Abstract
Retrogradation behavior is an important physicochemical property of starch during storage. A fast and sensitive method was developed for determining the retrogradation degree (RD) in corn starch by mid-infrared (MIR), Raman spectroscopy, and combination of MIR and Raman. MIR and Raman spectra were collected from different retrogradation starch and then processed by partial least squares (PLS), interval PLS (iPLS), synergy interval PLS (siPLS), and backward interval PLS (biPLS). Two different levels of fusion data extracted from MIR and Raman spectra were analyzed by PLS. The developed models demonstrated that both MIR and Raman techniques combined with chemometrics can be used to determine the RD in starch. The PLS model built by medium-level fusion approach achieved the most satisfied performance with a correlation coefficient of 0.9658. Integrating MIR and Raman technique combined with chemometrics improved the prediction performance of RD in comparison with a single technique.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Starch presenting in an enormous variety of food products acts as the main material to supply nutrition and energy, or as an additive to improve the quality of food. Starch retrogradation behavior is an important physicochemical property of starch during storage. Retrogradation could lead to deterioration of starch-based food during storage (Eliasson 2010), while retrogradation also could provides starch food with functional properties. Starch is beginning to retrograde after starch completely gelatinized. During retrogradation, molecular chains in starch begin to reassemble to develop an ordered structure (Ferrero et al. 1994). Generally, starch paste retrogradation is accompanied by gradual increases in rigidity and phase separation between polymer and solvent (Karim et al. 2000). Starch-based foods after retrogradation are indigestible by body enzymes and may make the consumer suffer from indigestion (Hayakawa et al. 1997). Therefore, a number of steps were attempted to study and prevent retrogradation (Liu et al. 2007). As we all know, one kind of resistant starch called RS3 is the retrograded starch formed during cooling of gelatinized starch. As a new resource of dietary fiber, retrogradation starch can provide functional properties and find applications in a variety of foods (Karim et al. 2000; Sajilata et al. 2006). Consumers prefer appropriate retrogradation to no retrogradation in starch-based products. Retrogradation is used to harden products and reduce product stickiness during the manufacturing process of breakfast cereals and parboiled rice (Karim et al. 2000). The retrogradation starch is often said that retrogradation deteriorates the quality of starch food. It is equally true that the suitable retrogradation starch is a benefit to gastrointestinal digestion. Therefore, the retrogradation degree (RD) in starch is a very important index for monitoring the quality of starch foods.
Various methods have been applied for studying retrogradation starch, such as rheological methods texture profile analysis (TPA) and rapid visco analyzer (RVA) (Mariotti et al. 2009; Olayinka et al. 2011), thermal analysis (differential scanning calorimetry (DSC), differential thermal analysis (DTA) and nuclear magnetic resonance (NMR))(Chang and Liu 1991). The most popular method is enzymatic methods based on acid or amylolytic enzymes (e.g., a-amylase and β-amylase) for determining RD in starch. Normally, the RD is measured by determining residual non-digestible starch which is not digested to glucose after incubation with amylolytic enzymes (Karim et al. 2000). These methods are complicated, laborious, and time-consuming. Rheological methods are used to evaluate the characteristics of retrogradation starch based on viscosity property, hardness, and elasticity (Karim et al. 2000). These properties can provide the qualitative description of starch retrogradation while the RD values were not determined specifically (Smits et al. 1998). The enthalpy in the melting endotherm resulting from thermal analysis was used as the index for evaluation of starch retrogradation (Paker and Matak 2016). Samples detected by thermal analysis, enzymatic methods, and rheological methods are not reusable by consumers (Chang and Liu 1991). Most NMR instruments are expensive and not available in many labs or industries (Monakhova and Diehl 2016).
Spectroscopic methods have been used in study of retrogradation as nondestructive methods, such as near-infrared (NIR) spectroscopy, mid-infrared (MIR) spectroscopy, Raman spectroscopy, and so on (de Peinder et al. 2008; Rocha et al. 2016; Thygesen et al. 2003). Each retrogradation starch has a unique and characteristic spectrum due to their particular molecular component and structure. NIR shows overtones and combination vibrations of the molecule when NIR beam irradiates into samples. The molecular bands observed in NIR spectra are very broad resulting in that it is difficult to ascribe specific bands to specific chemical components (Romano et al. 2016). Both MIR and Raman spectroscopy can generate bands linked to fundamental vibration and supply fingerprints of components that can be used for quantitative and qualitative characterization (Vankeirsbilck et al. 2002). They have been applied to characterize the molecular structural changes of retrogradation starch and are conducive to comprehending the changes of amylose and amylopectin (Flores-Morales et al. 2012). However, there is no study about quantitative analysis of RD in starch by MIR, Raman, and the combination of them. Different types of starch (from pure corn and cassava starch samples, as well with mixtures from both starch types) can be characterized by using Raman spectroscopy (Almeida et al. 2010). Besides Raman spectroscopy, MIR spectroscopy can also be used for quantitative analysis of RD in starch (Wu et al. 2016). Acting as complementary spectroscopic techniques, both types of measurements, Raman and MIR, can provide different molecular vibrations (Thygesen et al. 2003). Previous studies have demonstrated that data fusion technique based on MIR and Raman can increase the prediction ability of chemical components in food (Wu et al. 2016).
Therefore, the objectives of this paper were (1) to use MIR, Raman spectroscopy, and the combination of two techniques for investigating the retrogradation behavior in starch and (2) to establish a nondestructive and rapid method for measuring the quality, acceptability, and shelf life of starch-containing foods.
Materials and Methods
Retrogradation Starch Preparation
Corn starch was purchased from Runzhou Starch Company in Zhenjiang. A solution of corn starch (1 g) suspended in 19 ml of water was heated at 100 °C with constant stirring for 1 h in order to make starch completely gelatinized. The gelatinized starch paste was stored for different times (0, 1, 2, 3, 4, 5, 10, 15, 20 days) at 4 °C. After storage, retrogradation starch with different storage time was dried and kept in a desiccator. Three sets of samples were prepared from three independent experiments which were prepared with the same procedure. The first set of samples (96 samples) would be used for building models. Forty-eight samples prepared in the second and third experiments would act as prediction set for model predictive assessment. The samples from independent experiments would be used to test predictive performance of model for unknown samples.
MIR and Raman Spectroscopy
The MIR spectra of retrogradation starch were collected by Nicolet 380 FT-IR spectrometer (Thermo Electron Corporation, USA) in the spectral range of 650 to 4000 cm−1 at a resolution of 2 cm−1 (Flores-Morales et al. 2012). Raman spectra were recorded with DXR Laser micro-Raman spectrometer (Thermo Electron Corporation, USA) with 532-nm laser source. During collection of Raman spectra, time of integration is 5 s. For each spectrum, an average of 32 scans were performed at a resolution of 1 cm−1 over the 100–3200 cm−1 range (Xu et al. 2014). To obtain the most useful spectral information, multiple scans were performed in different points of the sample by moving the substrate on an X-Y stage. And the spectra from same sample were averaged into one spectra. Before collection, the Raman system was calibrated with a silicon semiconductor. The laser power irradiation over the samples was 4 mW. Finally, 144 and 144 spectra for MIR and Raman were obtained, respectively.
Reference Analysis of RD in Starch
The reference RD in starch was measured by the modified method of Tsuge et al. (Di Paola et al. 2003). A solution of 25 mg retrogradation starch in 8 ml distilled water was placed into a test tube. Five milliliter 0.1 mol l−1 phosphate buffer (pH 6.0, 0.3% NaC1) and 2 ml 3.5 u ml−1 α-amylase solution were then placed into the test tube. After incubation for 1 h at 37 °C, the enzymatic reaction was stopped by adding 5 ml of 4 mol L−1 NaOH. The pH of the solution was adjusted to neutrality with 4 mol L−1 HC1 and the volume was made up to100 ml with distilled water. Five milliliters of iodine solution (0.2% I2–2% KI) were added to l0 ml of the digested solution, and made up to l00 ml with distilled water. The absorbance of solution at 625 nm was measured after standing for 20 min. The RD (%) is calculated from equation described by Tsuge et al. The values of RD obtained would be used for the construction and validation of model (Kim et al. 1997).
Data Analysis
Different preprocessing techniques (standard normal variate (SNV), mean centering (MC), and multiplicative scatter correction (MSC), Savitzky–Golay smoothing (SG)) were applied for eliminating baseline shift and scatter effects, etc. By comparing results obtained from four preprocessing methods, SG is much better than SNV, MC, and MSC (Chen et al. 2011).
PCA was performed to show the clustering trend of retrogradation starch samples (Yeung and Ruzzo 2001). PCA is a well-known method for feature extraction in spectral analysis. It transforms the original independent variables into new variables (principal components (PCs)). The PCs are orthogonal and can be used as input variables for pattern recognition analysis (Haiyan et al. 2008).
Partial least squares (PLS) is used extensively for it is able to cope with high-dimensional data by extracting latent variables. So far, PLS has been widely used to build multivariate calibration models using the whole spectrum (WS) range (Lin et al. 2016). Therefore, WS-PLS models based on MIR spectra or Raman spectra were established. In application of PLS algorithm, the optimum number of latent variables (LVs) is a critical parameter in calibration model.
Interval variable selection algorithms (interval PLS (iPLS), synergy interval PLS (siPLS), and backward interval PLS (biPLS) proposed based on the PLS method were used to eliminate uncorrelated variables to improve PLS model performance (Chen et al. 2008). Literatures have discussion about the important variables selection or unimportant variables elimination (Zou et al. 2007; Ma et al. 2017). The principles of interval variable selection algorithms were described in various papers (Ma et al. 2017). In iPLS algorithm, the root mean square error of cross-validation (RMSECV) was calculated for every subinterval when the full spectrum was split into 40 intervals. The spectral region with the lowest RMSECV was chosen as the best interval for prediction of RD. In siPLS and biPLS algorithm, the combination of intervals with the lowest RMSECV is chosen (Chen et al. 2008). This enables us to select the best combination of intervals, generally providing better correlation coefficient (R) values and smaller prediction errors than iPLS (Nørgaard et al. 2000).
PLS models based on fusion data extracted from MIR and Raman spectra were investigated. Fusion data were carried out basically at three levels: low-, mid-, and high-level fusion (Borràs et al. 2015). High-level fusion has often provided worse results than the other two levels (Nunes et al. 2016). Thus, low and medium levels were investigated in this paper (Wu et al. 2016). Low-level fusion data consists of original variables of MIR and Raman after the preprocessing steps. Medium-level fusion extracts relevant features from MIR and Raman data separately and then merges them into a single matrix, which will be analyzed by chemometrics (Borràs et al. 2015). In this paper, the characteristic intervals of MIR and Raman were combined as fusion data which acted as input data for establishing models.
One hundred forty-four MIR spectra were divided into two subsets which were called calibration set and prediction set. The calibration set contained 90 spectra which were used for establishing model, and the remaining 54 spectra as prediction set were used to test the performance of predictive models. The 144 Raman spectra were processed like MIR spectra. In this study, WS-PLS, iPLS, siPLS, and biPLS models based on MIR or Raman spectra were obtained. The low- and medium-level fusion approaches were applied to combine of MIR and Raman. In all models, the optimum number of latent variables (LVs) was determined by root mean square error of cross-validation (RMSECV). The performance of the final models were evaluated in accordance with the correlated coefficient of determination (R), and root mean square error of prediction (RMSEP) values (Varliklioz Er et al. 2016). The data were processed in MATLAB software version 7.10 (Math Works, Natick, MA, USA).
Results and Discussion
RD in Starch Stored for Different Time
Corn starch was completely gelatinized as described in the experimental section and then the starch pastes stored at 4 °C for 0, 1, 2, 3, 4, 5, 10, 15, and 20 days were dried. Finally, nine kinds of retrogradation starch were obtained and named 0, 1, 2, 3, 4, 5, 10, 15, and 20d. RD of retrogradation starch was determined by the enzymatic method based on α-amylase. Average values of RD for nine kinds of retrogradation starch were 15.87, 45.64, 67.21, 77.39, 85.30, 91.05, 94.91, 97.07, and 98.73% shown in Fig. 1. The stand deviations of each kind retrogradation starch were 1.234, 1.322, 0.9814, 1.097, 1.501, 0.5618, 0.5222, 0.5189, and 0.3777. It is indicated that RD in starch increased with the storage time prolonged. RD at the first 5 days varied significantly from 15.81 to 85.30%. The speed of retrogradation declined gradually after 5 days. Particularly, the RD of 15d and 20d were similar. It can be concluded that the structure and component of retrogradation starch at 0, 1, 2, 3, 4, and 5d alter dramatically while that at 10, 15, and 20d alter slightly.
Spectral Analysis
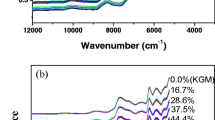
MIR and Raman spectroscopy can be used to detect properties of retrogradation starch, such as crystallinity and amorphization. Fig. 2a shows the average MIR spectra of retrogradation starch stored for 0, 1, 2, 3, 4, 5, 10, 15, and 20 days. Fig. 2b shows average Raman spectra for the retrogradation starch samples with different storage time.
MIR spectral patterns of retrogradation starch showed almost identical characteristic bands (Fig. 2a). The characteristic bands mainly contain 756, 820, 850, 880, 928, 949, 994, 1022, 1067, 1077, 1133, 1150, 1181, 1241, 1340, 1506, 1560, 1648, 1654, 1701, 2177, 2926, and 3275 cm−1. These bands mainly resulted from the vibrational modes of molecule in retrogradation starch. The bands around 1600 cm−1 are attributed to amorphous region of starch (Smits et al. 1998). The band at 1506 cm−1 is influenced by the skeletal mode vibration of a-1, 4 glycosidic linkage (C–O–C). The bands at 1022 and 850 cm−1 are sensitive to changes in crystallinity.
The nine Raman spectra for retrogradation starch also showed almost identical characteristic bands (Fig. 2b). The main vibrational bands of each spectrum are similar because different samples contain the same main components (polysaccharide). The vibrational bands mainly included 373, 408, 437, 480, 577, 860, 940, 952, 1051, 1082, 1126, 1260, 1338, 1382, 1462, 1518, 2116, 2870, and 2908 cm−1. The band at 2908 cm−1 is related to the symmetrical and antisymmetric CH stretching. The unobvious and sharped characteristic band at 2870 cm−1 can be attributed to the amylose and amylopectin presented in starch (Kizil et al. 2002). The region between 1200 and 1600 cm−1 contain a large supply of structural information. A majority of the bands in this region are due to coupled vibration involving hydrogen atoms. For instance, the band at 1462 is related to CH, CH2, and COH deformation. The feature at 1382 cm−1 corresponds to coupling of the CCH and COH deformation modes. The bands at 1260 and 1338 cm−1 can be mainly attributed to several vibrational modes, such as CO stretching, CC stretching, CCH deformation, COH deformation, and CCH deformation. The region between 1200 and 800 cm−1 is highly characteristic bands owing to CO stretching, CC stretching, and COC deformation modes, referring to the glycosidic bond (Mahdad-Benzerdjeb et al. 2007). This region is considered as the fingerprint or anomeric region and is discussed with high frequency in the previous papers (Baranska et al. 2005; Nikonenko et al. 2005; Yang and Zhang 2009).The vibrations originating from glycosidic linkages can be observed in the 920–960 cm−1 region. Particularly, the band observed at 940 cm−1 is assigned to the amylose α-1, 4 glycosidic linkage. Raman spectra of retrogradation starch exhibited complex vibrational modes at low wavenumbers (below 800 cm−1) due to the skeletal mode vibrations of the glucose pyranose ring. Among the Raman bands at 437, 480, and 577, a strong band at 480 cm−1 portraying the rate of polymerization in polysaccharides is one of the prominent and important indication of the presence of pyranose ring due to skeletal vibration mode (Kizil et al. 2002). Characteristic vibrational bands found in retrogradation starch are shown in Table 1 for both MIR and Raman.
Each spectrum of retrogradation starch is unique owing to its particular component and structure. MIR peaks at 1047 and 1022 cm−1 have been used for investigating changes in starch structure (organized starch and amorphous starch) during starch retrogradation (Flores-Morales et al. 2012). Previous studies have shown the most useful Raman bands which reflect the characteristics of retrogradation starch. For instance, according to Winter et al. (Winter and Kwak 1987), intensity of the Raman band at 480 cm−1 and the half-bandwidths of Raman bands at 2800–3000 cm−1 were used as excellent indexes for evaluating retrogradation starch. Besides the bands discussed in the previous work (Kizil et al. 2002), the intensities and shapes of other characteristic bands are also relevant to the starch component and structure. However, these bands were ignored for determination of retrogradation of starch. Therefore, the more spectral feature will be applied for determining retrogradation starch in further analysis.
General Discrimination of Samples
PCA was applied to MIR and Raman spectra to evaluate their ability to differentiate the retrogradation starch with different storage time. The scores plots of PCA for retrogradation starch stored 0, 1, 2, 3, 4, 5, 10, 15, 20 days were shown in Fig. 3a, b. Figure 3a shows the scores plot of PCA based on MIR spectra. The retrogradation starch samples were divided into two categories with some overlapping. The first category located at the left of the scores plot contained retrogradation starch samples stored 0, 1, 2, 3, and 4 days. The starch samples stored 5, 10, 15, and 20 days belonged to the second category located at the right of the scores plot. According to the RD determined by enzymatic method, the RD reached 85% when the starch was stored 4 days. The average growth rate of 1 day was 21.25%. The RD during the first 4 days increased rapidly. When the starch was stored for 20 days, the RD of starch achieved 99%. From the 5th to 20th day, the RD increased from 85 to 99% with growth rate of 14%. The average growth rate of every day was 0.875%. The RD increased slowly. The starch retrogradation may be divided into two stages, i.e., fast and slow stages. Figure 3b shows score plot of PCA based on Raman spectra. All retrogradation starch samples were divided into three categories. The three categories were not completely separated. One of the categories located at the middle of the scores plot contained retrogradation starch samples stored for 0, 1, 2, and 3d. The retrogradation starch samples stored 4 and 5 days belonged to the second category and the other retrogradation starch samples (stored 10, 15, and 20 days) constituted the third category. In the first category, the retrogradation starch stored for 0, 1, and 3d were separated well from each other except for 2d overlapping with 1d. According to the results by enzymatic method, their RD increased rapidly. The structure or component of retrogradation starch was very fast at the first 3 days. For the retrogradation starch samples stored for 4 and 5 days, their RD increased more slowly than the retrogradation starch sample stored 0, 1, 2, and 3d. The change of retrogradation starch was slow. The RD of retrogradation starch stored for 10, 15, and 20 days showed subtle difference by enzymatic method indicating that starch changed very slowly. Structure and component of retrogradation starch were beginning to stabilize. The aggregation of starch samples may be led by their similar component and structure. The starch retrogradation may be divided into three stage, i.e., fast, slow, and stable stages. The stage retrogradation starch categories could be discriminated by PCA. Whereas, the specific RD in starch sample was not determined. Therefore, the RD in starch would be determined in following research
Models for Determining RD
Results of Models Based on MIR or Raman Spectra
PLS, iPLS, siPLS, and biPLS algorithm were used in this paper for establishing models to determine RD in starch. The results of models based on MIR and Raman spectra were listed in Table 2. From Table 2, good performance was obtained from the PLS models based on MIR or Raman spectra. The prediction values of RD were between 10 and 100%. The lowest RMSECV value (8.4) was achieved by biPLS model based on Raman spectra. The corresponding correlation coefficient (R p ) of prediction set was 0.9252 which achieved the best performance. The worst model was iPLS model with highest RMSECV based on the single interval in Raman spectra. The optimal Raman spectral interval was the sixth interval in the spectral range of 407 and 485 cm−1 when the whole spectrum was split into 40 intervals. The characteristic interval variables were attributed to skeletal mode in starch. The MIR biPLS model based on intervals numbered 2, 40, 28, 34, 17, 33, 37, 35, and 38 achieved the best performance in all models of MIR. The selected intervals were located in the spectral ranges of 734–817, 3917–4000, 2911–2994, 3413–3497, 1989–2072, 3330–3413, 3665–3749, 3497–3581, 3749–3832 cm−1 (Fig. 4a). Figure 4b shows a correlation between RD measured by reference analysis and RD predicted by MIR biPLS in prediction set. For models of Raman, biPLS models also showed the best performance. The selected Raman intervals were numbered 1, 2, 5, 6, 9, 10, 15, 33, 37, and 38, corresponding to the wavenumbers in the range of 100–254, 410–564, 718–872, 1181–1258, 2570–2647, 2878–3033 cm−1 shown in Fig. 5a. Figure 5b shows a correlation between RD measured by reference analysis and RD predicted by Raman biPLS in prediction set.
Comparing the models based on MIR and Raman spectra, models based on Raman spectra were better than models based on MIR spectra except iPLS model. IPLS algorithm caused a decline of the model performance when applied to Raman spectra in comparison with full spectrum PLS model. The superiority of Raman spectroscopy may be attributed to specificity and sensitivity (Yuan et al. 2017). The two methods do not offer the identical information about the molecular vibrations and structure. MIR spectroscopy probe the molecular vibrations when the electrical dipole moment changes, while Raman spectroscopy detect molecular vibrations according to the changes of electrical polarizability (Thygesen et al. 2003). The difference between them indicates that molecules tend to be more sensitive to Raman spectroscopy than to MIR spectroscopy. For instance, the C–C or C=C bond is more sensitive to Raman spectroscopy than to MIR spectroscopy. According to the characteristic vibrational bands for MIR and Raman found in food system, the skeletal mode of starch lead to specific vibrations in the Raman spectral regions located at 900–800 and 500–400 cm−1 (Thygesen et al. 2003). MIR is strongly dependent on proper sample preparation and moisture content can seriously affect MIR spectra. Conversely, Raman is highly sensitive and do not require special sample treatment.
Results of Models Based on Fusion Data
The validity of data fusion method has been demonstrated in the literature. Even though the performance of models based on MIR or Raman spectra is satisfied, the models based on information extracted from MIR and Raman spectra were still established for verifying that Raman with aid of MIR would be better than Raman spectroscopy.
As listed in Table 2, the prediction performance of PLS model based on low-level data is not improved in comparison with that of models based on MIR or Raman. Our findings are in agreement with results obtained by other researchers (Nunes et al. 2016). This might be due to the fact that the fusion data from MIR and Raman spectra contained too much redundant information which significantly result in decline of PLS model performance.
To overcome too much redundant information, characteristic intervals of MIR and Raman which selected by biPLS in “Results of Models Based on MIR or Raman Spectra” were merged as medium-level fusion data. The process of medium-level data fusion was shown in Fig. 6. As a result, PLS model based on characteristic intervals of MIR and Raman (medium-level fusion data) achieved a better performance with the highest R p of 0.9658 than PLS model based on full raw variables (low-level fusion data). Low-level fusion is simple, it just uses merge raw spectra. But high data volume may contain a large number of noise or irrelevant information. The performance of predictive models would be influenced by the negative information. Some limitation of low-level fusion can be partially overcome by medium-level fusion. Characteristic intervals can decrease the data volume and eliminate noise or irrelevant information.
Retrogradation degree are determined only by Raman spectroscopy, and satisfied results are obtained (Table 2). But by combining MIR and Raman spectroscopy, more accurate and reliable models were obtained. The combination of MIR and Raman spectroscopy proved better in determination of retrogradation degree compared to single Raman spectroscopy. Though Raman spectroscopy and chemometric tools have been successfully used for exploratory analysis of pure corn, cassava starch samples, and mixtures of both starches, as well as for the quantification of amylose content in corn and cassava starch samples (Almeida et al. 2010). Both MIR and Raman can generate bands linked to fundamental vibration and supply fingerprints of components that can be used for quantitative and qualitative characterization. Even though both methods probe molecular vibrations and structure, they do not provide exactly the same information (Thygesen et al. 2003). Raman spectroscopy detect molecular vibrations according to the changes of electrical polarizability. While MIR spectroscopy probe the molecular vibrations when the electrical dipole moment changes. They are complementary techniques for the study of molecular vibrations and structure. For example, the C–C group has a strong Raman scattering band in Raman spectra but weak absorption bands in the mid-infrared. O–H vibration is very strong in MIR, but very weak in Raman (Yang and Irudayaraj 2002). The intensities of characteristic bands in Raman and MIR spectra collected from the same food are different and the information they contain are not identical (Flores-Morales et al. 2012). Due to their distinct advantages, data fusion, as an emerging technology, is an efficient way for the optimum utilization of data from different sources, and has been successfully used for the rapid measurement of retrogradation starch in this paper. Therefore, a useful methods based on MIR and Raman spectroscopy were developed for determining retrogradation starch.
Conclusion
The RD in starch stored for different storage times have been predicated by MIR and Raman spectroscopy combined with chemometrics. The PLS model based on medium-level fusion data of MIR and Raman spectra had the best prediction performance with correlation coefficient (R) of 0.9658. Retrogradation starch was obtained by chilling gelatinized corn starch for different times (0, 1, 2, 3, 4, 5, 10, 15, 20 days). The low- and medium-level fusion data extracted from MIR and Raman were also analyzed by PLS. In addition, The MIR and Raman spectra of retrogradation starch were analyzed by SG, PCA, PLS, iPLS, siPLS, and biPLS for determination of RD in starch. The results demonstrated that the prediction performance of models for Raman are better than those based on the MIR except iPLS model. Variables selection improved the performance of PLS models. PLS model based on medium-level fusion data achieved the best performance in comparison with the models. Prediction of the RD in starch based on combination of MIR and Raman spectroscopy are more accurate than that based on single technique. This indicates that the developed methodology may be able to forecast the quality, acceptability, and shelf life of starch products or starch-containing products easily damaged by starch retrogradation.
Reference
Almeida MR, Alves RS, Nascimbem LBLR, Stephani R, Poppi RJ, de LFC O (2010) Determination of amylose content in starch using Raman spectroscopy and multivariate calibration analysis. Anal Bioanal Chem 397:2693–2701. doi:10.1007/s00216-010-3566-2
Baranska M, Schulz H, Baranski R, Nothnagel T, Christensen LP (2005) Situ simultaneous analysis of polyacetylenes, carotenoids and polysaccharides in carrot roots. J Agric Food Chem 53:6565–6571. doi:10.1021/jf0510440
Borràs E, Ferré J, Boqué R, Mestres M, Aceña L, Busto O. (2015) Data fusion methodologies for food and beverage authentication and quality assessment—a review Analytica Chimica Acta 891:1–14 doi:10.1016/j.aca.2015.04.042
Chang S-M, Liu L-C (1991) Retrogradation of rice starches studied by differential scanning calorimetry and influence of sugars. NaCl and Lipids Journal of Food Science 56:564–566. doi:10.1111/j.1365-2621.1991.tb05325.x
Chen Q, Zhao J, Liu M, Cai J, Liu J (2008) Determination of total polyphenols content in green tea using FT-NIR spectroscopy and different PLS algorithms J Pharm Biomed Anal 46:568–573 doi:10.1016/j.jpba.2007.10.031
Chen H, Pan T, Chen J, Lu Q (2011) Waveband selection for NIR spectroscopy analysis of soil organic matter based on SG smoothing and MWPLS methods Chemom Intell Lab Syst 107:139–146 doi:10.1016/j.chemolab.2011.02.008
de Peinder P, Vredenbregt MJ, Visser T, de Kaste D (2008) Detection of Lipitor® counterfeits: a comparison of NIR and Raman spectroscopy in combination with chemometrics J Pharm Biomed Anal 47:688–694 doi:10.1016/j.jpba.2008.02.016
Di Paola RD, Asis R, Aldao MAJ (2003) Evaluation of the degree of starch gelatinization by a new enzymatic method starch. Stärke 55:403–409. doi:10.1002/star.200300167
Eliasson AC (2010) Gelatinization and retrogradation of starch in foods and its implications for food quality. In: Skibsted LH, Risbo J, Andersen ML (eds) Chemical deterioration and physical instability of food and beverages. Woodhead Publishing Limited, Oxford, U.K., pp 296–323
Ferrero C, Martino MN, Zaritzky NE (1994) Corn starch-xanthan gum interaction and its effect on the stability during storage of frozen gelatinized suspension starch. Stärke 46:300–308. doi:10.1002/star.19940460805
Flores-Morales A, Jiménez-Estrada M, Mora-Escobedo R (2012) Determination of the structural changes by FT-IR, Raman, and CP/MAS 13C NMR spectroscopy on retrograded starch of maize tortillas Carbohydr Polym 87:61–68 doi:10.1016/j.carbpol.2011.07.011
Haiyan Y, Xiaoying N, Yibin Y, Xingxiang P (2008) Non-invasive determination of enological parameters of rice wine by Vis-NIR spectroscopy and least-squares support vector machines. Paper presented at the 2008 Providence, Rhode Island, June 29 – July 2, 2008, St. Joseph, Mich.,
Hayakawa K, Tanaka K, Nakamura T, Endo S, Hoshino T (1997) Quality characteristics of waxy hexaploid wheat (Triticum aestivum L.): Properties of Starch Gelatinization and Retrogradation. Cereal Chemistry Journal 74:576–580. doi:10.1094/CCHEM.1997.74.5.576
Karim AA, Norziah MH, Seow CC (2000) Methods for the study of starch retrogradation Food Chem 71:9–36 doi:10.1016/S0308-8146(00)00130-8
Kim J-O, Kim W-S, Shin M-S (1997) A comparative study on retrogradation of rice starch gels by DSC, x-ray and α-amylase methods starch. Stärke 49:71–75. doi:10.1002/star.19970490207
Kizil R, Irudayaraj J, Seetharaman K (2002) Characterization of irradiated starches by using FT-Raman and FTIR spectroscopy. J Agric Food Chem 50:3912–3918. doi:10.1021/jf011652p
Lin Y-W, Deng B-C, Wang L-L, Xu Q-S, Liu L, Liang Y-Z (2016) Fisher optimal subspace shrinkage for block variable selection with applications to NIR spectroscopic analysis Chemometrics and Intelligent Laboratory Systems 159:196–204 doi:10.1016/j.chemolab.2016.11.002
Liu H, Yu L, Chen L, Li L (2007) Retrogradation of corn starch after thermal treatment at different temperatures Carbohydr Polym 69:756–762 doi:10.1016/j.carbpol.2007.02.011
Ma H-l, Wang J-w, Y-j C, Cheng J-l, Lai Z-t (2017) Rapid authentication of starch adulterations in ultrafine granular powder of Shanyao by near-infrared spectroscopy coupled with chemometric methods. Food Chem 215:108–115. doi:10.1016/j.foodchem.2016.07.156
Mahdad-Benzerdjeb A, Taleb-Mokhtari IN, Sekkal-Rahal M (2007) Normal coordinates analyses of disaccharides constituted by d-glucose, d-galactose and d-fructose units Spectrochim Acta A Mol Biomol Spectrosc 68:284–299 doi:10.1016/j.saa.2006.11.032
Mariotti M, Sinelli N, Catenacci F, Pagani MA, Lucisano M (2009) Retrogradation behaviour of milled and brown rice pastes during ageing J Cereal Sci 49:171–177 doi:10.1016/j.jcs.2008.09.005
Monakhova YB, Diehl BWK (2016) Authentication of the origin of sucrose-based sugar products using quantitative natural abundance 13C NMR. J Sci Food Agric 96:2861–2866. doi:10.1002/jsfa.7456
Nikonenko NA, Buslov DK, Sushko NI, Zhbankov RG (2005) Spectroscopic manifestation of stretching vibrations of glycosidic linkage in polysaccharides J Mol Struct 752:20–24 doi:10.1016/j.molstruc.2005.05.015
Nørgaard L, Saudland A, Wagner J, Nielsen JP, Munck L, Engelsen SB (2000) Interval partial least-squares regression (iPLS): a comparative chemometric study with an example from near-infrared. Spectroscopy Applied Spectroscopy 54:413–419
Nunes KM, Andrade MVO, Santos Filho AMP, Lasmar MC, Sena MM (2016)Detection and characterisation of frauds in bovine meat in natura by non-meat ingredient additions using data fusion of chemical parameters and ATR-FTIR spectroscopy Food Chem 205:14–22 doi:10.1016/j.foodchem.2016.02.158
Olayinka OO, Olu-Owolabi BI, Adebowale KO (2011) Effect of succinylation on the physicochemical, rheological, thermal and retrogradation properties of red and white sorghum starches. Food Hydrocoll 25:515–520. doi:10.1016/j.foodhyd.2010.08.002
Paker I, Matak KE (2016) Influence of pre-cooking protein paste gelation conditions and post-cooking gel storage conditions on gel texture. J Sci Food Agric 96:280–286. doi:10.1002/jsfa.7091
Rocha JTC et al (2016) Sulfur determination in Brazilian petroleum fractions by mid-infrared and near-infrared spectroscopy and partial least squares associated with variable selection. Methods Energy & Fuels 30:698–705. doi:10.1021/acs.energyfuels.5b02463
Romano G, Nagle M, Müller J (2016) Two-parameter Lorentzian distribution for monitoring physical parameters of golden colored fruits during drying by application of laser light in the Vis/NIR spectrum innovative Food Science & Emerging Technologies 33:498–505 doi:10.1016/j.ifset.2015.11.007
Sajilata MG, Singhal RS, Kulkarni PR (2006) Resistant starch—a review comprehensive. Reviews in Food Science and Food Safety 5:1–17. doi:10.1111/j.1541-4337.2006.tb00076.x
Smits ALM, Ruhnau FC, JFG V, JJG v S (1998) Ageing of starch based systems as observed with FT-IR and solid state NMR spectroscopy starch. Stärke 50:478–483. doi:10.1002/(SICI)1521-379X(199812)50:11/12<478::AID-STAR478>3.0.CO;2-P
Synytsya A, Čopı́Ková J, Matě jka P, Machovič V (2003) Fourier transform Raman and infrared spectroscopy of pectins. Carbohydr Polym 54:97–106
Thygesen LG, Løkke MM, Micklander E, Engelsen SB (2003) Vibrational microspectroscopy of food. Raman vs. FT-IR Trends Food Sci Technol 14:50–57 doi:10.1016/S0924-2244(02)00243-1
Vankeirsbilck T, Vercauteren A, Baeyens W, Van der Weken G, Verpoort F, Vergote G, Remon JP (2002) Applications of Raman spectroscopy in pharmaceutical analysis TrAC Trends Anal Chem 21:869–877 doi:10.1016/S0165-9936(02)01208-6
Varliklioz Er S, Eksi-Kocak H, Yetim H, Boyaci IH (2016) Novel spectroscopic method for determination and quantification of saffron adulteration. Food Analytical Methods:1–9 doi:10.1007/s12161-016-0710-4
Winter WT, Kwak YT (1987) Rapid-scanning Raman spectroscopy: a novel approach to starch retrogradation Food Hydrocoll 1:461–463 doi:10.1016/S0268-005X(87)80041-3
Wu Z, Xu E, Long J, Pan X, Xu X, Jin Z, Jiao A (2016) Comparison between ATR-IR, Raman, concatenated ATR-IR and Raman spectroscopy for the determination of total antioxidant capacity and total phenolic content of Chinese rice wine Food Chem 194:671–679 doi:10.1016/j.foodchem.2015.08.071
Xu W, Liu X, Xie L, Ying Y (2014) Comparison of Fourier transform near-infrared, visible near-infrared, mid-infrared, and Raman spectroscopy as non-invasive tools for transgenic Rice discrimination. Trans ASABE 57:141–150
Yang L, Zhang L-M (2009) Chemical structural and chain conformational characterization of some bioactive polysaccharides isolated from natural sources Carbohydr Polym 76:349–361 doi:10.1016/j.carbpol.2008.12.015
Yeung KY, Ruzzo WL (2001) Principal component analysis for clustering gene expression data. Bioinformatics 17:763–774. doi:10.1093/bioinformatics/17.9.763
Yuan J et al. (2017) A rapid Raman detection of deoxynivalenol in agricultural products Food Chem 221:797–802 doi:10.1016/j.foodchem.2016.11.101
Zou X, Zhao J, Li Y (2007) Selection of the efficient wavelength regions in FT-NIR spectroscopy for determination of SSC of ‘Fuji’ apple based on BiPLS and FiPLS models vibrational Spectroscopy 44:220–227 doi:10.1016/j.vibspec.2006.11.005
Acknowledgement
The authors gratefully acknowledge the financial support provided by the National Key Research and Development Program of China (2016YFD0401104), the National Science and Technology Support Program (2015BAD17B04), the National Natural Science Foundation of China (Grant No. 31671844), the natural science foundation of Jiangsu province (BK20160506, BE2016306), International Science and Technology Cooperation Project of Jiangsu Province (BZ2016013), Suzhou Science and Technology Project (SNG201503), Six talent peaks project in Jiangsu Province (GDZB-016) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Informed consent is not applicable for the nature of this study.
Rights and permissions
About this article
Cite this article
Hu, X., Shi, J., Zhang, F. et al. Determination of Retrogradation Degree in Starch by Mid-infrared and Raman Spectroscopy during Storage. Food Anal. Methods 10, 3694–3705 (2017). https://doi.org/10.1007/s12161-017-0932-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-017-0932-0