Abstract
By oligo microarray expression profiling, we identified a rice RING zinc-finger protein (RZFP), OsRZFP34, whose gene expression increased with high temperature or abscisic acid (ABA) treatment. As compared with the wild type, rice and Arabidopsis with OsRZFP34 overexpression showed increased relative stomata opening even with ABA treatment. Furthermore, loss-of-function mutation of OsRZFP34 and AtRZFP34 (At5g22920), an OsRZFP34 homolog in Arabidopsis, decreased relative stomata aperture under nonstress control conditions. Expressing OsRZFP34 in atrzfp34 reverted the mutant phenotype to normal, which indicates a conserved molecular function between OsRZFP34 and AtRZFP34. Analysis of water loss and leaf temperature under stress conditions revealed a higher evaporation rate and cooling effect in OsRZFP34-overexpressing Arabidopsis and rice than the wild type, atrzfp34 and osrzfp34. Thus, stomata opening, enhanced leaf cooling, and ABA insensitivity was conserved with OsRZFP34 expression. Transcription profiling of transgenic rice overexpressing OsRZFP34 revealed many genes involved in OsRZFP34-mediated stomatal movement. Several genes upregulated or downregulated in OsRZFP34-overexpressing plants were previously implicated in Ca2+ sensing, K+ regulator, and ABA response. We suggest that OsRZFP34 may modulate these genes to control stomata opening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants, being sessile organisms, cannot escape from their growing environments. Extremes in environmental factors can result in stressful conditions that inhibit plant growth and development. These detrimental effects inevitably limit crop productivity. Understanding how the plant responds to environmental stress is critical for increasing stress tolerance in crops through genetic engineering (Shinozaki and Yamaguchi-Shinozaki 2007).
Heat stress due to elevated temperature is the major environmental limitation of plant growth and development. High temperatures can create fluid membranes, reduce photosynthetic electron transport, and denature cellular proteins (Sun et al. 2002; Suzuki and Mittler 2006; Hüve et al. 2011). Plants have evolved a complex set of molecular responses to adapt to stressful high temperatures. For example, high temperature invokes the synthesis of heat shock proteins (HSPs), associated with acquired thermotolerance (Yeh et al. 1997; Kotak et al. 2007). Other pathways mediate thermotolerance through an HSP-independent mechanism. In Arabidopsis, mutation of MBF1c or Siz1, involved in the salicylic-acid signaling pathway, causes thermosensitivity but no alteration in expression of HSPs or heat shock (HS) factors (Yoo et al. 2006; Suzuki et al. 2008).
In additional to synthesizing protective molecules, plants may incorporate other physiological responses to cope with heat stress. For example, after exposure to high temperature, plant leaves can respond by actively regulating stomata opening to evaporate water, which results in heat dissipation from the leaves (Lewis and Nobel 1977). However, transpirational cooling involves water usage, which often forces plants to face water deficits under field conditions (Liu et al. 2005). Levels of the phytohormone abscisic acid (ABA) are triggered by water deficit to regulate necessary changes in gene expression and physiological adaptation (Chaves et al. 2002). One of the major adaptive responses mediated by ABA is stomata closure (Schroeder et al. 2001). Factors that function in ABA-mediated stomata closure include protein phosphatase 2C proteins such as ABI1 and ABI2, protein kinases such as SnRK2.6/OST1 and GHR1, transmembrane ATP binding cassette proteins such as ABCG22 and MRP5, and anion channels such as SLAC1 (Mustilli et al. 2002; Umezawa et al. 2009; Kuromori et al. 2011; Hua et al. 2012). ABA can mediate stomata closure by changing the cytosolic [Ca2+] and [H+] levels in guard cells, or enhancing the Ca2+ sensitivity for activation of SLAC1 and downregulation of inward-rectifying K+ channels KAT1 (Lebaudy et al. 2007; Siegel et al. 2009). In addition, stress can induce stomata closure through ABA-independent reactive oxygen species (ROS) homeostasis (Mustilli et al. 2002; Huang et al. 2009). These measures, although effective in preventing water loss, can ultimately hamper transpirational cooling. Plants must have mechanisms to balance these interconnected yet counteractive field-survival strategies, but the genetic determinants of the mechanisms are largely unclear.
Rice is a staple food crop consumed by more than half of the world’s population. Rice plants often face more than one stress during their growth in paddy fields. For instance, stomata close in rice plants grown under water deficit in arid regions, which leads to limited carbon uptake by leaves and increased leaf temperature under high irradiance (Hüve et al. 2011). Thus, discovery of genes involved in multiple environmental stress responses provides new insights to study the responses to multiple stresses and improve multiple stress tolerance in rice.
This study aimed to identify the genes involved in HS and ABA responses. We used oligo microarray expression profiling of leaves of rice (Oryza sativa L. cv. Tainung No. 67) to screen for an HS- and ABA-responsive novel gene and identified OsRZFP34 (cDNA accession no. AK068005). OsRZFP34 encodes a RING zinc-finger protein (RZFP, Freemont et al. 1991) (Supplementary Fig. S1). OsRZFP34 transcripts specifically accumulated in rice leaves at germination and vegetative stages after HS or ABA treatment. Gain- and loss-of-function analyses revealed that OsRZFP34 could have a critical role in mediating stomatal aperture and function in leaf cooling with exposure to high temperature and drought in rice.
Materials and methods
Plant materials
Seeds of rice (Oryza sativa L. cv. Tainung No. 67) were soaked in running water at 28 °C and when germinated, were transferred to soil and grown under a 12-h-light (28 °C)/12-h-dark (26 °C) cycle in a growth chamber with 70 % relative humidity. For HS treatment, 3-week-old rice plants were incubated in shaking buffer [1 % (w/v) sucrose in 5 mM potassium phosphate buffer, pH 6.8] at 41 °C for the indicated times. For ABA treatment, 3-week-old rice plants were incubated in shaking buffer with or without 100 μM ABA at 28 °C for the indicated times.
The Arabidopsis thaliana ecotype Columbia (Col) was used as the wild type. Arabidopsis plants were grown at 23 °C in a 16-h-light/8-h-dark cycle in a growth chamber with 60 % relative humidity. For HS and ABA treatment, 3-week-old Arabidopsis plants were used.
RNA isolation and first-strand cDNA synthesis
Total RNA was extracted as described (Guan et al. 2010). To remove genomic DNA contamination, total RNA for cDNA synthesis was first treated with DNase I (Promega, Madison, WI, USA) for 30 min at 37 °C. The first-strand cDNA was synthesized from 2 μg total RNA in a 20-μl reaction volume with use of the SuperScriptIII First-Strand Synthesis system (Invitrogen, Carlsbad, CA, USA). RNA and cDNA were quantified by optical density measurement with use of a spectrophotometer (NanoDrop 1000, Thermo Scientific, Waltham, MA, USA).
Analysis of OsRZFP34 transcripts by RT-PCR
For analysis of OsRZFP34 in rice plants, leaves collected from seed germination, seedling, tillering, booting, heading, and ripening stage rice plants were incubated in shaking buffer at 41 °C and with or 100 μM ABA at 28 °C for the indicated times, respectively. RT-PCR was used to analyze transcript levels of OsRZFP34. PCR amplification conditions were 24 cycles of 30 s at 95 °C, 30 s at 48 °C and 30 s at 72 °C, then 5 min at 72 °C. Primer sequences for OsRZFP34 were forward (5′-GCATGTGTTGAAGGAGCG-3′) and reverse (5′-CTGGCACTTGTGCGCAAT-3′). DNA from 15 μl of each PCR reaction was fractionated by 1.2 % (w/v) agarose gel electrophoresis. ImageJ for Windows (http://rsbweb.nih.gov/ij/) was used to quantify the intensity of the ethidium-bromide–stained DNA bands. The expression of 18S rRNA was an internal control.
Microarray analysis
To analyze the genes involved in the response to HS and ABA, Agilent 60-mer rice oligonucleotide microarrays (Agilent Technologies, CA, USA) were used to study the transcriptome profiles of ABA- and HS-treated 3-week-old rice leaves. The hybridizations were conducted as suggested by the manufacturer (Agilent Technologies, CA, USA). We performed two independent biological replicates of hybridizations.
For microarray analysis of OsRZFP34-overexpressing rice plants, we used the Rice Whole Genome OneArray v1.1 (Phalanx Biotech Group, Taiwan), which contains 22,003 DNA oligonucleotide probes, and each probe is a 60-mer designed in the sense direction. Among the probes, 21,179 probes correspond to the annotated genes in RGAP v.6.1 and BGI (2008) database. In addition, we included 824 control probes. The detailed descriptions of the gene array list are available from http://www.phalanx.com.tw/products/RiOA_Probe.php. Fluorescent antisense RNA (aRNA) targets were prepared from 1 μg total RNA samples with the OneArray Amino Allyl aRNA Amplification Kit (Phalanx Biotech Group, Taiwan) and Cy5 dyes (Amersham Pharmacia, Piscataway, NJ, USA). Fluorescent targets were hybridized to the Human Whole Genome OneArray with Phalanx hybridization buffer by the Phalanx Hybridization System. The hybridizations were conducted as suggested by the manufacturer (Phalanx Biotech Group, Taiwan). The slides were dried by centrifugation and scanned by use of an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA, USA). The Cy5 fluorescent intensities of each spot were analyzed by use of GenePix 4.1 (Molecular Devices). Normalized spot intensities were transformed to gene expression log2 ratios between control and treatment groups. The spots with log2 ratio ≥1 or ≤−1 and P value <0.001 were tested for further analysis. Two independent biological replicates of hybridizations were performed.
Quantitative real-time RT-PCR (qRT-PCR)
To analyze transcript levels of OsRZFP34-upregulated genes, qRT-PCR involved use of the iCycler- iQ5 Multicolor Real time PCR Detection System and iQ SYBR Green Hot-start Supermix (Bio-Rad, Hercules, CA, USA) with the primer sequences in Supplementary Table S1. Real-time PCR reaction involved 60 ng cDNA with each set of primers and the iQ™ SYBR® Green Supermix (Bio-Rad). PCR cycling included an initial step at 95 °C for 5 min, then 40 cycles of 10 s at 95 °C, 30 s at 56 °C, and 20 s at 72 °C. The comparative CT method was used to determine the relative amount of each sample, with the expression of 18S rRNA as an internal control.
Preparation of DNA constructs and transformation
To generate the construct for overexpressing OsRZFP34, we synthesized the coding region of OsRZFP34 by PCR amplification of the first-strand HS-induced rice cDNAs with the primer sequences 5′-ACTAGTATTCAAGATGGGCGCCAT-3′ and 5′-GCGGCCGCTTCGCCACTTCAGATCTG-3′ with built-in SpeI and NotI restriction sequences, respectively. The PCR product was subcloned into the SpeI and NotI sites of the pCYH8 vector (Guo and Ho 2008) in the sense orientation downstream of the ubiquitin 1 promoter (Ubi promoter). The resulting Ubi::OsRZFP34 construct was cloned into the pPZP/35H Ti-based vector by inserting 35S-hph-tml as a HindIII fragment into the HindIII site of pPZP200 (Hajdukiewicz et al. 1994), which was introduced into Agrobacterium tumefaciens (strain EHA105). Transformation of Arabidopsis involved the floral-dip method (Weigel and Glazebrook 2002). Rice calli (Oryza sativa L. cv. Tainung No. 67) were transformed by Agrobacterium-mediated transformation as described (Hiei et al. 1994). Transformed rice or Arabidopsis seedlings were selected on medium that contained 50 μg/ml hygromycin. The hygromycin-resistant T1 seedlings were transferred to soil and grown to maturity. Homozygous T2 plants were selected by growth of their T3 generations on medium that contained hygromycin. T3 seeds derived from homozygous T2 plants were used for subsequent tests.
Stomata analysis
For imaging rice stomata, leaves of 3-week-old plants were fixed as described (Hurng et al. 1988). Stomata were observed under a Hitachi S-4700 scanning electron microscope. For ABA treatment, detached rice leaves were incubated in stomata-opening solution (OS) containing 10 mM KCl, 100 μM CaCl2, and 10 mM MES, pH 6.1, for 2 h and then transferred to OS supplemented with 5 μM ABA for 30 min. To measure rice stomatal aperture under high temperature, 3-week-old rice seedlings were subjected to 38 °C for the indicated times and leaves were immediately cut and fixed. More than 30 guard cells from each sample were used to measure stomatal aperture. For imaging Arabidopsis stomatal aperture, mature leaves of 3-week-old Arabidopsis plants grown at 23 °C for 16 h of light were used. 3 M clear tape was applied to the abaxial surface of each leaf to peel off the epidermal layer. Epidermal strips were immediately mounted on glass slides and observed under a Nikon SMZ-1500 stereoscopic zoom microscope. For ABA treatment, detached leaves were treated as described above. More than 100 guard cells from each sample were used to measure stomatal aperture.
Thermal imaging
Thermal imaging of stressed plantlets was as described (Mustilli et al. 2002). Arabidopsis and rice plants were grown at 23 or 28 °C for 16 h of light for 14 or 21 days, respectively. Drought stress was induced by withholding watering as described above and transferring the pots to a drier atmosphere (24 °C, 50 % relative humidity, 16-h photoperiod). Thermal images were obtained by use of an Avio TVS-200EX infrared camera (Nippon Avionics, Japan).
Stress treatment of transgenic Arabidopsis
For water loss measurement, the aerial parts of 4-week-old plants grown in soil were separated from roots and allowed to dry (25 °C, 60 % relative humidity). The fresh weight of each sample was recorded at 30-min regular intervals.
Statistical analysis
Data are shown as mean ± SE from 3 independent experiments. Statistical differences were analyzed by Student’s t test or Duncan’s multiple range test. A P < 0.05 was considered statistically significant.
Results
Cloning and expression analysis of OsRZFP34
Among the genes with >twofold difference in expression between nonstress control and HS- and ABA-treated rice leaves (CY Hong, unpublished results), we selected and further characterized OsRZFP34 because it showed the highest HS responsiveness among the ABA responsive genes. The predicted protein of OsRZFP34 consists of 302 amino acids of 34 kDa. Multiple sequence alignment was performed by ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) (Fig. 1a). The deduced amino acid sequence is mostly identical to that of OsRFP1 (OsRFP1; GenBank accession no. AY574990) but differs in amino acids 15 (R in OsRZFP34 and H in OsRFP1) and 265 (D in OsRZFP34 and G in OsRFP1) (Zhou et al. 2008) (Fig. 1a). A search of rice DNA sequences revealed that these 2 proteins were derived from the same gene, so the differences may be due to polymorphism between different cultivars. To understand the evolutionary relationship among plant RZFPs, we constructed a phylogenetic tree with the full-length amino acid sequences using the UPGMA method of Mega5 (http://megasoftware.net/). The members of the OsRZFP34 clade exist in Ananas comosus, Arabidopsis thaliana, Glycine max, Oryza sativa, Petunia hybrida, Populus trichocarpa, Ricinus communis, Selaginella moellendorffii, Sorghum bicolor, Triticum aestivum, Vitis vinifera, and Zea mays (Supplementary Fig. S2), which suggests that they may have conserved molecular functions in these plants, although many of their physiological roles are not identical.
a Alignment of OsRZFP34 and other plant RZFPs based on deduced amino acid sequences. ClustalW analysis showed that they all contain conserved CHY zinc finger and RING-finger. The conserved CHY zinc finger domain (CHY-zf) and the RING zinc finger domain (RING-zf) were underlined, respectively. The different amino acids between OsRZFP34 and OsRFP1 were indicated (*). b RT-PCR analysis of OsRZFP34 in rice. mRNA expression of OsRZFP34 in the shoot and root of rice seedlings under HS and ABA stresses. Expression of OsRZFP34 in leaves from seed germination, seedling, tillering, booting, heading, and ripening stage of rice plants treated with c 41 °C and d 100 μM ABA for the indicated times. Bars are mean expression relative to that of 18S rRNA from 3 independent experiments
To further confirm the responsiveness of OsRZFP34 to heat and ABA, we analyzed its transcript levels in various rice tissues exposed to HS and ABA treatments. OsRZFP34 mRNA level in leaves of rice seedlings was markedly higher with HS and ABA than control treatment (Fig. 1b), which suggests that OsRZFP34 is implicated in the rice leaf response to thermal and ABA conditions. In contrast, OsRZF34 mRNA was not detectable in seedling roots, even under HS and ABA treatments.
Under HS, transcripts of OsRZFP34 were detected in leaves from germination, seedling, tillering, and booting but not heading or ripening stages (Fig. 1c). OsRZFP34 transcripts were induced by ABA in leaves from germination, seedling, and tillering stages but were low or not detected in booting, heading, and ripening stages (Fig. 1d). Therefore, the expression of OsRZFP34 depends on developmental stage and is tissue specific.
Constitutive expression of OsRZFP34 increases rice stomata opening and leaf cooling
We used both loss- and gain-of-function strategies to examine the biological function of OsRZFP34. We obtained an OsRZFP34 rice mutant, M0028121 (osrzfp34), with a transfer DNA (T-DNA) insertion in the last intron, from the Taiwan Rice Insertion Mutant (TRIM) pool; OsRZFP34 was not expressed in osrzfp34 even under HS stress (Fig. 2a). Also, we constructed 3 independent T3 homozygous transgenic rice lines constitutively expressing OsRZFP34:OsOE1, OsOE2, and OsOE3 (Fig. 2a). We compared agronomic traits between the wild-type (WT) and the transgenic rice plants grown in a growth chamber under nonstress control conditions. Among the traits, plant height of the OsOE1, OsOE2, and OsOE3 was significantly lower than that of the WT (16–20 % reduction) at the heading stage (data not shown). We did not find significant difference in panicle length, number of tillers per plant, numbers of panicles, spikelets per panicles, filling rate, and total grain weight of 1,000 grains among the WT, OsOE1, OsOE2, OsOE3, and osrzfp34 plants (data not shown). Since OsRZFP34 was induced by heat and ABA, we tested whether OsRZFP34 is involved in transpirational cooling of rice leaves, which is an important aspect to stressful high temperatures and ABA-dependent regulation. We used infrared thermal imaging to measure leaf surface temperature of the transgenic rice plants with high temperature treatment (Mustilli et al. 2002). The results revealed that leaf temperature was significantly lower in OsOE1, OsOE2, and OsOE3 than WT plants, and the difference was up to 1–2 °C within 60 min (Fig. 2b, c). Conversely, leaf temperature was higher in osrzfp34 than in the WT under high temperature (Fig. 2b, c). OsRZFP34 may play an important role in regulating leaf temperature under heat stress.
Analysis of cooling effect in OsRZFP34-overexpressing rice plants. a Expression of OsRZFP34 in OsRZFP34-overexpression and-knockout rice plants. b Leaf temperature of heat-stressed rice plants. Thermal images of the 4th leaf of 3-week-old soil-grown seedlings of wild-type (WT), OsOE1, OsOE2, OsOE3, and osrzfp34 plants exposed to 38 °C for 60 min. c Data are mean ± SE temperature of the 4th leaf from 3 independent experiments (5 plants in each test)
Stomatal movement in response to high temperature is critical for leaf cooling. Thus, we used scanning electron microscopy to observe stomata. Under nonstress control conditions, stomatal aperture was significantly greater in rice plants constitutively expressing OsRZFP34 than in WT plants (Fig. 3a, b, c). Of note, in response to ABA, the mean stomata opening was significantly greater in OsRZFP34 transgenic plants than the WT (Fig. 3a, b), which suggests that OsRZFP34 may mediate stomata opening even with ABA treatment in rice. Furthermore, during heat stress, the stomatal aperture was larger in OsRZFP34-overexpressing rice than the WT, and elevated temperature did not enhance stomata opening in osrzfp34 (Fig. 3c). This finding correlates well with the increased cooling effect in OsRZFP34 transgenic plants and increased leaf temperature in osrzfp34.
Analysis of stomatal aperture in OsRZFP34-overexpressed rice plants. a The 4th leaf of 3-week-old soil-grown seedlings of WT, OsOE1, OsOE2, and OsOE3 was detached and incubated under light in stomata-opening solution (OS) for 2 h and then treated with 5 μM ABA for 30 min (OS → ABA). Scanning electron microscopy of stomatal aperture. Scale bar = 1 μm. b The stomata aperture of each plant incubated in OS for 150 min. c 3-week-old soil-grown seedlings of WT, OsOE1, OsOE2, OsOE3, and osrzfp34 were subjected to 38 °C heat stress. Data are mean ± SE from 3 independent experiments (30 stomata in each test). Bars with the same letter are not significantly different at P < 0.05
OsRZFP34 and AtRZFP34 are functional orthologs
To gain better insight into the function of OsRZFP34, we overexpressed OsRZFP34 in Arabidopsis thaliana ecotype Col plants. We selected three independent T3 homozygous transgenic lines AtOE2, AtOE3, and AtOE5, which constitutively expressed OsRZFP34 transcripts (Supplementary Fig. S3), to examine stomatal movement phenotypes. Stomatal aperture was measured in the focal planes of the outer edges of guard cells in epidermal strips of 3-week-old leaves. As compared with Col WT leaves, AtOE2, AtOE3 and AtOE5 leaves showed a 21–37 % increase in stomatal aperture under nonstress control conditions (Fig. 4a), which supports the expression of OsRZFP34 possibly related to stomata opening. We did not find significant difference in plant height, leaf morphology, and flowering time among the WT, AtOE2, AtOE3, and AtOE5 (data not shown).
OsRZFP34 transgenic Arabidopsis show increased stomata opening. a Col WT; OsRZFP34 transgenic plants AtOE2, AtOE3, and AtOE5; atrzfp34; and the complemented atrzfp34 lines atrzfp34 + OsRZFP34-2 and atrzfp34 + OsRZFP34-3 were grown on soil for 3 weeks, and abaxial epidermal imprints were observed by light microscopy. b Detached leaves were incubated under light in OS for 2 h, then treated with ABA for 30 min (OS → ABA). Data are mean ± SE from 3 independent experiments. Bars with the same letter are not significantly different at P < 0.05
To confirm that OsRZFP34 was responsible for regulating stomatal aperture, we used the deduced amino acid sequence to search for its homologs in the Arabidopsis genome database. We identified an Arabidopsis RZFP gene, At5g22920 (AtRZFP34), sharing 66 % amino acid identity with OsRZFP34, and obtained an allelic mutant, salk_017562 (atrzfp34), with a T-DNA insertion in its promoter region (Supplementary Fig. S4). After confirmation of abolished AtRZFP34 expression (Supplementary Fig. S4), homozygous mutant plants derived from a single T-DNA insertion were checked for stomatal aperture. As compared with leaves of the Col WT, those of atrzfp34 showed significantly decreased (~15 % reduction) stomatal aperture under nonstress control conditions (Fig. 4a). To test whether OsRZFP34 complements the mutation of AtRZFP34, we characterized the phenotype of 2 independent T3 homozygous complemented mutants, atrzfp34 + OsRZFP34-2 and atrzfp34 + OsRZFP34-3 (Supplementary Fig. S5). The OsRZFP34 transgene could restore stomata opening in the atrzfp34 mutant (Fig. 4a). Loss-of-function and complementation results indicated that these two highly similar proteins shared similar function in mediating stomatal aperture. Furthermore, we measured the stomatal aperture of mutant plants in response to ABA. After ABA treatment, the mean stomata opening was significantly greater for OsRZFP34 transgenic plants and OsRZFP34-complemented mutants than the Col WT and atrzfp34 (Fig. 4b). The expression of OsRZFP34 in transgenic Arabidopsis plants may increase stomata opening even under ABA treatment.
Ectopic expression of OsRZFP34 can elevate water loss and reduce leaf temperatures during stress
To test the effect of increased stomatal aperture in OsRZFP34 transgenic Arabidopsis plants, we determined the rate of water loss by measuring the fresh weight of shoots detached for 5 h. The fresh weight of detached shoots of AtOE2, AtOE3, and AtOE5 was reduced to <40 % of the initial weight, but that of the Col WT and atrzfp34 was maintained at 45 and 52 %, respectively (Fig. 5a). In addition, the change in fresh shoot weight was similar between atrzfp34 + OsRZFP34-2 and the Col WT during drought stress (Fig. 5a). Thus, increased stomatal aperture caused by the expression of OsRZFP34 is associated with enhanced water loss, which is an early response to high temperature stress (Chaves et al. 2002).
Analysis of water loss and cooling effect in OsRZFP34-overexpressing Arabidopsis plants. a Water loss analysis. The aerial parts of 4-week-old plants were allowed to dry under the fume hood. Water loss is expressed as the percentage of initial fresh weight. Data are mean ± SE from 3 independent experiments (10 plants in each test). b Leaf temperature of heat-stressed Arabidopsis plants. Twenty-day-old seedlings were subjected to up to 30 °C heat stress. Data are mean ± SE temperature of the 8th, 9th, and 10th leaf from quantification of infrared thermal images of 3 independent experiments (5 plants in each test). c Leaf temperature of drought-stressed Arabidopsis plants. Water was withheld from 12-day-old seedlings for 5 days. Data are mean ± SE temperature from 3 independent experiments (5 plants in each test). Bars with the same letter are not significantly different at P < 0.05
Using infrared thermography, we investigated the change in leaf temperature with high temperature treatment. AtOE2, AtOE3, and AtOE5 leaves maintained a significantly lower temperature within 60 min at 30 °C temperature as compared with Col WT leaves, whereas the temperature was higher for atrzfp34 than Col WT leaves (Fig. 5b). In contrast, leaf temperatures did not significantly differ among the Col WT, AtOE3, and atrzfp34 after 150 min of heat treatment. These results confirm the role of OsRZFP34 in a cooling effect during heat stress. Next, we examined the leaf temperature of plants under drought. After withholding water for 5 days, the leaf temperature of atrzfp34 was ~1 °C higher than that of the Col WT, which reflects reduced cooling from leaves of this mutant (Fig. 5c). This result is consistent with the lower water-loss rate of atrzfp34 (Fig. 5a). However, leaf temperature of OsRZFP34-overexpressing Arabidopsis was not significantly lower than that of Col WT with drought treatment. Nevertheless, these results clearly show that under heat and drought stress, OsRZFP34 was able to complement the atrzfp34 mutation and the leaf cooling effect.
Identification of genes upregulated by overexpression of OsRZFP34
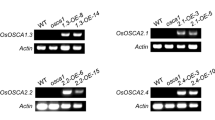
To further identify the target genes regulated by OsRZFP34 and elucidate the molecular mechanism mediated by this gene, we performed a transcriptome analysis of leaves using the Rice Whole Genome OneArray v1.1 (Phalanx Biotech Group, Taiwan) containing 22,003 DNA oligonucleotide probes. Relative change was calculated by comparing the data for OsOE1 against those for WT plants grown at 28 °C, with relative change of two-fold or more considered significantly different. We found 549 upregulated and 478 downregulated genes in OsOE1 plants as compared with WT plants (Supplementary Tables S2, S3). Calmodulin binding proteins, CAMKs, serine/threonine-protein phosphatase, serine/threonine-protein kinases, protein phosphatase 2C, potassium transporter genes, ABC transporter genes, and OsCML27, candidate components involved in ABA, K+, or Ca2+ signaling, comprised a significant portion of the differentially expressed genes (Table 1) (Sanders et al. 2002; Zhang and Lu 2003; Nagy et al. 2009; Siegel et al. 2009; Sirichandra et al. 2009; Umezawa et al. 2009). We also found transcripts of OsWRKYs, suggested to function in abiotic stresses including ABA stress, significantly upregulated in OsOE1 plants (Table 1) (Ross et al. 2007). qRT-PCR was performed to confirm the expression of 24 upregulated genes, and the results suggested that all of the chosen genes showed elevated expression levels in OsOE1 plants (Fig. 6), although the absolute values of the fold changes showed some variation for some genes between microarray and qRT-PCR analysis.
qRT-PCR analysis of upregulated genes in RZFP34-overexpression rice. WT and OsRZFP34 transgenic plants OsOE1 were grown in soil for 3 weeks. Leaves were harvested for extraction of total RNA. The mRNA expression of transcript levels of OsRZFP34-upregulated genes. The expression of genes in each sample is relative to that of the WT control, and the expression of 18S rRNA was an internal control. Data are mean ± SE from 3 independent experiments
Discussion
RING motifs, functioning in protein–protein interaction and ubiquitination, can be classified into 2 subgroups by presence of a cysteine or histidine in the fifth position: C3HC4 (RING-HC) and C3H2C3 (RING-H2) fingers. OsRZFP34 contains 2 types of zinc-finger domains, a Pfam:zf-CHY and a RING domain, and is one of the C3H2C3-type RZFPs (Supplementary Fig. S1). Most studies of the function of RZFP genes suggest a link between their expression and the stress response in plants. One RZFP, HOS1, has been shown to be a negative regulator of cold-responsive gene expression and physically interacts with ICE1 to mediate the ubiquitination of ICE1 and downregulate cold-induced genes such as CBF1 and COR15a (Lee et al. 2001; Dong et al. 2006). Others, such as SDIR1 and XERICO, are positive regulators of drought tolerance by altering the homeostasis of the ABA signaling pathway (Ko et al. 2006; Zhang et al. 2007). From microarray results, we identified a rice RZFP gene, OsRZFP34, that responds to ABA and heat. Phylogenetic analysis of RZFPs indicated the existence of OsRZFP34 and its homologs in higher land plants (Supplementary Fig. S2). Further analysis revealed that OsRZFP34 was expressed before the early flowering stage and transcripts were detected mainly in leaves (Fig. 1b, c, d), which is not often found with the expression patterns of RZFP genes (Zhang et al. 2007). Thus, OsRZFP34 may have a specific physiological function in rice leaves for the effect induced by heat and ABA stress.
High temperature and ABA treatment have opposite effects on mediating stomatal aperture; heat stress induces stomata opening for transpirational cooling to lower leaf temperature, but an increase in ABA triggers stomata closure to reduce water loss. Under natural growth conditions of plants, heat-induced rapid and continuous evaporation or a decrease in water content of the leaf causing immediate desiccation results in accumulation of ABA and stomata closure. Overexpression of OsRZFP34 may result in increased water loss even under drought treatment (Fig. 5a). Thus, the ABA responsiveness of OsRZFP34 may be contrary to ABA-induced stomata closure and coordinate transpiration cooling in response to elevated ambient temperatures. We further confirmed that transgenic rice and Arabidopsis overexpressing OsRZFP34 with larger stomata aperture can have a better cooling effect and reduce leaf temperature during the early stages of the heat condition (Figs 2c, 3c, 5b) or under drought (Fig. 5c). By contrast, null mutation of OsRZFP34 and AtRZFP34 conferred smaller stomata opening and limited evaporative cooling. Thus, OsRZFP34 and its homolog AtRZFP34 may ensure stomata opening when heat dissipation is more urgent than water preservation. Recent studies found that rice yield significantly declined for each 1 °C rise in the growing season minimum temperature in some low-latitude regions (Peng et al. 2004; Welch et al. 2010), which implied a negative impact on rice yields from increasing in night temperature during the vegetative phase. Consequently, it would be interesting to test whether OsRZFP34, which functions in lowering leaf temperature, may ameliorate the negative effects of thermal stress on the yield of irrigated rice.
During the heading stage, which in rice is the most sensitive stage to temperature, upregulation of stress-related proteins such as small HSPs (sHSPs) may protect metabolic activities and enhance heat tolerance (Jagadish et al. 2010). However, we found the survival of OsRZFP34 transgenic plants comparable to that of WT plants after HS treatment (unpublished observations in this lab), which suggests that OsRZFP34 cannot suppress HS-induced cellular damage. In addition, HS-responsive OsRZFP34 transcripts were less abundant in the heat-sensitive flowering stage (Fig. 1c). These results are consistent with our suggestion that OsRZFP34 functions in cooling of vegetative leaves during heat stress.
A number of signaling compounds, including ABA, ROS, nitric oxide and Ca2+ ions are involved in the regulation of stomatal aperture. The ABA signaling in guard cells has been reported to engage a complex network of both positive and negative regulating protein kinases and serine/threonine-protein phosphatase (Hirayama and Shinozaki 2007; Umezawa et al. 2009), and cytosolic Ca2+ elevation or sensitivity in Arabidopsis (Sanders et al. 2002; Zhang and Lu 2003; Siegel et al. 2009). However, little is known about the HS-induced signals for stomata opening and transpiration cooling. In this study, our microarray analysis showed the changed expression of serine/threonine-protein phosphatase and kinase genes, protein phosphatase 2C genes, calmodulin binding proteins, CAMKs, and OsCML27 in OsRZFP34-overexpressing rice plants as compared with WT plants (Table 1). Meanwhile, 2 potassium transporter genes, 3 ABC transporter genes, and 3 OsWRKY genes were upregulated in OsRZFP34-overexpressing rice plants. In Arabidopsis, a calcium-modulated serine/threonine-protein phosphatase 2C, AtABI1, is suggested to integrate ABA and Ca2+ signals to regulate stomata aperture in leaves (Leung et al. 1994); an ABC transporter, AtMRP5, might regulate Ca2+ and K+ release into the cytosol and promote stomata opening (Suh et al. 2007; Nagy et al. 2009); a plasma membrane K+ inward-rectifying channel, and AtKAT1, is extremely efficient in K+ uptake in the guard cells (Lebaudy et al. 2007). In addition, among the OsRZFP34-upregulated OsWRKY genes, OsWRKY80 shows ABA responsiveness (Ricachenevsky et al. 2010). Therefore, the role of overexpression of OsRZFP34 as a pleiotropic regulator may lead to enhanced stomatal opening by mediating the expression of these genes to result in leaf cooling (Figs. 2c, 5b, c). The direct involvement of OsRZFP34 protein in transpiration cooling would still need to be clarified by, for instance, characterization of the roles of these specific genes in mediating stomatal movement and the interaction of OsRZFP34 with these genes. However, the features of gain- and loss-of-function of OsRZFP34 raise attractive possibilities regarding its putative role in mediating transpiration cooling described above.
In conclusion, we identified an RZFP, OsRZFP34, as an important regulator of the response of heat and ABA treatments in rice plants. OsRZFP34 may increase stomatal opening and function in transpiration cooling when rice plants are exposed to high temperatures. Higher transpirational cooling lowers leaf temperature and thus provides an early avoidance strategy of heat stress to irrigated rice grown at supra-optimal temperatures. OsRZFP34 may be a good candidate for understanding the signaling pathways for transpiration cooling.
Abbreviations
- ABA:
-
Abscisic acid
- Col:
-
Arabidopsis thaliana ecotype Columbia
- HS:
-
Heat shock
- HSPs:
-
Heat shock proteins
- OS:
-
Opening solution
- ROS:
-
Reactive oxygen species
- RZFP:
-
RING zinc-finger protein
- sHSPs:
-
Small heat shock proteins
- TRIM:
-
Taiwan rice insertion mutant
- WT:
-
Wild type
References
Chaves MM, Pereira JS, Maroco J, Rodrigues ML, Ricardo CPP, Osório ML, Carvalho I, Faria T, Pinheiro C (2002) How plants cope with water stress in the field. Photosynthesis and growth. Ann Bot 89:907–916
Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Nat Acad Sci 103:8281–8286
Freemont PS, Hanson IM, Trowsdale J (1991) A novel cysteine-rich sequence motif. Cell 64:483–484
Guan JC, Yeh CH, Lin YP, Ke YT, Chen MT, You JW, Liu YH, Lu CA, Wu SJ, Lin CY (2010) A 9 bp cis-element in the promoters of class I small heat shock protein genes on chromosome 3 in rice mediates L-azetidine-2-carboxylic acid and heat shock responses. J Exp Bot 61:4249–4261
Guo WJ, Ho THD (2008) An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147:1710–1722
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:343–351
Hua D, Wang C, He J, Liao H, Duan Y, Zhu Z, Guo Y, Chen Z, Gong Z (2012) A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24:2546–2561
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23:1805–1817
Hurng WP, Lin TL, Ren SS, Chen JC, Chen YR, Kao CH (1988) Senescence of rice leaves XVIII. Changes of stomatal aperture during senescence. Plant Cell Physiol 29:27–31
Hüve K, Bichele I, Rasulov B, Niinemets Ü (2011) When it is too hot for photosynthesis: heat-induced instability of photosynthesis in relation to respiratory burst, cell permeability changes and H2O2 formation. Plant Cell Environ 34:113–126
Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61:143–156
Ko JH, Yang SH, Han KH (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47:343–355
Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10:310–316
Kuromori T, Sugimoto E, Shinozaki K (2011) Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J 67:885–894
Lebaudy A, Anne-Alie’nor Véry A-A, Sentenac A (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581:2357–2366
Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15:912–924
Leung J, Bouvier-Durand M, Morris P-C, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264:1448–1452
Lewis DA, Nobel PS (1977) Thermal energy exchange model and water loss of a barrel cactus, Ferocactus acanthodes. Plant Physiol 60:609–616
Liu FL, Jensen CR, Shahanzari A, Andersen MN, Jacobsen SE (2005) ABA regulated stomatal control and photosynthetic water use efficiency of potato (Solanum tuberosum L.) during progressive soil drying. Plant Sci 168:831–836
Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14:3089–3099
Nagy R, Grob H, Weder B, Green P, Klein M, Frelet-Barrand A, Schjoerring JK, Brearley C, Martinoia E (2009) The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J Biol Chem 284:33614–33622
Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG (2004) Rice yields decline with higher night temperature from global warming. Proc Nat Acad Sci 101:9971–9975
Ricachenevsky FK, Sperotto RA, Menguer PK, Fett JP (2010) Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol Biol Rep 37:3735–3745
Ross CA, Liu Y, Shen QJ (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49:827–842
Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14:S401–S417
Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410:327–330
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Siegel RS, Xue S, Murata Y, Yang Y, Nishimura N, Wang A, Schroeder JI (2009) Calcium elevation-and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and Kin channels in Arabidopsis guard cells. Plant J 59:207–220
Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J (2009) The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot 60:1439–1463
Suh SJ, Wang YF, Frelet A, Leonhardt N, Klein M, Forestier C, Mueller-Roeber B, Cho MH, Martinoia E, Schroeder JI (2007) The ATP binding cassette transporter AtMRP5 modulates anion and calcium channel activities in Arabidopsis guard cells. J Biol Chem 282:1916–1924
Sun W, van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochem Biophys Acta 1577:1–9
Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126:45–51
Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R (2008) The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283:9269–9275
Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci 106:17588–17593
Weigel D, Glazebrook J (2002) Arabidopsis: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Welch JR, Vincent JR, Auffhammer M, Moya PF, Dobermann A, Dawe D (2010) Rice yields in tropical/subtropical Asia exhibit large but opposing sensitivities to minimum and maximum temperatures. Proc Natl Acad Sci 107:14562–14567
Yeh CH, Chang PFL, Yeh KW, Lin WC, Chen YM, Lin CY (1997) Expression of a gene encoding a 16.9-kDa heat shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci 94:10967–10972
Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142:1548–1558
Zhang L, Lu YT (2003) Calmodulin-binding protein kinases in plants. Trends Plant Sci 8:123–127
Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xiea Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Zhou S, Zhao W, Li H, Guo Z, Peng YL (2008) Characterization of a novel RING finger gene OsRFP1, which is induced by ethylene, salicylic acid and blast fungus infection in rice. J Phytopathol 156:396–402
Acknowledgments
We are grateful to Drs. Tong-Seung Tseng, Tzung-Meng Wu, and Kuo-Chen Yeh for comments and suggestions on the manuscript; and Dr. Yee-Yung Charng for technical assistance with thermal imaging. We thank the electron microscopy laboratory of Tzu Chi University. We also thank ABRC and TRIM for providing seeds. This work was supported by the National Science Council, Taiwan, ROC (NSC96-2317-B-008-003 and NSC98-2324-B-008-002 to C.H. Yeh).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hsu, KH., Liu, CC., Wu, SJ. et al. Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol Biol 86, 125–137 (2014). https://doi.org/10.1007/s11103-014-0217-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0217-6