Abstract
Nucleolin plays important roles in chromatin structure, rDNA transcription, rRNA maturation, nucleocytoplasmic transport, and ribosome assembly. Although it has been shown to be anti-apoptotic, the underlying mechanisms remain unclear. In the current study, we first examined endogenous nucleolin expression in response to oxidative stress-induced apoptosis in human umbilical vascular endothelial cells (HUVECs). Flow cytometry and caspase activity assays showed that H2O2 treatment caused apoptosis of the cells; reverse-transcription polymerase chain reaction and Western blotting revealed the downregulation of nucleolin expression and increased protein cleavage during this process. Overexpression of nucleolin protein by transfecting cells with the full-length nucleolin cDNA inhibited apoptosis, but nucleolin deficiency brought about by transfection with antisense oligonucleotide increased apoptosis of HUVECs. Concurrently, the expression of the apoptotic protein gene Bax was also downregulated following nucleolin overexpression. All these results indicate an important negative regulatory role for nucleolin in the apoptosis of endothelial cells, likely involving the Bax pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
As a multifunctional DNA- and RNA-binding protein, nucleolin (C23) is abundantly expressed in both normal and cancerous cells (Derenzini et al. 1995) in the nucleolus, nucleus, and cytoplasm of the cell as well as at the cell surface (Hovanessian et al. 2000). Nucleolin has been shown to regulate many aspects of DNA and RNA metabolisms. For example, it is involved in the regulation of RNA polymerase I-mediated transcription and is required for newly synthesized pre-rRNA folding, maturation, ribosome assembly, and transport outside the nuclei (Ginisty et al. 1998). Due to its widespread distribution and broad range of involvement and the cross-talking among different cellular processes, dissection of nucleolin’s functions is particularly challenging.
Recently, nucleolin has also been implicated in apoptosis. All-trans retinoic acid-induced apoptosis results in nucleolin downregulation and bcl-2 mRNA instability; overexpression of nucleolin in chronic lymphocytic leukemia cells induces the stabilization of bcl-2 mRNA (Otake et al. 2005, 2007). Our previous study (Wang et al. 2004) also showed that oxidative stress can induce nucleolin cleavage and apoptosis. Although nucleolin cleavage and apoptosis occurred simultaneously, it remains unclear whether nucleolin plays a direct role in the oxidative stress-induced apoptosis. In this study, we investigated the expression and translocation of endogenous nucleolin in human umbilical vein endothelial cells (HUVECs) in response to H2O2-induced apoptosis and further studied the effects of nucleolin overexpression or ablation on this apoptosis induced by H2O2.
2 Materials and methods
Cell culture
HUVECs were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences and routinely grown in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal calf serum at 37°C under 5% CO2.
Plasmid constructs
A recombinant plasmid pEGFP-C1-C23 carrying the full-length human nucleolin cDNA was a generous gift from Professor Michael B. Kastan at St. Jude Children’s Research Hospital, USA. Oligonucleotide primers designed to amplify the cDNA were as follows: 5′-CGC GGA TCC CAT GGT GAA GCT CGC-3′ (forward) and 5′-CGG GGT ACC TAT TCA AAC TTC GTC TTC T-3′ (reverse). The PCR products were electrophoresed in a 0.9% agarose gel and the DNA fragment was purified with a Gel Extraction Kit (Qiagen). The fragment was then inserted into the pcDNA3.1 vector (Strategene) and the insert sequence was confirmed by sequencing (Invitrogen).
Gene transfection
Transfection of HUVECs was carried out with LIPOFECTAMINE 2000™ according to the manufacturer’s instructions (Invitrogen, US; Jiang et al. 2005a). Briefly, 10 μg plasmid DNA pcDNA3.1-C23 (experimental) or the pcDNA3.1 empty vector (control) was mixed with 20 μL lipofectamine in serum- and antibiotics-free DMEM, and DNA/lipofectamine mixture was added into the cell culture medium and incubated at 37°C in a CO2 incubator for 6 h. The medium was then replaced with fresh Rapid Prototyping and Manufacturing Institute-1640 containing 10% fetal bovine serum. After 18 h in culture, the cells were then treated with H2O2 following the experiment procedures as described in “Results” section.
Loss-of-function assay using morpholino oligos
A nucleolin morpholino antisense oligonucleotide (Wang et al. 2005) was designed to target the initiation site for nucleolin translation (AS: ACC TGC CTT CGC GAG CTT CAC CAT) and was synthesized commercially (Invitrogen). Morpholinos were transfected into HUVECs with lipofectamine according to the manufacturer’s instructions (LIPOFECTAMINE 2000™, Invitrogen) 24 h after plating. To determine the specificity of the antisense oligo, a sense oligo (S: CAT GGT GAA GCT CGC GAA GGC AGG T) and a random invalid oligo (R: CTA CGA GAC TGC CTC CAC TGC TTC G) were used to transfect cells, and lipofectamine solution (Ctrl) without any DNA was also used to mock-transfect the cells as controls. Cell samples were collected 48 h after transfection and analyzed for nucleolin mRNA and protein levels by reverse-transcription polymerase chain reaction (RT-PCR) and Western blot analysis, respectively.
Reverse-transcription PCR
Total RNA was extracted by TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. One microgram of total RNA was reverse-transcribed (Fermentas), and the cDNA was used as template to amplify genes of interest by PCR in an iCycler Apparatus (Biometra) with the following gene-specific primers: glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 22 cycles), 5′-ACC CAG AAG ACT GTG GAT GG-3′ (forward) and 5′-TTC TAG ACG GCA GGT CAG GT-3′ (reverse); nucleolin (25 cycles), 5′-CGC GGA TCC CAT GGT GAA GCT CGC-3′ (forward) and 5′-CGG GGT ACC TAT TCA AAC TTC GTC TTC T-3′ (reverse); and Bcl-2-associated X protein (Bax; 25 cycles), 5′-GTG CAC CAA GGT GCC GGA AC-3′ (forward) and 5′-TCA GCC CAT CTT CTT CCA GA-3′ (reverse).
Western blot analysis
After various treatments, the cells were lysed in lysis buffer (62.5 mM Tris-HCl, pH 6.8; 2% sodium dodecyl sulfate (SDS), 10% glycerine, and 50 mM dithiothreitol). Total proteins in the whole-cell lysates were resolved on 10% SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred onto polyvinylidene fluoride (PVDF) membranes (Schleicher & Schuell). After blocking overnight in phosphate-buffered saline containing 10% nonfat dry milk and 0.5% Tween-20, membranes were then incubated with mouse anti-CC23 or anti-GAPDH monoclonal antibody (1:1,000, Santa Cruz Biotechnology) at room temperature for 2 h. After washing with phosphate buffered saline (PBS), the blot was incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (1:1,000, Boster Biological Technology, China). The immunoreactive bands were visualized using DAB reagent (Boster Biological Technology, China). The protein level of the housekeeping gene GAPDH was also detected in parallel for normalization of protein loadings and the normalized values were used to calculate the relative induction ratio.
Preparation of cellular extracts
At indicated time points after the treatment, cells were harvested and washed twice with cold PBS and nuclear and cytoplasmic extracts were prepared according to the method of Schreiber et al. (Schreiber et al. 1989).
Flow cytometry
All groups of cells in culture were collected at confluence. After washing twice with PBS, the cells were fixed in 70% ethanol for 18 h at 4°C. All the fixed cell samples were delivered to Beijing Dingguo Biotechnology for evaluation of the cell cycle progression and apoptosis using Coulter Epics XL.
Caspase-3 activity assay
Caspase-3 fluorescent assay kit (NanJing KeyGen Biotech, China) was used to detect caspase activity as described previously (Jiang et al. 2005b). In brief, cells were cultured in 60-mm dishes and treated according to the protocol described in Results. Cells were lyzed in the lysis buffer and centrifuged at 10,000×g for 1 min, and then the supernatants were collected. With bovine serum albumin as the standard, equal amounts of protein samples were reacted with the synthetic fluorescent substrates at 37°C for 1.5 h and the reactions were read at 405 nm in a microplate reader (Biorad). Fold-increase in caspase-3 activity was determined with values obtained from the treatment samples divided by those from the controls.
Statistical analysis
Each experiment was performed for three times, and the data were expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed using a two-tailed Student’s t test. P < 0.05 was considered statistically significant.
3 Results
3.1 Apoptosis of HUVEC induced by H2O2
First, HUVECs were treated with hydrogen peroxide (H2O2) at an apoptosis-inducing dose previously determined in our laboratory (0.5 mmol/L; Wang et al. 2004) for various time periods. As shown by flow cytometry in Fig. 1a, H2O2 treatment caused apoptosis of HUVECs, and the apoptosis was accompanied by an increase of caspase-3 activity (Fig. 1b).
Apoptosis of HUVECs induced by H2O2. HUVECs were stimulated with H2O2 at the indicated dose over a period of time, and cells were harvested at various time points for different assays. a Percentages of apoptotic HUVECs were determined by flow cytometry. b Caspase-3 activities were determined by caspase-3 activity detection assay. The results were expressed as mean ± SEM of relative values from three experiments. *P < 0.05, statistically significant versus control group (0 h)
3.2 Nucleolin expression and translocation in apoptotic HUVECs
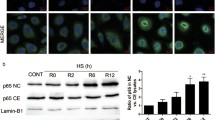
Subsequently, nucleolin expression was determined during the apoptosis of HUVECs. As shown in Fig. 2a, following H2O2 treatment, nucleolin mRNA levels showed an initial rapid decrease and then gradual recovery from 8 to 12 h but did not reach the pretreatment level in the control even at the 12-h time point. At the protein levels, as shown in Fig. 2b, nucleolin was degraded into a fragment of 80 kD after H2O2 treatment, and the amount of the 80-kD fragment significantly increased after 24 h. This result suggests that nucleolin cleavage might contribute to apoptosis of these cells. In addition, by Western blot analysis with fractionated protein samples, translocation of nucleolin in apoptotic HUVECs was observed. As shown in Fig. 2c, under normal conditions without treatment, nucleolin protein was mostly found in the nuclei with trace amount in the cytosol. Exposure to H2O2 resulted in significant increase of the protein in the cytosol fraction, indicating the release of nucleolin from the nuclei into the cytosol.
Expression and translocation of nucleolin in apoptotic HUVECs. a The mRNA levels of nucleolin were determined by RT-PCR. b The protein levels of nucleolin were determined by Western blot analysis. The results were expressed as mean ± SEM of relative values from three experiments in duplicates. **P < 0.01, statistically significant versus the control group (0 h). c The cells were treated with 0.5 mmol/L H2O2 for 1 and 3 h, respectively, and cytosolic (Cyto) and nucleolar (Nuc) extracts were separated by 12% SDS-PAGE and analyzed by immunoblotting with anti-C23, anti-PCNA, and anti-GAPDH
3.3 Apoptosis of HUVECs with nucleolin overexpression
To investigate the effect of nucleolin overexpression on H2O2-induced apoptosis, we transfected HUVECs with pcDNA3.1-C23 construct using the empty vector pcDNA3.1 as the control. After the overexpressed nucleolin protein was confirmed (Fig. 3a), the cells showed dramatic decreases in both the percentage of apoptotic cells (Fig. 3b) and the overall caspase-3 activity (Fig. 3c) following H2O2 treatment (0.5 mmol/L) for 24 h. These results indicated that overexpression of nucleolin inhibited apoptosis of HUVECs.
Apoptosis of HUVECs with nucleolin overexpression. a Expression levels of nucleolin protein in HUVECs transfected with the nucleolin construct and the empty vector pcDNA3.1 were determined by Western blot analysis. *P < 0.05, statistically significant versus the vector control group. b Percentages of apoptotic HUVECs were determined by flow cytometry. **P < 0.01, statistically significant versus the vector control group without treatment (pcDNA3.1); ##P < 0.01, statistically significant versus the vector control group with H2O2 treatment (+H2O2). c Caspase-3 activities in HUVECs were determined by caspase-3 activity detection assay. **P < 0.01, statistically significant versus the vector control group (pcDNA3.1); #P < 0.05, statistically significant versus the vector control group with H2O2 treatment (pcDNA3.1+H2O2). The results were expressed as mean ± SEM of relative values from three experiments
3.4 Apoptosis of nucleolin-deficient HUVECs
Next is to investigate the effect of nucleolin deficiency on apoptosis. The morpholino antisense oligonucleotides of nucleolin were transfected into HUVECs, and the inhibition of nucleolin expression was confirmed by RT-PCR and Western blot (Fig. 4a, b) at the mRNA and protein levels, respectively. The nucleolin deficiency resulted in significant increases in the percentage of apoptotic cells and also caspase-3 activity (Fig. 4c, d). Although sense treatment showed a slight increase in caspase activity but this increase was not statistically significant, possibly caused by a nonspecific apoptotic effect of transfection. These results further demonstrated the anti-apoptotic effect of nucleolin on HUVECs.
Apoptosis of nucleolin-deficient HUVECs. Nucleolin deficient cells were subjected to RT-PCR (a) and Western blot analysis (b) to measure the residual expression of nucleolin; the percentages of apoptotic cells were determined by flow cytometry (c) and caspase-3 activities were determined by caspase-3 activity detection assay (d). Ctrl: HUVECs were only treated with lipofectamine; S: HUVECs were treated with sense oligonucleotide of nucleolin; AS: HUVECs were transiently transfected with antisense oligonucleotide against nucleolin; R: HUVECs were transiently transfected with a random oligonucleotide nonspecific to nucleolin. The results were expressed as mean ± SEM of relative values from three experiments. *P < 0.05, statistically significant versus the control group (Ctrl). **P < 0.01, statistically significant versus the control group (Ctrl)
3.5 Bax expressions in HUVEC with overexpression or deficiency of nucleolin
Since Bax is a well-known apoptotic protein marker, we further investigated the changes of Bax gene expression in HUVECs with overexpression or deficiency of nucleolin. As shown in Fig. 5, nucleolin deficiency upregulated while nucleolin overexpression downregulated Bax expression in HUVECs, suggesting a potentially negative effect of nucleolin on Bax expression in HUVECs (Fig. 5). Furthermore, after H2O2 treatment, Bax protein levels markedly increased in the control cells transfected with the empty vector alone, which is consistent with the function of Bax during apoptosis; however, overexpression of nucleolin by transfection with pcDNA-C23 plasmid (C23/C) under H2O2 treatment significantly inhibited this apoptotic protein expression and caused an overall decrease of Bax protein levels in the same cells, similar to the untreated cells (Fig. 6).
Expressions of Bax in HUVECs with nucleolin overexpression or deficiency. Effects of nucleolin overexpression or deficiency on the levels of Bax were measured in HUVECs by RT-PCR (a) and Western blot analysis (b). Ctrl: HUVECs were treated with lipofectamine alone; AS: HUVECs were transiently transfected with antisense oligonucleotide against nucleolin; p-C23: HUVECs were transiently transfected with pcDNA3.1-C23. * and #P < 0.05, statistically significant versus the control group (Ctrl). ** and ##P < 0.01, statistically significant versus the control group (Ctrl)
Bax protein analysis in C23-transfected cells during H2O2 exposure. HUVECs were transfected with pcDNA vector (pcDNA/C) or pcDNA-C23 (C23/C). After treatment with H2O2 (0.5 mmol/L) for 12 h, cells were harvested and protein lysates were prepared. Total cellular proteins (10 μg) were separated by 10% SDS-PAGE and blotted onto PVDF membrane. The blots were incubated with anti-Bax or anti-GAPDH antibody for 2 h at room temperature. Immunoreactivity was determined by enhanced chemiluminescence
4 Discussion
Apoptosis is involved in the pathogenesis of many diseases including ischemia–perfusion injury and myocardial infarction. In particular, apoptosis of endothelial cells is involved in many common cardiovascular diseases such as atherosclerosis, renal diseases, and cardiac death. Protection of endothelial cells from apoptosis is therefore critical to the prevention and treatment of cardiovascular diseases.
Nucleolin is a major nucleolar protein of eukaryotic cells. It induces chromatin decondensation after binding to intranucleolar chromatin and preribosomal particles, as well as histone H1. It also plays roles in pre-rRNA transcription, ribosome assembly, and transcriptional elongation. Previous experiments have demonstrated that downregulation and protein cleavage of nucleolin are related to apoptosis (Kito et al. 2005; Otake et al. 2005). Here, we show that H2O2 treatment induces apoptosis of HUVECs, but the same treatment inhibits nucleolin expression and causes the protein to degrade into a smaller 80-kD fragment. These results confirm the protective role of nucleolin in apoptosis.
The negative regulation of nucleolin on H2O2-induced apoptosis is further confirmed by assays of apoptosis and caspase-3 activity in HUVECs following the overexpression or inhibition of nucleolin protein. Although caspase is a commonly used apoptosis index of cells, in Fig. 3a, b, overexpression of nucleolin caused a 75% decline in apoptosis by H2O2, but only about 40% of decrease in caspase-3 activity. First, it is not totally surprising to have different results by assessing apoptosis with different cellular indexes in different methods. Second, although overexpression of nucleolin affects both the overall function such as dysregulation of cell cycles and deteriorate cell morphology, as well as the activity of cellular proteins, these changes cannot be always synchronized in real time. They can differ not only in the degree and magnitude of the changes but also the time courses of the changes.
As an RNA binding protein, nucleolin contains four consensus RNA-binding domains, and the molecular function of this protein is likely reflected by its RNA-binding properties. Nucleolin has been shown to control p53 translation and induction by binding to the 5′-untranslated region of p53 mRNA after DNA damage (Takagi et al. 2005). Nucleolin may function to stabilize bcl-2 mRNA since recombinant human nucleolin can slow down the rate of bcl-2 mRNA decay upon addition to normal B cell extracts; on the other hand, siRNA knockdown of nucleolin decreased the bcl-2 mRNA and protein levels in MCF-7 cells. Since Bcl-2 suppresses the initiation of the cell-death process (Chao and Korsmeyer 1998), stabilization of Bcl-2 may be one mechanism by which nucleolin inhibits apoptosis. The Bcl-2-associated X protein gene is the first identified pro-apoptotic member of the Bcl-2 protein family (Oltvai et al. 1993). We show here that the overexpression of nucleolin downregulates Bax and nucleolin deficiency upregulates the gene. However, whether nucleolin binds to Bax mRNA still needs to be further studied. We also show the translocation of nucleolin from the nucleus to the cytoplasm during apoptosis in these cells, which might be related to the regulatory mechanism of nucleolin on the target genes.
In conclusion, our study showed that H2O2 induces apoptosis of HUVECs and nucleolin is downregulated during this process. Nucleolin overexpression leads to the inhibition of the H2O2-induced apoptosis and concurrently the reduction in the expression of the pro-apoptotic factor Bax. Together, these results demonstrate a role of nucleolin in the protection of the endothelial system from apoptosis.
References
Chao DT, Korsmeyer SJ (1998) BCL-2 family: regulators of cell death. Annu Rev Immunol 16:395–419
Derenzini M, Sirri V, Trere D, Ochs RL (1995) The quantity of nucleolar proteins nucleolin and protein B23 is related to cell doubling time in human cancer cells. Lab Invest 73(4):497–502
Ginisty H, Amalric F, Bouvet P (1998) Nucleolin functions in the first step of ribosomal RNA processing. Embo J 17(5):1476–1486
Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng JS, Krust B (2000) The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res 261(2):312–328
Jiang B, Xiao W, Shi Y, Liu M, Xiao X (2005a) Heat shock pretreatment inhibited the release of Smac/DIABLO from mitochondria and apoptosis induced by hydrogen peroxide in cardiomyocytes and C2C12 myogenic cells. Cell Stress Chaperones 10(3):252–262
Jiang B, Xiao W, Shi Y, Liu M, Xiao X (2005b) Role of Smac/DIABLO in hydrogen peroxide-induced apoptosis in C2C12 myogenic cells. Free Radic Biol Med 39(5):658–667
Kito S, Morimoto Y, Tanaka T, Haneji T, Ohba T (2005) Cleavage of nucleolin and AgNOR proteins during apoptosis induced by anticancer drugs in human salivary gland cells. J Oral Pathol Med 34(8):478–485
Oltvai ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74(4):609–619
Otake Y, Sengupta TK, Bandyopadhyay S, Spicer EK, Fernandes DJ (2005) Retinoid-induced apoptosis in HL-60 cells is associated with nucleolin down-regulation and destabilization of Bcl-2 mRNA. Mol Pharmacol 67(1):319–326
Otake Y, Soundararajan S, Sengupta TK et al (2007) Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood 109(7):3069–3075
Schreiber E, Matthias P, Muller MM, Schaffner W (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 17:6419
Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123(1):49–63
Wang KK, Jiang L, Yi YX et al (2004) Effect of heat shock response on the cleavage of nucleolin induced by oxidative stress. Zhong Nan Da Xue Xue Bao Yi Xue Ban 29(05):504–508
Wang KK, Jiang L, E SM, Liu K, Zhang LL, Liu MD, Xiao XZ (2005) Effect of nucleolin down-regulation on the proliferation and apoptosis in C2C12 cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban 30(2):125–129
Acknowledgments
This work was supported by fundings from the National Natural Science Foundation of China (30700290), the Major National Basic Research Program of China (2007CB512007), and the Opening Fund of The State Key Laboratory Of Trauma Burns and Combined Injury (SKLKF200806).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Bin Zhang and Haiyun Wang contributed equally to the study.
Rights and permissions
About this article
Cite this article
Zhang, B., Wang, H., Jiang, B. et al. Nucleolin/C23 is a negative regulator of hydrogen peroxide-induced apoptosis in HUVECs. Cell Stress and Chaperones 15, 249–257 (2010). https://doi.org/10.1007/s12192-009-0138-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-009-0138-5