP1 a Brassica rapa C3HC4-type RING zinc finger protein involved in cold, salt and dehydration stress. Plant Biol 15:274–283
Kam J, Gresshoff P, Shorter R, Xue GP (2007) Expression analysis of RING zinc finger genes from Triticum aestivum and identification of TaRZF70 that contains four RING-H2 domains and differentially responds to water deficit between leaf and root. Plant Sci 173:650–659
Kosarev P, Mayer KF, Hardtke CS (2002) Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. https://doi.org/10.1186/gb-2002-3-4-research0016
Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol 42:313–349
Lim SD, Yim WC, Moon JC, Kim DS, Lee BM, Jang CS (2010) A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co-expressed genes. Plant Mol Biol 72:369–380
Liu X, Huang B (2000) Heat stress injury in relation to membrane lipid peroxidation in creeping bentgrass. Crop Sci 40:503–510
Miller J, McLachlan AD, Klug A (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4:1609–1614
Min DH, Zhao Y, Huo DY, Li LC, Chen M, Xu ZS, Ma YZ (2013) Isolation and identification of a wheat gene encoding a zinc finger protein (TaZnFP) responsive to abiotic stresses. Acta Physiol Plant 35:1597–1604
Pepper AE, Chory J (1997) Extragenic suppressors of the Arabidopsis Det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics 145:1125–1137
Porter JR, Semenov MA (2005) Crop responses to climatic variation. Philos Trans R Soc Lond Ser B Biol Sci 360:2021–2035
Prestele J, Hierl G, Scherling C, Hetkamp S, Schwechheimer C, Isono E, Weckwerth W, Wanner G, Gietl C (2010) Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc Natl Acad Sci U S A 107:14915–14920
Reynolds MP, Nagarajan S, Razzaque MA, Ageeb OAA (2001) Heat tolerance. Appl Physiol Wheat Breed 124–135
Schraudner M, Moeder W, Wiese C, Van Camp W, Inze D, Langebartels C, Sandermann H (1998) Ozone-induced oxidative burst in the ozone biomonitor plant, tobacco Bel W3. Plant J 16:235–245
Shah NH, Paulsen GM (2003) Interaction of drought and high temperature on photosynthesis and grain-filling of wheat. Plant Soil 257:219–226
Sharkey TD (2005) Effects of moderate heat stress on photosynthesis: importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ 28:269–277
Stone SL (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137:13–30
Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci CMLS 54:582–596
von Arnim AG, Deng XW (1993) Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J Biol Chem 268:19626–19631
Wang Y-S, Pi L-Y, Chen X, Chakrabarty PK, Jiang J, Leon ALD, Liu G-Z, Li L, Benny U, Oard J et al (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18:3635–3646
Xu R, Li QQ (2003) A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Mol Biol 53:37–50
Xu T, Gu L, Choi MJ, Kim RJ, Suh MC, Kang H (2014) Comparative functional analysis of wheat (Triticum aestivum) zinc finger-containing glycine-rich RNA-binding proteins in response to abiotic stresses. PLoS One 9:e96877
Xu T, Lee HJ, Sy ND et al (2015) Wheat (Triticum aestivum) zinc finger-containing glycine-rich RNA-binding protein TaRZ1 affects plant growth and defense response in Arabidopsis thaliana. Plant Growth Regul 76:243–250
Yamane Y, Kashino Y, Koike H, Satoh K (1998) Effects of high temperatures on the photosynthetic systems in spinach: oxygen-evolving activities, fluorescence characteristics and the denaturation process. Photosynth Res 57:51–59
Yang Y, Ma C, Xu Y, Wei Q, Imtiaz M, Lan H, Gao S, Cheng L, Wang M, Fei Z et al (2014) A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26:2038–2054
Zeba N, Isbat M, Kwon N-J, Lee MO, Kim SR, Hong CB (2009) Heat-inducible C3HC4 type RING zinc finger protein gene from Capsicum annuum enhances growth of transgenic tobacco. Planta 229:861–871
Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive Abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Acknowledgements
This work was funded by the Department of Biotechnology, Government of India, New Delhi and the Council of Scientific and Industrial Research, New Delhi which awarded the Senior Research Fellowship.
Author information
Authors and Affiliations
Contributions
PA and PK planned experiments. PA performed experiments. PA & PK analyzed data and PA wrote the manuscript. PK reviewed the manuscript.
Corresponding author
Electronic supplementary material
Table S1
(XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Agarwal, P., Khurana, P. Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring heat stress tolerance in Arabidopsis . Cell Stress and Chaperones 23, 253–267 (2018). https://doi.org/10.1007/s12192-017-0838-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-017-0838-1