Abstract
Apart from their roles as chaperones, heat shock proteins are involved in other vital activities including apoptosis with mammalian Hsp60 being ascribed proapoptotic as well as antiapoptotic roles. Using conditional RNAi or overexpression of Hsp60D, a member of the Hsp60 family in Drosophila melanogaster, we show that the downregulation of this protein blocks caspase-dependent induced apoptosis. GMR-Gal4-driven RNAi for Hsp60D in developing eyes dominantly suppressed cell death caused by expression of Reaper, Hid, or Grim (RHG), the key activators of canonical cell death pathway. Likewise, Hsp60D-RNAi rescued cell death induced by GMR-Gal4-directed expression of full-length and activated DRONC. Overexpression of Hsp60D enhanced cell death induced either by directed expression of RHG or DRONC. However, the downregulation of Hsp60D failed to suppress apoptosis caused by unguarded caspases in DIAP1-RNAi flies. Furthermore, in DIAP1-RNAi background, Hsp60D-RNAi also failed to inhibit apoptosis induced by RHG expression. The Hsp60 and DIAP1 show diffuse and distinct granular overlapping distributions in the photoreceptor cells with the bulk of both proteins being outside the mitochondria. Depletion of either of these proteins disrupts the granular distribution of the other. We suggest that in the absence of Hsp60D, DIAP1 is unable to dissociate from effecter and executioner caspases, which thus remain inactive.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The Hsp60 family, a major group of the heat shock proteins (Nover 1984), includes stress inducible and constitutively expressed members (McMullin and Hallberg 1988). Hsp60 members are believed to be predominately mitochondrial, although some are also reported in cytosol and in extracellular compartments (Gupta 1990; Retzlaff et al. 1994; Sarkar et al. 2006). As a molecular chaperon, Hsp60 helps in the folding of nascent polypeptides and in the transport of proteins from cytoplasm to organelles (Fink 1999). In addition to typical chaperon function, these proteins are also implicated in other diverse activities like amino acid transport, signal transduction, peptide presentation, regulation of immune system, apoptosis, etc. (Ikawa and Weinberg 1992; Jones et al. 1994; Wells et al. 1997; Woodlock et al. 1997; Sarkar et al. 2006).
Hsp60 family member is generally not induced by heat shock in Drosophila tissues (Tissieres et al. 1974) except in Malpighian tubules (Lakhotia and Singh 1989, 1996). The Berkeley Drosophila Genome Project has reveled four Hsp60 genes, named as Hsp60A, Hsp60B, Hsp60C, and Hsp60D, respectively (Sarkar and Lakhotia 2005). Studies in our and other laboratories have shown that the Hsp60A, Hsp60B, and Hsp60C proteins have distinct functions in normal development (Kozlova et al. 1997; Perezgasga et al. 1999; Timakov and Zhang 2001; Sarkar and Lakhotia 2005, 2008; Sarkar et al. 2006).
In the present study, we examined the possible functions of the Hsp60D (CG16954) gene. This gene is located at the 34C1 band, spans a 1.9-kb long region, and codes for a putative protein of 448aa, which shows sequence homology with the GroEL of bacteria and human Hsp60 (HSPD1). Transcripts of this gene are ubiquitously found in larval tissues like imaginal discs, salivary glands, etc. (Arya and Lakhotia, unpublished manuscript). In the absence of availability of a mutant allele of this gene, we generated transgenic lines for its conditional RNAi or overexpression. Initial studies showed that ubiquitous ablation or overexpression of Hsp60D using Act5C-Gal4 or Tub-Gal4 drivers caused varying degrees of larval or pupal lethality but the surviving flies appeared normal. For tissue-specific expression of the transgenes, we used various Gal4 drivers, including GMR-Gal4, which expresses (Ellis et al. 1993; Hay et al. 1994) in developing eye cells posterior to the morphogenetic furrow (MF). It is known that flies homozygous for the GMR-Gal4 transgene by themselves show some degree of degeneration in eyes (Kramer and Staveley 2003). Unexpectedly, however, the expression of Hsp60D-RNAi in the presence of two copies of GMR-Gal4 transgenes was found to rescue the eye degeneration. Because the eye degeneration in GMR-Gal4 flies is due to elevated incidence of apoptosis and because Hsp60 proteins have been implicated to have proapoptotic and antiapoptotic roles (Sarkar et al. 2006; Arya et al. 2007), we undertook the present set of experiments to see if Hsp60D protein of Drosophila melanogaster has a role in cell death pathway/s.
Programmed cell death by apoptosis is a well-known mechanism through which unwanted cells are removed (Vaux and Korsmeyer 1999). Topmost regulators of the canonical cell death pathway in fly are Reaper (Rpr), Hid, and Grim, collectively often referred to as RHG proteins, which inactivate the inhibitors of caspases and thus trigger apoptosis when expressed (White et al. 1994; Grether et al. 1995; Chen et al. 1996), (Hay and Guo 2006; Arya et al. 2007). The caspases normally remain inactive as unprocessed zymogens and/or bound to inhibitors like inhibitor of apoptosis protein or IAP (Hawkins et al. 1998; Deveraux and Reed 1999; Rodriguez et al. 2002; Salvesen and Duckett 2002). Upon apoptotic stimuli, upstream activators cause the release of inactive caspases from their inhibitors and bring about their proteolytic processing to make active caspases available (Kumar and Colussi 1999; Nicholson 1999). Of the seven caspases known in Drosophila, DRONC, DREDD, and STRICA are initiators whereas DRICE, DCP-1, DECAY, and DCP2/DAMM are effectors (Riedl and Shi 2004; Hay and Guo 2006). A number of baculovirus IAP-related proteins have been identified in Drosophila, the most extensively studied among them being DIAP1 and DIAP2 (Hay et al. 1995; Duckett et al. 1996; Jones et al. 2000). Ectopic expression of DIAP1 or DIAP2 can suppress cell death induced by RHG proteins or DRONC (Hay et al. 1995; Meier et al. 2000).
In the present paper, we show that the modulation of Hsp60D levels through conditional RNAi or overexpression affects induced apoptosis. Our results suggest that Hsp60D may be necessary for apoptosis through its interaction with DIAP1. A preliminary account of these findings was presented earlier (Arya et al. 2007).
2 Materials and methods
2.1 Generation of Hsp60D-RNAi and UAS-Hsp60DWT transgenic flies
The base sequence of the Hsp60D gene carries short but significant homologies with other Hsp60 genes of Drosophila. Bioinformatic analysis showed that the sequence of a 671-bp BglII–EcoRI (902–1,573bp) fragment from the Hsp60D cDNA clone (AT04835, MRC Gene Services, UK) was unique to Hsp60D. Further bioinformatics analysis was undertaken to check the possibility of “off target effects” (Kulkarni et al. 2006; Perrimon and Mathey-Prevot 2007) of the siRNA fragments produced from the 671-bp dsRNA in the Hsp60D-RNAi transgenic line using two online softwares (http://www.dkfz.de/signaling2/e-rnai/ and http://www.shigen.nig.ac.jp/fly/nigfly/). These analyses confirmed that the 671-bp BglII–EcoRI fragment of the Hsp60D cDNA clone can cause specific RNAi for Hsp60D mRNA without any other mRNA being significantly targeted (data not presented). Therefore, the 671-bp BglII–EcoRI fragment from the Hsp60D cDNA clone (AT04835) was inserted between two oppositely oriented UAS promoters in the Sym-pUAST vector (Giordano et al. 2002) to generate the Hsp60D-RNAi transgene.
For the overexpression of Hsp60D, the AT04835 Hsp60D cDNA clone was partially digested with EcoRI and BglII to isolate a 2,010-bp fragment, which carries the full coding region of the Hsp60D gene. This fragment was cloned in the pUAST vector (Brand and Perrimon 1993) to generate UAS-Hsp60DWT overexpressing transgene.
To generate transgenic flies, the above two constructs were microinjected separately in y w; +; Δ2–3 Sb ki/Δ2–3 Sb ki embryos (Cooley et al. 1988). Stable transgenic insertion lines were established by removing the transposase source through appropriate crosses. From among the several independent lines established for the Hsp60D-RNAi and the UAS-Hsp60DWT transgenes, two lines of each were selected for further studies (see the “Results” section).
2.2 Fly stocks and crosses
Fly cultures were maintained at 23 ± 1°C on standard food containing agar, maize powder, yeast, and sugar. Oregon-R+ and Canton-S were used as wild-type strains. Various transgenic lines such as GMR-rpr, GMR-hid, UAS-pro-dronc w, UAS-pro-dronc s, UAS-ΔNdronc, GMR-p35 (Meier et al. 2000), GMR-grim (Muro et al. 2006), UAS-DIAP1-RNAi (Leulier et al. 2006), UAS-eiger (Igaki et al. 2002), GMR-argos (Freeman 1994), and Mito-GFP (Pilling et al. 2006; Goyal et al. 2007) were obtained from various laboratories. GMR-Gal4 and Act5C-Gal4 stocks were obtained from the Bloomington stock center. For genetic interaction studies of Hsp60D with various cell death pathway members, the Hsp60D transgenic lines (UAS-Hsp60DWT or UAS-Hsp60D-RNAi), generated as above, were independently introgressed with the GMR-Gal4 driver and used for various crosses to generate the desired genotypes. Most of the genetic interactions were carried out with a single copy of the UAS-Hsp60DWT or UAS-Hsp60D-RNAi transgene unless mentioned otherwise.
2.3 Confirmation of transgenic lines
2.3.1 DNA isolation and Southern hybridization
Genomic DNA from 50 flies of each genotype of interest was isolated following Sambrook et al. (1989) and digested with BamH1 restriction enzyme, electrophoretically separated on agarose gel, transferred onto positively charged nylon membrane (Boehringer, Germany), and hybridized with the Hsp60D cDNA (EcoRI fragment from the AT04835 clone) probe labeled with digoxigenin by random priming following standard methods. After stringent washes, the blot was processed for colorimetric detection of hybridization as per the manufacturer’s instructions (Roche, Germany).
2.3.2 RNA isolation and semiquantitative reverse transcription polymerase chain reaction
Total RNA was isolated from ten adult flies of control (Act5C-Gal4/CyO) and transgenic lines expressing two copies of Hsp60D-RNAi or UAS-Hsp60DWT under Act5C-Gal4 driver (Act5C-Gal4/CyO; Hsp60D-RNAi/Hsp60D-RNAi and Act5C-Gal4/CyO; UAS-Hsp60DWT/UAS-Hsp60DWT, respectively) using Trizol as per the manufacturer’s (Life Technology, USA) instructions. The RNA samples were treated with 2U of RNase-free DNAse I (MBI Fermentas, USA) at 37°C for 30min to remove any residual DNA. Semiquantitative reverse transcription polymerase chain reaction (RT-PCR) was performed as per the manufacturer’s (Amersham) instructions. Briefly, for cDNA synthesis, 2μg of total RNA, 80pmol of oligo(dT)17 primer, 2U of RNase inhibitor, 500μM of dNTP mix, and 100U of MMLV reverse transcriptase (Amersham) were added to make the final 20μl reaction volume. For PCR amplification, 1/10 volume of the RT product was used for each reaction. Hsp60D-specific primers (forward primer: 5′-TGCGATGCTCTAGGTGAGAT-3′ and reverse primer: 5′-ACATCGTAGATAGGCGGTTC-3′) and glycerol-3-phosphate dehydrogenase (G3PDH) primers (forward primer: 5′-CCACTGCCGAGGAGGTCAACTA-3′ and reverse primer: 5′-GCTCAGGGTGATTGCGTATGCA-3′) were added for PCR. G3PDH is a house-keeping enzyme and has been used in earlier studies (Kanuka et al. 1999) as internal control for RT-PCR. The thermal cycling program included initial denaturation at 94°C for 5min followed by 35 cycles at 94°C for 30s, 60°C for 40s, and 72°C for 1min. Final extension was carried out at 72°C for 5min. The PCR products were run on a 2% agarose gel with 100bp ladder as a molecular weight marker.
2.3.3 RNA–RNA in situ hybridization in third instar larval eye discs
The AT04835 cDNA clone was digested with HindIII and EcoRI and the 613-bp HindIII–EcoRI fragment (from 960 to 1573bp position of the AT04835 clone), which is unique to the Hsp60D gene (see above), was eluted and ligated with the HindIII–EcoRI-digested pGEM-3 vector. The resulting subclone, named as p34C-hsp60D.613, was linearized by EcoRI and used for synthesis of Hsp60D-specific digoxigenin-labeled (Roche, Germany) antisense riboprobe with T7 RNA polymerase. RNA–RNA in situ hybridization (RISH) with intact larval tissues was performed essentially as described earlier (Lakhotia et al. 2001). As a negative control, some of the tissues were treated with RNase (20μg/ml) for 1h at 37°C before hybridization with the Hsp60D riboprobe.
2.3.4 Assay for apoptosis
Acridine orange (AO) staining was used to assay the extent of apoptosis (Spreij 1971; Abrams et al. 1993) in third instar eye imaginal discs from larvae of desired genotype. The discs were dissected in PBS (130mM NaCl, 7mM Na2HPO4, 3mM KH2PO4, pH7.2) and immediately stained, without any fixation, with 1μg AO/ml of PBS for 3min, following which the discs were washed twice in PBS and mounted in PBS for viewing in LSM510 Meta Zeiss confocal microscope.
2.4 Pseudopupil analysis
Adult flies of the desired genotypes were decapitated, and the heads were arranged on a microscope slide. To see the photoreceptor arrangement, corneal neutralization was achieved by adding one drop of immersion oil on the heads (Franceschini 1972). The pseudopupils were viewed in Nikon Ellipse 800 microscope with a ×60 plan-Apo oil immersion objective (NA 1.4) by illuminating the back of head with a narrow beam of bright light. The images were captured with a Nikon DXM 1200 digital camera.
2.5 Nail polish imprints
The nail polish imprints of adult eye surfaces were prepared as described earlier (Arya and Lakhotia 2006). These were examined under DIC optics in a Nikon Ellipse 800 microscope.
2.5.1 Immunostaining and confocal microscopy
For antibody staining, the eye discs were dissected from late third instar larvae of the desired genotypes and fixed in freshly prepared 4% paraformaldehyde in PBS for 20min and processed for immunostaining as described earlier (Prasanth et al. 2000). For detection of the protein/s of interest, the desired primary antibody/antibodies (anti-Hsp60: SPA805 [1:100] and SPA806 [1:50] of Stressgen, Canada; anti-αDIAP1 [1:1,000] from Dr. K. White; anti-GRP75 [1:200] from Dr. S. Ganesh) was/were added singly or in the desired combination to eye discs at the indicated dilution/s. Primary antibody binding was detected with 1:200 dilution of antirabbit or antimouse secondary antibody labeled with Alexa-Fluor 488 (Molecular Probes, USA) or with Cy3 (Sigma), as required. Confocal imaging was carried out on LSM510 Meta Zeiss confocal microscope using appropriate dichroics and filters. All images were assembled using the Adobe Photoshop software.
3 Results
3.1 Hsp60-RNAi and UAS-Hsp60D lines
Among the several transgenic lines generated by us, we selected two each of the Hsp60D-RNAi (Hsp60D-RNAi 36.d.2 on chromosome 2 and Hsp60D-RNAi 41.d.2 on chromosome 3) and UAS-Hsp60D (UAS-Hsp60DWT 3 on chromosome 2 and UAS-Hsp60WT 8.a on chromosome 3) lines for further studies. To ascertain that the Hsp60D-RNAi and UAS-Hsp60DWT flies carry the expected transgene, genomic Southern hybridization was carried out. Oregon-R and Canton-S flies were used as controls for identification of the endogenous Hsp60D fragments. Because microinjections of transgenic constructs were performed in Canton-S strain eggs and because the Hsp60D gene is located on chromosome 2, the second chromosome transgene stocks carry the chromosome 2 of Canton-S origin, while in the case of third chromosome transgenic lines, the second chromosome was replaced with the chromosome 2 of Oregon-R background during establishment of the transgenic stocks. Therefore, genomic DNAs from both Oregon-R and Canton-S flies were used as controls. As shown in Fig. 1, BamH1-digested genomic DNAs of Oregon-R and Canton-S flies showed different-sized Hsp60D fragments (8.3 and 6.2kb, respectively), reflecting a polymorphism of genomic organization in the two fly strains. In addition to the endogenous Hsp60D fragment corresponding to either Oregon-R or Canton-S controls, the Hsp60D-RNAi lines released a single band (3.095kb) whereas the UAS lines liberated two (1.96 and 1.18kb) transgene-specific bands due to a BamHI site existing within the Hsp60D cDNA. These results confirmed that the transgenes in these four lines were as expected. To rule out the possibility of insertions other than the desired constructs in these transgenic lines, we performed Southern hybridization using a P-element-specific miniwhite probe. While Hsp60D-RNAi 36.d.2, Hsp60D-RNAi 41.d.2, and UAS-Hsp60DWT 8.a revealed the presence of only a single P-element insertion, in the case of UAS-Hsp60DWT 3, two P-element-specific signals were seen (data not shown). Another Southern blot using an Hsp60D-specific probe confirmed that both the P-element transposons inserted in UAS-Hsp60DWT 3 correspond to Hsp60D only (data not shown). These results showed that the transgenic lines carry the expected transgene as a single insertion in the Hsp60D-RNAi 36.d.2, Hsp60D-RNAi 41.d.2, and UAS-Hsp60DWT 8.a lines but the UAS-Hsp60DWT 3 line carried two insertions of the transgene and, therefore, was not used further. None of these transgene insertions by themselves, i.e., in absence of any Gal4 driver, showed any phenotype in heterozygous or homozygous conditions. The two Hsp60D-RNAi transgenes (Hsp60D-RNAi 36.d.2 and Hsp60D-RNAi 41.d.2) showed comparable effects on phenotypes when driven with any Gal4 driver and, therefore, in the studies reported in this paper, the Hsp60D-RNAi 41.d.2 and UAS-Hsp60DWT 8.a lines were used and, in the following discussion, these two lines are referred to as Hsp60D-RNAi and UAS-Hsp60DWT, respectively.
Genomic DNAs from wild-type Oregon-R (lane 1) and Canton-S (lane 2), and from Hsp60D-RNAi 36.d.2 (lane 3), Hsp60D-RNAi 41.d.2 (lane 4), UAS-Hsp60DWT 3 (lane 5), UAS-Hsp60DWT 8.a (lane 6) homozygous transgenic flies were hybridized with the Hsp60D-specific dig-labeled probe. The 8.3 kb (Oregon-R-specific) or 6.2 kb (Canton-S-specific) bands in different lanes correspond to the endogenous Hsp60D gene. The second and third chromosome Hsp60D-RNAi homozygotes (lanes 3 and 4, respectively) show the expected 3.09 kb transgene-specific fragment while the UAS-HSp60D lines with transgene insertion on second or third chromosome (lanes 5 and 6, respectively) show the expected two transgene-specific bands (1.96 and 1.18 kb)
3.2 Levels of Drosophila Hsp60D transcripts in different transgenic lines are affected as expected
To confirm that the transgenic lines modulate the levels of the Hsp60D transcripts as expected, they were driven with the ubiquitous Act5C-Gal4 driver (Ekengren et al. 2001). RT-PCR was performed with RNA isolated from adult flies. As shown in Fig. 2, compared with Act5C-Gal4/CyO;+/+ (Fig. 2, lane 1) flies, a distinct increase in transcript level was seen in UAS-Hsp60DWT flies (Act5C-Gal4/CyO; UAS-Hsp60DWT/UAS-Hsp60DWT; Fig. 2, lane 2). On the other hand, the level of Hsp60D transcripts (long arrow) is detectably reduced following Hsp60D-RNAi expression (Act5C-Gal4/CyO; Hsp60D-RNAi/Hsp60D-RNAi; Fig. 2, lane 3). Nearly equal levels of G3PDH amplicons in all the lanes (Fig. 2, short arrow) showed that the alterations in the levels of Hsp60D transcripts were not due to variations in loading of samples.
Modulation of Hsp60D transcript levels following the expression of the UAS-Hsp60DWT or Hsp60D-RNAi transgenes with the ubiquitous Act5C-Gal4 driver. RT-PCR for Hsp60D mRNA produces the 876-bp fragment (long arrow) whereas RT-PCR amplicon for the G3PDH mRNA is 140 bp (small arrow). Lane 1 is control (Act5C-Gal4/CyO; +/+) showing the normal level of Hsp60D mRNA in adult flies. Lane 2 is Act5C-Gal4/CyO; UAS-Hsp60DWT/UAS-Hsp60DWT showing increase in Hsp60D mRNA levels and lane 3 is Act5C-Gal4/CyO; Hsp60D-RNAi/Hsp60D-RNAi showing reduction in levels of Hsp60D. Amplicons for G3PDH, used as loading control, show equal loading in all lanes. The M lane shows a 100-bp ladder molecular size marker. This is a negative image of an ethidium bromide-stained agarose gel
3.3 Hsp60D is essential for normal development
To see the effect of global ablation of Hsp60D or its overexpression, the Hsp60D-RNAi or the UAS-Hsp60D transgenes were expressed with the Act5C-Gal4 driver after introgression of either of the transgenes (inserted on chromosome 3) in the Act5C-Gal4/CyO stock. Eggs were collected from each of the three stocks, viz., (1) Act5C-Gal4/CyO; +/+, (2) Act5C-Gal4/CyO; Hsp60D-RNAi/Hsp60D-RNAi, and (3) Act5C-Gal4/CyO; UAS-Hsp60D/UAS-Hsp60D, and their survival to adult stage counted (Table 1). It may be noted that Act5C-Gal4/Act5C-Gal4 and CyO/CyO progeny die at embryonic or larval stages and thus only 50% of the eggs (Act5C-Gal4/CyO) are expected to survive to adult stage. The actual survival of Act5C-Gal4/CyO progeny was about 43%, which is significantly less (P < 0.001 for χ 2 test) than the expected 50%. It is interesting to note that Act5C-Gal4-driven expression of two copies of Hsp60D-RNAi transgene further reduced the survival of Act5C-Gal4/CyO progeny to 27.6%. On the other hand, overexpression of Hsp60D was somewhat less deleterious (Table 1). The surviving flies in either case had no morphological phenotypes ascribable to ablation or overexpression of the Hsp60D. However, the reduced viability following RNAi for Hsp60D is indicative of an essential role of this gene in normal development.
3.4 Hsp60D is expressed in a characteristic pattern in third instar larval eye discs
RISH revealed that besides a low level of ubiquitous presence of Hsp60D transcripts in the cytoplasm of third instar larval eye discs, the transcripts showed a prominent accumulation at the MF and in the differentiating photoreceptor cells posterior to it (Fig. 3a). The specificity of hybridization of the riboprobe with Hsp60D transcripts in eye cells was confirmed by treating the discs with RNase preceding RISH for Hsp60D. These discs did not show any staining (inset in Fig. 3a′). Chromogenic and fluorescent detection of RISH showed that heat shock at 37°C to third instar larval eye discs for 1h did not induce Hsp60D transcripts beyond the control levels (not shown).
Localization of Hsp60D transcripts by RISH (a, b) and Hsp60 protein by immunostaining (c–k) in larval eye discs. The Hsp60D transcript levels are high at the MF and the photoreceptor cells posterior to it (a). b is a higher magnification image of part of the disc in a showing patterned distribution of Hsp60D transcripts in photoreceptor units. a′ (inset) shows a disc treated with RNase before RISH. Immunostaining with rabbit anti-Hsp60 (green; c, d, f) and mouse anti-Hsp60 (red; e, f) antibodies also shows comparable distribution of Hsp60 proteins in eye discs with distinct Hsp60-positive granules in photoreceptor cells, especially in their apical regions (arrows). f is a merged image of d and e. Hsp60C 1 homozygous eye discs (g) show only a slight reduction in Hsp60 proteins recognized by the rabbit and mouse anti-Hsp60 antibodies. On the other hand, the eye discs from GMR-Gal4; Hsp60DRNAi larvae (h) show greatly reduced diffuse staining with both antibodies while the mouse antibody-recognized (green) Hsp60 granules are much less affected than the rabbit antibody-recognized (red) ones. Hsp60 granules recognized by either the mouse anti-Hsp60 (red; i) or the rabbit anti-Hsp60 antibody (red; j) do not show any significant localization with mitochondria marked by mito-GFP (green; i, j). The mouse anti-Hsp60 (red) and rabbit antimitochondrial Grp75 (green) also do not colocalize (k). Images in c–k are single confocal optical sections. Scale bars represent 10 μm (bar in h is common for d–h and that in k is common for i–k)
3.5 Hsp60 family proteins in eye discs form distinct cytoplasmic granules
For immunofluorescence localization of Hsp60 in eye discs, we used two commercially available anti-Hsp60 antibodies, a rabbit polyclonal antibody against Hsp60 of Heleothis viridis (SPA805, Stressgen) and a mouse monoclonal antibody against human Hsp60 (SPA806, Stressgen). It is expected that both the antibodies identify all the four Hsp60 forms in D. melanogaster (Lakhotia et al. 2002).
The Hsp60 immunostaining pattern generally resembled the Hsp60D transcript distribution pattern in wild-type eye discs (compare Fig. 3a and b with c). Examination of the immunofluorescently stained discs at higher magnification revealed that in addition to the general diffuse cytoplasmic distribution, both antibodies recognized distinct brightly fluorescing cytoplasmic granules, which were more abundant in the apical regions of the developing photoreceptor units (Fig. 3d,e). To see if the immunofluorescence patterns generated by the two antibodies were identical, we costained wild-type eye discs with both the antibodies. This revealed that while the general cytoplasmic staining was similar (Fig. 3d,e), the granules in photoreceptor cells identified by each antibody were distinct, although mostly adjacent and partially overlapping (Fig. 3f).
The above finding raised the possibility that the two anti-Hsp60 antibodies may recognize different Hsp60 forms present in D. melanogaster. It is known that Hsp60A is expressed ubiquitously but at low levels in most cell types of Drosophila (Kozlova et al. 1997) while the Hsp60B is not expressed in any somatic cell (Timakov and Zhang 2001; Srivastava 2004). The Hsp60C is expressed ubiquitously, but at low levels, in most cells of larval eye discs (Sarkar and Lakhotia 2005). Therefore, we examined coimmunostaining with both the antibodies in eye discs from Hsp60C 1 homozygous larvae, which have depleted Hsp60C, and from GMR-Gal4/GMR-Gal4; Hsp60D-RNAi/Hsp60D-RNAi larvae, which have depleted Hsp60D (present study). Costaining with the two Hsp60 antibodies showed only a slight reduction in the overall intensity of both the antibodies in Hsp60C 1 homozygous eye discs although the patterns of diffuse and granular staining were generally similar to those of the wild-type (Fig. 3g). On the other hand, discs expressing the Hsp60D-RNAi transgene consistently displayed significantly reduced general diffused staining with both the antibodies. It is interesting to note that while the rabbit anti-Hsp60 antibody reacting Hsp60 granules in their photoreceptor cells were nearly absent, those recognized by the mouse antibody were less affected (Fig. 3h).
3.6 The Hsp60 proteins are not restricted to mitochondria in photoreceptor cells
The Hsp60 family proteins are generally believed to be mitochondrial (Ellis and van der Vies 1991; Martin et al. 1995; Bukau and Horwich 1998). Therefore, to check if the Hsp60-positive granules in the cytoplasm of eye disc cells were colocalizing with mitochondria, we used two mitochondrial markers in combination with either of the two Hsp60 antibodies. In one case, larvae expressing GFP in mitochondria (Pilling et al. 2006) were used (Fig. 3i,j), while in the other set, eye discs from wild-type larvae were immunostained with a polyclonal anti-Grp75 antibody (Fig. 3k), which recognizes the Hsp70 family protein localizing in mammalian mitochondrial matrix (Mukamel and Kimchi 2004). As seen in Fig. 3i–k, the Hsp60-rich granules, identified by either of the Hsp60 antibodies, rarely showed any association with mitochondria.
3.7 RNAi of Hsp60D rescues degeneration seen in eyes of GMR-Gal4 homozygous adult flies
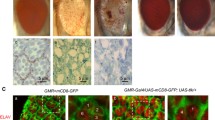
With a view to understand possible function(s) of the Hsp60D gene in Drosophila eye development, we used one copy of the GMR-Gal4 driver to either overexpress or ablate the Hsp60D in developing eyes but did not notice any visible phenotypic consequence following misexpression of this gene. It is interesting to note that, however, we noticed that the eye degeneration (Fig. 4a–c) caused by two copies of GMR-Gal4 by themselves (Kramer and Staveley 2003) was rescued following ablation of Hsp60D transcripts because the eyes of GMR-Gal4/GMR-Gal4; Hsp60D-RNAi/Hsp60D-RNAi flies were indistinguishable from those of the wild-type eyes, externally and in their rhabdomere organization (Fig. 4d–e). On the other hand, overexpression of Hsp60D enhanced the eye degeneration caused by two copies of GMR-Gal4 resulting in rougher eyes (compare a–b with g–h in Fig. 4). To identify apoptotic cells in eye discs from larvae of different genetic backgrounds, AO staining was carried out as described in the “Materials and methods” section. In agreement with the observed adult eye phenotypes, compared to only GMR-Gal4 homozygous eye discs, fewer AO-positive cells were seen in the GMR-Gal4-driven Hsp60D-RNAi-expressing discs (compare c with f in Fig. 4). On the other hand, the AO-positive cells were more frequent in Hsp60D-overexpressing discs (Fig. 4i). These observations raised the possibility that Hsp60D may have some role in cell death pathway.
Hsp60D levels modulate eye phenotypes of GMR-Gal4 homozygous flies. a, d, and g are nail polish imprints while b, e, and h are pseudopupil images of adult eyes; c, f, and i are images of AO-stained third instar larval eye discs. a–c are from GMR-Gal4/GMR-Gal4:+/+, d–f from GMR-Gal4/GMR-Gal4; Hsp60D-RNAi/Hsp60D-RNAi, and g–i from GMR-Gal4/GMR-Gal4; UAS-Hsp60DWT/UAS-Hsp60DWT genotypes. Images in c, f, and i are projections of confocal optical sections
3.8 RNAi of Hsp60D strongly suppresses cell death caused by RHG family proteins
To elucidate the possible role of Hsp60D in apoptosis, the effects of varying levels of Hsp60D on cell death caused by the expression of typical apoptosis-inducing RHG proteins were examined. Unless otherwise mentioned, only single copies of the Hsp60D-RNAi or the UAS-Hsp60DWT transgenes were driven in the desired genetic backgrounds to see the effects of depletion or overabundance of the Hsp60D protein. As reported earlier (Hay et al. 1994; White et al. 1994; Grether et al. 1995; Chen et al. 1996) and shown in Fig. 5a,d,g,j,m, and q, ectopic expression of any of the RHG proteins under the control of the GMR-promoter caused excessive cell death resulting in varyingly malformed and reduced eyes in adult flies. It is interesting to note that coexpression of a single copy of Hsp60D-RNAi significantly rescued the eye phenotypes (Fig. 5, compare a, d, g, j, m, q with b, e, h, k, n, r, respectively). GMR-driven expression of Hid caused maximum degeneration while that of Grim caused the least (Fig. 5 compare m, q with g, j); correspondingly, the recovery following Hsp60D-RNAi coexpression was maximum with Grim and least with Hid (Fig. 5, compare h, k with n, r). Overexpression of Hsp60D further enhanced the eye phenotype caused by GMR-hid (Fig. 5o,s) resulting in narrower eyes, whereas it had less perceptible effect on Reaper- or Grim-induced eye phenotypes (Fig. 5c,f,i,l).
Hsp60D levels modulate cell death caused by expression of RHG proteins. Eyes of flies of different genotypes as seen by light microscopy (a–c, g–i, m–p) or in nail polish imprints ( d–f, j–l, q–t ) of GMR-rpr (a, d), GMR-grim (g, j), or GMR-hid (m, q) adults not expressing any of the Hsp60D transgenes (column 1) or coexpressing GMR-Gal4-driven Hsp60D-RNAi (column 2) or UAS-Hsp60DWT (column 3) are shown. Images in the fourth column (p, t) are from flies expressing GMR-hid together with GMR-Gal4-driven Hsp60D-RNAi and UAS-Hsp60DWT transgenes
In agreement with above, a simultaneous overexpression and ablation of Hsp60D transgenes through the GMR-Gal4 driver in GMR-hid background nullified the rescuing effect of Hsp60D RNAi because the eyes in GMR-Gal4/GMR-hid; UAS-Hsp60DWT/UAS-Hsp60D-RNAi flies showed nearly as much degeneration as in GMR-hid/+ flies (Fig. 5p,t). This observation confirmed that the modulations of eye phenotypes by UAS-Hsp60DWT or Hsp60D-RNAi transgenes are indeed due to alterations in the levels of endogenous Hsp60D protein.
3.9 Hsp60D-RNAi suppresses apoptosis caused by expression of full-length and processed DRONC
It is known that, under normal conditions, the caspases remain inactive because of binding with DIAP1 and, upon receiving a death signal, the DIAP1 is removed so that the activated caspases bring about cell death. Conditional overexpression of full-length or processed caspases results in increased cell death (Meier et al. 2000), presumably because the inhibitory effects of cellular DIAPs becomes limiting. To check if RNAi of Hsp60D has any effect on cell death caused by overexpression of full-length or activated caspases, we used transgenic lines expressing full-length UAS-pro-dronc w (weak), UAS-pro-dronc s (strong) or processed UAS-deltaN-dronc transgenes. As reported by Meier et al. (2000), eyes of flies (N = 83) expressing the UAS-pro-dronc w under the GMR-Gal4 driver showed near-regular arrays of ommatidia but with an uneven pigmentation (Fig. 6a,e); internally, however, these eyes were degenerate because no pseudopupil image was formed (not shown). GMR-Gal4-driven overexpression of strong (N = 430) or processed DRONC (N = 208) resulted in more severely degenerated and depigmented eyes with reduced head size (Fig. 6c,g and 7a,c, respectively). As already reported (Meier et al. 2000), majority of those expressing GMR-Gal4-driven strong (419 out of 430 examined) or processed DRONC (203 out of 208) died as pharates due to their inability to open the pupal case. It is interesting to note that all pupae coexpressing Hsp60D-RNAi with UAS-pro-dronc s (N = 161) or UAS-deltaN-dronc (N = 97) eclosed with normal head size. Moreover, coexpression of Hsp60D-RNAi with the weak (N = 127, Fig. 6b,f,i) or the strong full-length DRONC (Fig. 6d,h,j) or the processed deltaN-dronc (Fig. 7b,d) transgene resulted in normal eyes comparable to those of wild-type flies. AO staining of larval eye discs revealed that a large number of dying cells were present in deltaN-dronc-expressing third instar eye discs (Fig. 7e), but their number was significantly reduced when the Hsp60D-RNAi transgene was coexpressed (Fig. 7h).
RNAi of Hsp60D suppresses DRONC-mediated eye degeneration. All transgenes, UAS-Pro-dronc w (a, b, e, f, i), UAS-Pro-dronc s (c, d, g, h, j), and/or Hsp60D-RNAi (b, d, f, h, i, j), were expressed under control of GMR-Gal4. a–d are photomicrographs, e–h are nail polish imprints, and i and j are pseudopupil images of adult eyes from flies of different genotypes as indicated on the top of each column
Hsp60D-RNAi effectively suppresses cell death caused by active DRONC. Photomicrographs (a, b) or nail polish imprints (c, d) of eyes of GMR-Gal4/+; UAS-deltaN-dronc/+ (a, c) or GMR-Gal4/+; UAS-deltaN-dronc/Hsp60D-RNAi (b, d) flies. e and f show AO-stained eye discs from GMR-Gal4/+; UAS-deltaN-dronc/+ (e) or GMR-Gal4/+; UAS-deltaN-dronc/Hsp60D-RNAi (f) third instar larvae
3.10 Downregulation of Hsp60D is unable to rescue cell death caused by DIAP1-RNAi
Because Hsp60D-RNAi suppressed cell death caused by activated caspases, we further checked if it could also suppress apoptosis following DIAP1-RNAi. Many caspases, including DRONC, become active in the absence of enough DIAP1 and, therefore, cause massive cell death (Leulier et al. 2006; Muro et al. 2006). Overexpression of DIAP1 in developing eyes rescues the small eye phenotypes caused by RHG proteins (Hay et al. 1995) or overexpression of DRONC (Meier et al. 2000). In agreement with earlier reports, we observed that Drosophila eyes expressing DIAP1-RNAi were reduced, deformed, and partially depigmented (Fig. 8a,d). Intriguingly, unlike in previous cases, Hsp60D-RNAi failed to rescue the reduced and deformed eye phenotype resulting from DIAP1-RNAi (Fig. 8b,e). Even two copies of Hsp60D-RNAi transgenes failed to improve the phenotype of DIAP1-RNAi-expressing eyes (not shown). Overexpression of Hsp60D mildly enhanced the eye degeneration caused by DIAP1-RNAi (Fig. 8c,f).
Furthermore, ablation of DIAP1 enhanced the eye phenotypes of GMR-Hid or GMR-grim due to increased apoptosis along with pupal lethality, but this was not suppressed by coexpression of Hsp60D-RNAi (not shown). These results strongly suggest that the presence of DIAP1 is necessary for the inhibitory effect of the Hsp60D-RNAi on apoptosis.
3.11 Hsp60 and DIAP1 show adjacent or overlapping distribution in eye disc cell cytoplasm
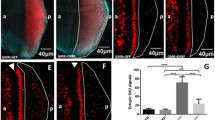
Coimmunostaining of wild-type eye discs with the mouse anti-Hsp60 antibody and rabbit anti-DIAP1 antibody revealed that like the Hsp60 (see above, Fig. 3d–f), DIAP1 is also present in diffuse manner through the cytoplasm and in greater concentrations in distinct cytoplasmic granules in the apical regions of each developing ommatidial unit (Fig. 9a). It is interesting to note that the Hsp60 granules, recognized by the mouse anti-Hsp60 antibody, and DIAP1 granules were usually adjacent with occasional partial overlap (see Fig. 9b,b′). Because the Hsp60 granules identified by the rabbit anti-Hsp60 antibody also showed a comparable adjacent and sometimes partially overlapping localization with the mouse anti-Hsp60 antibody (Fig. 3f), it is possible that the Hsp60 granules identified by the rabbit antibody may actually show a greater colocalization with the DIAP1 granules. However, because the DIAP1 antibody is also raised in rabbit, it could not be used for conventional coimmunostaining with the rabbit anti-Hsp60 antibody. To circumvent this limitation, to some extent at least, we used an indirect approach to see if the Hsp60 identified by the rabbit anti-Hsp60 antibody actually colocalized or overlapped with DIAP1. Eye discs from wild-type late third instar larvae were first incubated with the rabbit anti-Hsp60 antibody, washed, and incubated with antirabbit Alexa-Fluor 488 conjugated secondary antibody. After washing, the Hsp60-stained discs were divided into two sets. In one set, the discs were directly incubated with antirabbit Cy3 conjugated secondary antibody. In the second set, the Hsp60-stained eye discs were incubated with the rabbit anti-DIAP1 antibody followed by washing and incubation with antirabbit Cy3 conjugated secondary antibody. The first set of discs would show green fluorescence reflecting Hsp60 recognized by the rabbit anti-Hsp60 antibody while the red fluorescence (excitation at 561nm) in these discs would reflect the extent of Cy3-labeled antirabbit secondary antibody binding with the residual primary rabbit anti-Hsp60 antibody that was not occupied by the antirabbit Alexa-Fluor 488 conjugated secondary antibody. For examining the second set of discs, which were exposed to the two primary rabbit (anti-Hsp60 and anti-DIAP1) and two secondary antirabbit antibodies, the gain of the channel selected for recording red (Cy3) fluorescence was reduced to a level which just eliminated the signal generated by the antirabbit Cy3 conjugated antibody in the first set of discs (Fig. 9d). This reduction in gain was thus expected to remove, in the second set of discs, any signal generated by the antirabbit Cy3 conjugated secondary antibody that binds with the anti-Hsp60 primary antibody while any additional signal in this channel would be expected to represent the binding of this secondary antibody with the anti-DIAP1 primary antibody. The results of this double immunostaining are shown in Fig. 9c–h. It is seen that double staining with rabbit anti-Hsp60 antibody and rabbit anti-DIAP1 antibody resulted in almost complete colocalization of the diffuse and the granular forms of Hsp60 and DIAP1 (Fig. 9h). Granules showing only red but no green fluorescence, indicating the absence of Hsp60, were not seen in the second set of discs (Fig. 9h). Comparable results were obtained when the first primary antibody used was the rabbit anti-DIAP1 antibody and the second primary antibody was the rabbit anti-Hsp60 antibody (not shown). These results suggested that all granular forms of DIAP1 in these cells colocalized with those recognized by the rabbit anti-Hsp60 antibody.
Colocalization of Drosophila Hsp60 and DIAP1 in developing photoreceptor cells. Confocal optical sections of wild-type (a–h), GMR-Gal4/GMR-Gal4; Hsp60D-RNAi/Hsp60D-RNAi (i), and GMR-Gal4 DIAP1-RNAi/CyO (j) larval eye discs following immunostaining with rabbit anti-DIAP1 (red; a, b, b′ [inset], g, h–j), mouse anti-Hsp60 (green; b, b′, i, and j) or rabbit anti-Hsp60 (green; c, e, f, and h) primary antibodies. d shows the confocal image at 561 nm excitation of a wild-type disc incubated with rabbit anti-Hsp60 primary antibody followed sequentially by binding with antirabbit Alexa-Fluor 488 and antirabbit Cy3 secondary antibodies; the gain in red channel was set at a level which just eliminated any red fluorescence. The same gain setting was used for recording the image in g and h. Images in e and h are merges of c and d and f and g, respectively. b, b′, i, and j are also merged images for green and red channels. The scale bars, representing 10 μm, are common for each row except the bar in b′, which represents 1 μm
To check if the above-noted adjacent/overlapping distribution of these two proteins is of functional significance, we examined the effect of depleting Hsp60D or DIAP1 on their distributions. Coimmunostaining of the eye discs for both the proteins following GMR-Gal4-driven expression of DIAP1-RNAi or Hsp60D-RNAi transgenes showed that the reduction in the level of DIAP1 also affected the Hsp60 protein granules, which were smaller and fewer in such cells (Fig. 9i). Likewise, ablation of Hsp60D by RNAi also resulted in significant reduction of DIAP1 granules (Fig. 9j).
3.12 Downregulation of Hsp60D rescues JNK and EGFR mutant eye phenotypes resulting from cell death
Following the above results that RNAi of Hsp60D prevented cell death caused by the direct activation of caspases, we checked if the downregulation of Hsp60D could also suppress the canonical caspase-mediated cell death in JNK or EGFR mutants. Argos is a negative regulator of Drosophila EGFR (Schweitzer et al. 1995; Sawamoto et al. 1996). GMR-Argos flies showed reduced eyes with irregularly arranged ommatidia (Fig. 10a,c) due to hampered cellular differentiation and activation of cell death (Freeman 1994; Sawamoto et al. 1994; Sawamoto et al. 1998). Coexpression of Hsp60D-RNAi suppressed the GMR-Argos-induced eye degeneration and resulted in regularly arrayed ommatidial units (Fig. 10b,d).
Hsp60D-RNAi effectively rescues JNK and EGFR mediated cell death. Photomicrographs (a, b, e, f) and nail polish imprints (c, d, g, h) of eyes of GMR-Gal4/GMR-argos; +/+ (a, c), GMR-Gal4/GMR-argos; Hsp60D-RNAi/+ (b, d), GMR-Gal4/+; UAS-Eiger/+ (e, g), and GMR-Gal4/+; UAS-Eiger/Hsp60D-RNAi (f, h) flies
Eiger is an extracellular activator of Jun-N-terminal kinase (JNK) pathway (Igaki et al. 2002). When expressed ectopically in Drosophila eyes, it causes JNK-mediated cell death through the involvement of Hid, Dark, and DRONC (Moreno et al. 2002). As reported by other authors (Igaki et al. 2002; Moreno et al. 2002; Kauppila et al. 2003), ectopic expression of Eiger through GMR-Gal4 in developing Drosophila eye discs resulted in severely damaged eyes (Fig. 10e,g). In this case also, ablation of Hsp60D in Eiger-expressing eye discs resulted in eyes comparable to those of the wild-type (Fig. 10f,h).
4 Discussion
A general notion about Hsp60 family proteins, as also for the other families of Hsps or heat shock proteins, is that their synthesis is enhanced following cell stress. This is also reflected in the description of the different Hsp60 genes in D. melanogaster genome available at the databases like http://flybase.org. However, as noted earlier, in the case of Drosophila larvae, Hsp60 is induced by heat shock only in the Malpighian tubules (Lakhotia and Singh 1989, 1996). In agreement with this, the present results show that the level of Hsp60D transcripts is not enhanced in heat-shocked eye discs.
Transcripts of Hsp60A and Hsp60C genes show low abundance in eye disc cells (Kozlova et al. 1997; Sarkar and Lakhotia 2005) while Hsp60B is reported to express only in male germ cells (Timakov and Zhang 2001). However, information available in the databases on the expression of specific genes in different tissues of D. melanogaster (e.g., http://flybase.org or http://www.ncbi.nih.gov/geo/ (GDS196)), suggest that the Hsp60A gene transcripts are abundant in larval eye discs while those of the other Hsp60 genes are much less common or absent. On the other hand, our results of RISH clearly show a significant level of Hsp60D transcripts in subsets of differentiating eye discs. A possible reason for this discrepancy may be the significant level of homology in the base sequences of the four Hsp60 genes of D. melanogaster (Sarkar and Lakhotia 2005). Therefore, unless gene-specific probes are used, all Hsp60 transcripts may be ascribed to the Hsp60A gene because that is presumed to be the ancestral one among the four Hsp60 genes in the D. melanogaster genome (Sarkar and Lakhotia 2005). Thus, notwithstanding the information given in the databases, our results with the Hsp60D gene-specific riboprobe show that this gene’s transcripts have high abundance in third instar larval eye discs, especially in the MF and differentiating photoreceptor cells. This is paralleled by the pattern of immunostaining with either of the two anti-Hsp60 antibodies. Together, these suggest that Hsp60D contributes significantly to the pool of Hsp60 family proteins in eye discs. It is intriguing, however, that the two different anti-Hsp60 antibodies (SPA805, polyclonal antibody raised in rabbit, and SPA806, a mouse monoclonal antibody; Stressgen), while revealing similar diffuse distribution of Hsp60 in the cytoplasm, decorated different subsets of more brightly fluorescing Hsp60 granules, especially in the apical regions of differentiating photoreceptor cells (Fig. 3d–f). This was unexpected because the two antibodies are believed to detect all the four Hsp60s in Drosophila (Lakhotia et al. 2002). Our further studies with these two anti-Hsp60 antibodies in conjunction with conditions that would deplete either Hsp60C (Hsp60C 1 homozygous condition) or Hsp60D (through RNAi) revealed that the Hsp60 granules identified by the rabbit anti-Hsp60 antibody were more sensitive to Hsp60D RNAi. Notwithstanding this differential staining of Hsp60 granules by the two antibodies in eye discs and their differential sensitivity of Hsp60D-RNAi, we do not think that the rabbit anti-Hsp60 antibody specifically recognizes the Hsp60D protein because in several other tissues like the trachea, testis, and ovary, the same antibody also recognizes the Hsp60C protein which is depleted by homozygosity of the Hsp60C 1 allele (Sarkar and Lakhotia 2005, 2008). It appears more likely that the two antibodies recognize all the four Hsp60 proteins, as expected on their epitope specificities (Lakhotia et al. 2002), but some conformational changes under specific conditions or in specific cells may differentially affect binding of the two antibodies with one or the other Hsp60 form. This needs further analysis.
Conventionally, the Hsp60 family proteins in animal cells are believed to be mostly mitochondrial, although several studies have shown their presence outside the mitochondria as well (Soltys and Gupta 1999; Pfister et al. 2005; Sarkar et al. 2006; Arya et al. 2007). In the present study, we used two well-known mitochondrial markers but found that none of the Hsp60 granules colocalized or associated with mitochondria, although the more diffusely distributed Hsp60 may overlap with mitochondria. The close proximity of the two types of Hsp60 granules with each other in differentiating photoreceptor cells suggests that they may be part of a larger complex, the identity of which remains to be determined. The apparent colocalization of granular DIAP1 with the rabbit anti-Hsp60 antibody-recognized Hsp60 granules further suggests that DIAP1 is localized in part of this structure. It is possible that conformation of the Hsp60 in the two components of this presumed bipartite structure is different, which makes them differentially recognizable by the two Hsp60 antibodies.
Our present studies reveal a novel role of the Hsp60D protein of D. melanogaster in apoptosis. It was seen that apoptosis induced by the expression of Reaper, Grim, or Hid protein by the GMR-promoter in developing eye discs could be dominantly blocked by the depletion of Hsp60D through the coexpression of the Hsp60D-RNAi transgene. We have further seen that Hsp60D RNAi dominantly suppressed RHG-induced apoptosis in other cell types as well because Scabrous-Gal4-driven expression of Reaper affects the macrochaetae in fly thorax (Igaki et al. 2002) but coexpression of Hsp60D-RNAi rescues the phenotype (data not presented). It is significant that depletion of Hsp60D also dominantly inhibited apoptosis induced either by excess of procaspases or activated caspases or by JNK or EGFR mutants. These observations indicate that Hsp60D is essential for some step/s in the chain of the canonical cell death pathway.
Reaper, Hid, and Grim proteins are believed to induce apoptosis by removing inhibitors of caspases like DIAP1 from the procaspases/caspases (Vernooy et al. 2000). Modes of actions of the three RHG proteins are different in some respects and each of them may also have some distinct roles in apoptosis (Yoo et al. 2002). In this context, it is interesting that the recovery of adult eye structure in Hsp60D-RNAi background was proportional to the initial damage caused by these three proapoptotic factors. This suggests that the presence of Hsp60D protein is essential for the canonical caspase-driven apoptosis but may not be essential for some other paths that lead to apoptosis. Because overexpression of Hsp60D by itself did not result in significantly enhanced apoptosis in developing eye discs, it appears that Hsp60D has a negative role in the progression of apoptosis such that its presence is essential at one or more steps in apoptosis, but this protein by itself cannot bring about apoptosis.
Hsp60 family members have been suggested to have proapoptotic and prosurvival roles in diverse mammalian cell types (Arya et al. 2007; Chandra et al. 2007). In several cell types, Hsp60 promotes caspase activation and thus has a proapoptotic role (Samali et al. 1999; Xanthoudakis et al. 1999; Chandra et al. 2007) so that the absence of Hsp60 affects maturation of procaspases. In case of prosurvival function, absence of Hsp60 promotes caspase activation (Chandra et al. 2007; Lanneau et al. 2008). Because the downregulation of Hsp60D prevents apoptosis in eye disc cells of Drosophila larvae, it may appear that Hsp60D resembles the prodeath role of some mammalian Hsp60 members. However, our results further show that the apparent prodeath role of Hsp60D of D. melanogaster differs from that of the mammalian cells. Unlike the mammalian prodeath Hsp60, depletion of Hsp60D blocked apoptosis triggered not only by upstream signals like JNK or EGFR or by excess of procaspase, but also by the expression of activated caspases. In this context, it is significant that apoptosis caused by the absence of functional DIAP1 could not be prevented by the depletion of Hsp60D. DIAP1 associates with procaspases and with activated caspases and thus prevent their downstream activity (Hay and Guo 2006). Keeping in view these observations, we suggest that the removal of DIAP1 from procaspases or activated caspases, following the apoptotic signal, requires the presence of Hsp60D so that when Hsp60D is depleted by RNAi, the procaspases or the activated caspases cannot be released from DIAP1 and thus cannot execute cell death. Our observations on subcellular localization of Hsp60 and DIAP1 indeed suggest a spatially close association of these two proteins. A functional interaction of these two proteins is also indicated by the loss of granular distribution of either of these proteins when any one of these (Hsp60D or DIAP1) is depleted by RNAi.
Lethality associated with global ablation or overexpression of this gene’s transcripts through the Act5C-Gal4 driver does suggest some essential role of this gene in normal development. However, in the absence of any detectable phenotype of adult eyes following its depletion or overexpression through the GMR-Gal4 driver, except the recovery of damage caused by two copies of the GMR-Gal4 transgene by Hsp60D-RNAi, this gene’s developmental role in eye discs could not be specifically addressed. It is unlikely that this protein serves only as a promoter of apoptosis as and when a cell is triggered to enter the death pathway. Several of the nonapoptotic functions of caspases in Drosophila development, like border cell migration during oogenesis, sperm individualization, shaping of aristae, dendrite pruning, development of sensory organ precursor, etc. (Kuranaga et al. 2006) are mediated via the DIAP1. The DmIKKɛ degrades DIAP1 which in turn regulates actin dynamics and thus cell morphology, movement, and differentiation of sensory organ precursor cells (Montell 2006; Oshima et al. 2006). The close association of Hsp60 with DIAP1 in granular structures appears to be significant also in Hsp60D gene’s developmental roles in photoreceptor cells. We have seen (data not presented) that the Hsp60 (and thus DIAP1) granules are present immediately below and abutting the F-actin layer, which is present at the apical regions of photoreceptor cells in late larval eye discs (Arikawa et al. 1990). The close proximity of DIAP1 with F-actin, which plays a critical role in photoreceptor morphogenesis (Benlali et al. 2000; Tepass and Harris 2007), suggests that, as reported in other cell types (Kuranaga et al. 2006; Montell 2006; Oshima et al. 2006), a nonapoptotic function of DIAP1 in eye discs may relate to actin dynamics and thus photoreceptor morphogenesis. Other studies in our laboratory (Sarkar and Lakhotia 2008) have revealed an essential role of Hsp60C in organizing F-actin and other cytoskeletal structures in follicle and germ cells in developing egg chambers. It is, therefore, possible that in differentiating photoreceptor cells, the Hsp60D, together with DIAP1 may regulate the cytoskeletal remodeling of the specific architecture of these highly specialized cells. Absence of an eye phenotype following the targeted depletion or overexpression of Hsp60D in eye disc cells may indicate the existence of alternative paths, e.g., recruitment of other Hsp60 forms and/or critical threshold levels which may not be disrupted following our experimental conditions. The presence of survivors with globally ablated or overexpressed Hsp60D transcripts without any apparent morphological phenotypes also suggests the existence of other mechanisms that can buffer the altered Hsp60D levels.
Further studies are required to understand the specific interaction between Hsp60D and DIAP1 and the mechanism through which one regulates the other in normal development and during induced apoptosis. It will also be interesting to examine if such multiple forms with specialized functions of Hsp60 exist in genomes of other species of Drosophila so that their evolutionary significance can be better addressed.
References
Abrams JM, White K, Fessler LI, Steller H (1993) Programmed cell death during Drosophila embryogenesis. Development 117:29–43
Arikawa K, Hicks JL, Williams DS (1990) Identification of actin filaments in the rhabdomeral microvilli of Drosophila photoreceptors. J Cell Biol 110:1993–1998. DOI 10.1083/jcb.110.6.1993
Arya R, Lakhotia SC (2006) A simple nail polish imprint technique for examination of external morphology of Drosophila eyes. Curr Sci 90:1179–1180
Arya R, Mallik M, Lakhotia SC (2007) Heat shock genes—integrating cell survival and death. J Biosci 32:595–610. DOI 10.1007/s12038-007-0059-3
Benlali A, Draskovic I, Hazelett DJ, Treisman JE (2000) act up controls actin polymerization to alter cell shape and restrict hedgehog signaling in the Drosophila eye disc. Cell 101:271–281. DOI 10.1016/S0092-8674(00)80837-5
Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366. DOI 10.1016/S0092-8674(00)80928-9
Chandra D, Choy G, Tang DG (2007) Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. J Biol Chem 282:31289–31301. DOI 10.1074/jbc.M702777200
Chen P, Nordstrom W, Gish B, Abrams JM (1996) grim, a novel cell death gene in Drosophila. Genes Dev 10:1773–1782. DOI 10.1101/gad.10.14.1773
Cooley L, Kelley R, Spradling A (1988) Insertional mutagenesis of the Drosophila genome with single P elements. Science 239:1121–1128. DOI 10.1126/science.2830671
Deveraux QL, Reed JC (1999) IAP family proteins—suppressors of apoptosis. Genes Dev 13:239–252. DOI 10.1101/gad.13.3.239
Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB (1996) A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J 15:2685–2694
Ekengren S, Tryselius Y, Dushay MS, Liu G, Steiner H, Hultmark D (2001) A humoral stress response in Drosophila. Curr Biol 11:1479. DOI 10.1016/S0960-9822(01)00452-3
Ellis RJ, van der Vies SM (1991) Molecular chaperones. Ann Rev Biochem 60:321–347. DOI 10.1146/annurev.bi.60.070191.001541
Ellis MC, O’Neill EM, Rubin GM (1993) Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119:855–865
Fink AL (1999) Chaperone-mediated protein folding. Physiol Rev 79:425–449
Franceschini N (1972) Pupil and pseudopupil in the compound eye of Drosophila. In: Wehner R (ed) Information processing in the visual system of Drosophila. Springer, Berlin, pp 75–82
Freeman M (1994) The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech Dev 48:25–33. DOI 10.1016/0925-4773(94)90003-5
Giordano E, Rendina R, Peluso I, Furia M (2002) RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics 160:637–648
Goyal G, Fell B, Sarin A, Youle RJ, Sriram V (2007) Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell 12:807–816. DOI 10.1016/j.devcel.2007.02.002
Grether ME, Abrams JM, Agapite J, White K, Steller H (1995) The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 9:1694–1708. DOI 10.1101/gad.9.14.1694
Gupta RS (1990) Microtubules, mitochondria, and molecular chaperones: a new hypothesis for in vivo assembly of microtubules. Biochem Cell Biol 68:1352–1363
Hawkins CJ, Ekert PG, Uren AG, Holmgreen SP, Vaux DL (1998) Anti-apoptotic potential of insect cellular and viral IAPs in mammalian cells. Cell Death Differ 5:569–576. DOI 10.1038/sj.cdd.4400389
Hay BA, Guo M (2006) Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol 22:623–650. DOI 10.1146/annurev.cellbio.21.012804.093845
Hay BA, Wolff T, Rubin GM (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development 120:2121–2129
Hay BA, Wassarman DA, Rubin GM (1995) Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83:1253–1262. DOI 10.1016/0092-8674(95)90150-7
Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M (2002) Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J 21:3009–3018. DOI 10.1093/emboj/cdf306
Ikawa S, Weinberg RA (1992) An interaction between p21ras and heat shock protein hsp60, a chaperonin. Proc Natl Acad Sci U S A 89:2012–2016. DOI 10.1073/pnas.89.6.2012
Jones M, Gupta RS, Englesberg E (1994) Enhancement in amount of P1 (hsp60) in mutants of Chinese hamster ovary (CHO-K1) cells exhibiting increases in the A system of amino acid transport. Proc Natl Acad Sci U S A 91:858–862. DOI 10.1073/pnas.91.3.858
Jones G, Jones D, Zhou L, Steller H, Chu Y (2000) Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J Biol Chem 275:22157–22165. DOI 10.1074/jbc.M000369200
Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M (1999) Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell 4:757–769. DOI 10.1016/S1097-2765(00)80386-X
Kauppila S, Maaty WS, Chen P, Tomar RS, Eby MT, Chapo J, Chew S, Rathore N, Zachariah S, Sinha SK, Abrams JM, Chaudhary PM (2003) Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene 22:4860–4867. DOI 10.1038/sj.onc.1206715
Kozlova T, Perezgasga L, Reynaud E, Zurita M (1997) The Drosophila melanogaster homologue of the hsp60 gene is encoded by the essential locus l(1)10Ac and is differentially expressed during fly development. Dev Genes Evol 207:253–263. DOI 10.1007/s004270050113
Kramer JM, Staveley BE (2003) GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet Mol Res 2:43–47
Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B (2006) Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods 3:833–838
Kumar S, Colussi PA (1999) Prodomains–adaptors–oligomerization: the pursuit of caspase activation in apoptosis. Trends Biochem Sci 24:1–4. DOI 10.1016/S0968-0004(98)01332-2
Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, Miura M (2006) Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 126:583–596. DOI 10.1016/j.cell.2006.05.048
Lakhotia SC, Singh AK (1989) A novel heat shock polypeptide in Malpighian tubule of Drosophila melanogaster. J Genet 68:129–268
Lakhotia SC, Singh BN (1996) Synthesis of a ubiquitously present new HSP60 family protein is enhanced by heat shock only in the Malpighian tubules of Drosophila. Experientia 52:751–756. DOI 10.1007/BF01923984
Lakhotia SC, Rajendra TK, Prasanth KV (2001) Developmental regulation and complex organization of the promoter of the non-coding hsr(omega) gene of Drosophila melanogaster. J Biosci 26:25–38. DOI 10.1007/BF02708978
Lakhotia SC, Srivastava P, Prasanth KV (2002) Regulation of heat shock proteins, Hsp70 and Hsp64, in heat-shocked Malpighian tubules of Drosophila melanogaster larvae. Cell Stress Chaperones 7:347–356. DOI 10.1379/1466-1268(2002)007<0347:ROHSPH>2.0.CO;2
Lanneau D, Brunet M, Frisan E, Solary E, Fontenay F, Garrido C (2008) Heat shock proteins: essential proteins for apoptosis regulation. J Cell Mol Med (in press)
Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D, Zachariou A, Pichaud F, Ueda R, Meier P (2006) Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ 13:1663–1674. DOI 10.1038/sj.cdd.4401868
Martin CS, Flores AI, Cuezva JM (1995) Cpn60 is exclusively localized into mitochondria of rat liver and embryonic Drosophila cells. J Cell Biochem 59:235–245. DOI 10.1002/jcb.240590212
McMullin TW, Hallberg RL (1988) A highly evolutionarily conserved mitochondrial protein is structurally related to the protein encoded by the Escherichia coli groEL gene. Mol Cell Biol 8:371–380
Meier P, Silke J, Leevers SJ, Evan GI (2000) The Drosophila caspase DRONC is regulated by DIAP1. EMBO J 19:598–611. DOI 10.1093/emboj/19.4.598
Montell DJ (2006) A kinase gets caspases into shape. Cell 126:450–452. DOI 10.1016/j.cell.2006.07.017
Moreno E, Yan M, Basler K (2002) Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol 12:1263–1268. DOI 10.1016/S0960-9822(02)00954-5
Mukamel Z, Kimchi A (2004) Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. J Biol Chem 279:36732–36738. DOI 10.1074/jbc.M400041200
Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, Guo M, Baehrecke EH, Hay BA (2006) The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133:3305–3315. DOI 10.1242/dev.02495
Nicholson DW (1999) Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ 6:1028–1042. DOI 10.1038/sj.cdd.4400598
Nover L (1984) Heat shock response of eukaryotic cells. Springer, Berlin, pp 1–78
Oshima K, Takeda M, Kuranaga E, Ueda R, Aigaki T, Miura M, Hayashi S (2006) IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr Biol 16:1531–1537
Perezgasga L, Segovia L, Zurita M (1999) Molecular characterization of the 5′ control region and of two lethal alleles affecting the hsp60 gene in Drosophila melanogaster. FEBS Lett 456:269–273. DOI 10.1016/S0014-5793(99)00963-1
Perrimon N, Mathey-Prevot B (2007) Off targets and genome scale RNAi screens in Drosophila. Fly 1:1–5
Pfister G, Stroh CM, Perschinka H, Kind M, Knoflach M, Hinterdorfer P, Wick G (2005) Detection of HSP60 on the membrane surface of stressed human endothelial cells by atomic force and confocal microscopy. J Cell Sci 118:1587–1594. DOI 10.1242/jcs.02292
Pilling AD, Horiuchi D, Lively CM, Saxton WM (2006) Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell 17:2057–2068. DOI 10.1091/mbc.E05-06-0526
Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC (2000) Omega speckles—a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci 113(Pt 19):3485–3497
Retzlaff C, Yamamoto Y, Hoffman PS, Friedman H, Klein TW (1994) Bacterial heat shock proteins directly induce cytokine mRNA and interleukin-1 secretion in macrophage cultures. Infect Immun 62:5689–5693
Riedl SJ, Shi Y (2004) Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 5:897–907. DOI 10.1038/nrm1496
Rodriguez A, Chen P, Oliver H, Abrams JM (2002) Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J 21:2189–2197. DOI 10.1093/emboj/21.9.2189
Salvesen GS, Duckett CS (2002) IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol 3:401–410. DOI 10.1038/nrm830
Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S (1999) Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J 18:2040–2048. DOI 10.1093/emboj/18.8.2040
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA
Sarkar S, Lakhotia SC (2005) The Hsp60C gene in the 25F cytogenetic region in Drosophila melanogaster is essential for tracheal development and fertility. J Genet 84:265–281. DOI 10.1007/BF02715797
Sarkar S, Lakhotia SC (2008) Hsp60C is required in follicle as well as germline cells during oogenesis in Drosophila melanogaster. Dev Dyn 237:1334–1347. DOI 10.1002/dvdy.21524
Sarkar S, Arya R, Lakhotia SC (2006) Chaperonins: in life and death. In: Sreedhar AS, Srinivas UK (eds) Stress responses: a molecular biology approach. Signpost, Trivandrum, India, pp 43–60
Sawamoto K, Okano H, Kobayakawa Y, Hayashi S, Mikoshiba K, Tanimura T (1994) The function of argos in regulating cell fate decisions during Drosophila eye and wing vein development. Dev Biol 164:267–276. DOI 10.1006/dbio.1994.1197
Sawamoto K, Okabe M, Tanimura T, Mikoshiba K, Nishida Y, Okano H (1996) The Drosophila secreted protein Argos regulates signal transduction in the Ras/MAPK pathway. Dev Biol 178:13–22. DOI 10.1006/dbio.1996.0194
Sawamoto K, Taguchi A, Hirota Y, Yamada C, Jin M, Okano H (1998) Argos induces programmed cell death in the developing Drosophila eye by inhibition of the Ras pathway. Cell Death Differ 5:548. DOI 10.1038/sj.cdd.4400398
Schweitzer R, Shaharabany M, Seger R, Shilo BZ (1995) Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev 9:1518–1529. DOI 10.1101/gad.9.12.1518
Soltys BJ, Gupta RS (1999) Mitochondrial-matrix proteins at unexpected locations: are they exported? Trends Biochem Sci 24:174–177. DOI 10.1016/S0968-0004(99)01390-0
Spreij TE (1971) Cell death during the development of the imaginal discs of Calliphora erythrocephala. Neth J Zool 21:221–264. DOI 10.1163/002829670X00295
Srivastava P (2004) Studies on the constitutively expressed members of Hsp60 and Hsp70 families in Drosophila melanogaster. Ph.D. Thesis, Banaras Hindu University, Varanasi
Tepass U, Harris KP (2007) Adherens junctions in Drosophila retinal morphogenesis. Trends Cell Biol 17:26–35. DOI 10.1016/j.tcb.2006.11.006
Timakov B, Zhang P (2001) The hsp60B gene of Drosophila melanogaster is essential for the spermatid individualization process. Cell Stress Chaperones 6:71–77. DOI 10.1379/1466-1268(2001)006<0071:THGODM>2.0.CO;2
Tissieres A, Mitchell HK, Tracy UM (1974) Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol 84:389–98. DOI 10.1016/0022-2836(74)90447-1
Vaux DL, Korsmeyer SJ (1999) Cell death in development. Cell 96:245–254. DOI 10.1016/S0092-8674(00)80564-4
Vernooy SY, Copeland J, Ghaboosi N, Griffin EE, Yoo SJ, Hay BA (2000) Cell death regulation in Drosophila: conservation of mechanism and unique insights. J Cell Biol 150:F69–F76. DOI 10.1083/jcb.150.2.F69
Wells AD, Rai SK, Salvato MS, Band H, Malkovsky M (1997) Restoration of MHC class I surface expression and endogenous antigen presentation by a molecular chaperone. Scand J Immunol 45:605–612. DOI 10.1046/j.1365-3083.1997.d01-436.x
White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H (1994) Genetic control of programmed cell death in Drosophila. Science 264:677–683. DOI 10.1126/science.8171319
Woodlock TJ, Chen X, Young DA, Bethlendy G, Lichtman MA, Segel GB (1997) Association of HSP60-like proteins with the L-system amino acid transporter. Arch Biochem Biophys 338:50–56. DOI 10.1006/abbi.1996.9798
Xanthoudakis S, Roy S, Rasper D, Hennessey T, Aubin Y, Cassady R, Tawa P, Ruel R, Rosen A, Nicholson DW (1999) Hsp60 accelerates the maturation of pro-caspase-3 by upstream activator proteases during apoptosis. EMBO J 18:2049–2056. DOI 10.1093/emboj/18.8.2049
Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA (2002) Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 4:416–424. DOI 10.1038/ncb793
Acknowledgements
We thank Drs. M. Freeman (Cambridge, UK), B. Hay, and I. Muro (California Institute of Technology, USA), P. Meier and G.I. Evan (Breakthrough Toby Robins Breast Cancer Research Centre, UK), M. Miura (Brain Science Institute, RIKEN), Dr. V. Sriram (National Centre for Biological Sciences, Bangalore, India) and the Bloomington Fly Stock Centre (Indiana, USA) for providing different fly stocks used in this study. We also thank Dr. K. White (CBRC, Massachusetts General Hospital) and Dr. S. Ganesh (IIT Kanpur, India) for providing DIAP1 and Grp75 antibodies, respectively. We thank Dr. L. S. Shashidhara for his help in the generation of the Hsp60D transgenic flies at the Centre for Cellular and Molecular Biology (Hyderabad). This work was supported by a research grant from the Department of Biotechnology, Government of India, N. Delhi, to SCL. The Laser Scanning Confocal Microscope Facility is supported by the Department of Science and Technology, Government of India, N. Delhi. RA is supported by a research fellowship from the Council of Scientific and Industrial Research, N. Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arya, R., Lakhotia, S.C. Hsp60D is essential for caspase-mediated induced apoptosis in Drosophila melanogaster . Cell Stress and Chaperones 13, 509–526 (2008). https://doi.org/10.1007/s12192-008-0051-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-008-0051-3