Abstract
To develop effective alternatives for detecting genetically modified organisms (GMOs), we reported one optimized visual loop-mediated isothermal amplification (LAMP) method for the detection of exogenous DNA targets from two GM soybean events in this study. This isothermal amplification can be performed within 40 min without polymerase chain reaction (PCR) equipment and the derived LAMP products can be directly observed by naked eye employing SybrGreen I dye instead of conventional gel electrophoresis analysis. The limits of detection of these established visual LAMP assays were about 4 copies of haploid soybean genomic DNA, and which were much higher than those of reported conventional PCR assays. Furthermore, the high specificity of LAMP assays was determined. All the results demonstrated that the developed visual LAMP assays are convenient, cost-efficient, and rapid for on spot detection of GMOs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past two decades, the recombinant DNA technique has been widely applied in modern agriculture and around 150 kinds of genetically modified (GM) crops with new traits have been produced and approved for commercialization in global area Andersen et al. (2006). By the end of 2008, the global area of GM crops being planted has reached 125 million hectares. GM soybean is by far the world's most cultivated GM crop, and its planted area was 65.8 million hectares in 2008, occupying 53% of GM crops worldwide (James 2008). However, since the consumers concern GM food safety, series of laws have been issued for GMOs regulation and labeling in China, European Union (EU), Brazil, Korea, Japan, and Australia, etc.
Currently, various molecular methodologies based on protein or nucleic acid analysis have been developed and used for GMOs detection (Hernández et al. 2005; Rodríguez-Lázaro et al. 2007). In particular, conventional and real-time quantitative polymerase chain reaction (PCR) methods have been widely accepted as standards for identifying and determining the GM contents in food and feeds because of their high efficiency, sensitivity, and stability (Bellocchi et al. 2008; Holst-Jensen et al. 2003; Singh et al. 2008; Marmiroli et al. 2008; Zel et al. 2008). Several conventional and quantitative real-time PCR methods for GM soybean GTS 40-3-2 have been developed and validated (Yang et al. 2008; Taverniers et al. 2005; Berdal and Holst-Jensen 2001; Huang and Pan 2005). However, the requirements of either a high-precision instrument for PCR amplification or complicated procedures for PCR analysis are their main disadvantages, which limit these methods being widely used on spot detection.
Loop mediated isothermal amplification (LAMP) is one isothermal nucleic acids amplification technique developed by Notomi et al. (Notomi et al. 2000). The LAMP assay is performed using Bst DNA polymerase large fragment (New England Biolabs) with strand displacement activity under isothermal conditions ranging from 60 to 65 °C within shorter time. In LAMP assay, a set of four specially designed primers, termed inner primers (FIP and BIP) and outer primers (F3 and B3) were used, which can recognize at least six independent target DNA sequences, providing the exceptionally higher specificity of amplification than conventional PCR method. The LAMP products are typical ladder-like pattern and could be judged by agarose gel electrophoresis or real-time monitored according to the turbidity using turbidimeter (Mori et al. 2001). Alternatively, the LAMP reaction is determined visually by adding SybrGreen I dye to the reaction mixture: the color of the solution would change to green in the presence of LAMP amplicons, while it remains orange for the mixture with no amplification (Iwamoto et al. 2003). LAMP technique has been used successfully to detect bacterial microorganisms (Enosawa et al. 2003; Misawa et al. 2007), pathogenic virus (Poon et al. 2005; Imai et al. 2007), and parasites (Han et al. 2007), etc. However, there are few reports on the plant genomic DNA amplification using the LAMP method because of larger and more complex genomic DNA. Also, few applications using LAMP technique have been reported on GMOs detection (David et al. 2009).
In this study, the optimized visual LAMP methods for the detection of GM soybean events, GTS 40-3-2 and MON89788, have been established. The amplified efficiencies, limits of detection (LODs) and specificities of these systems indicated that the developed LAMP assays are more specific, effective, and sensitive than conventional PCR assays, and which can be used for GMOs detection on spot.
Materials and Methods
Plant Materials
The GM plant events (GTS 40-3-2 and MON89788 soybeans, MON810 maize, GT73 canola, and MON531 cotton) were developed by Monsanto Company, and the certified reference materials of these GM events were purchased from Sigma-Aldrich Trading Co., Ltd. (Shanghai, China) or the American Oil Chemists' Society. The seeds of GM Huafan No. 1 tomato were kindly supplied by Huazhong Agricultural University, China. Non-transgenic seeds of soybean, sweet pepper, wheat, and rice were purchased from local market in Shanghai, China.
DNA Extraction and Purification
The plant genomic DNA was extracted and purified using the Plant DNA Mini-Prep kit (Ruifeng Agro-tech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. The quantity and quality of purified DNA samples were measured and evaluated using the NanoDrop 1000 UV/Vis Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). Also, the quality of DNA was analyzed by 1% (w/v) agarose gel electrophoresis in 0.5 × TBE with GelRed staining.
DNA Oligonucleotides
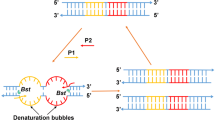
The scheme of the LAMP primers design for GM soybeans was shown in Fig. 1a, and a set of four primers containing two inner primers (FIP and BIP) and two outer primers (F3 and B3) were designed for each target sequence. FIP contains the F1c (complementary to F1) and the F2 sequence. BIP contains the B1c sequence (complementary to B1) and the B2 sequence. All of the LAMP primers used in this work were newly designed using the specific software of Primer Explorer V4 (http://primerexplorer.jp/elamp4.0.0/index.html). The conventional PCR primers (89788-F/R) were used according to the previous report (Liu et al. 2009). All primers used in this study were listed in Table 1 and synthesized by Invitrogen Co. Ltd. (Shanghai, China). The detailed locations of LAMP primers in target DNA sequence were shown in Fig. 1b.
Conventional PCR Assay
In conventional PCR, the reaction was carried out in a 30 μL volume, including 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.2 μM each primer, 1.5 U Taq DNA polymerase (TaKaRa biotechnology Co., Ltd, Dalian, China) and 5 μL template DNA. The conventional PCR was performed in PTC-100 Thermalcycler (MJ Research, Watertown, MA, USA) according to the following program: an initial denaturation at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 7 min. The PCR products were analyzed by 2% (w/v) agarose electrophoresis in 0.5 × TBE with GelRed staining.
LAMP Assay
LAMP assay was performed in a 25 μL total reaction mixture containing 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Triton X-100, 0.5 M betaine (Sigma), 1.2 μM each FIP and BIP, 0.2 μM each F3 and B3, 400 μM each dNTP. After adding 5 μL template DNA, the mixture was incubated for 5 min at 95 °C and cooled on ice, and then 8 U Bst DNA polymerase large fragment (New England Biolabs) were added. The mixture was incubated at 63 °C for 60 min in a conventional heating block. LAMP assay was carried out in triplicate for each template DNA, and the no template control (NTC) contained water instead of the template.
Analysis of LAMP Products
In general, 10 μL amplified products were analyzed by 2% agarose gel electrophoresis in 0.5 × TBE with GelRed staining after the LAMP amplification. Alternatively, the LAMP amplified products were directly observed by the naked eye through adding 2 μL 1,000 × SybrGreen I (Generay Biotech Co., Ltd., Shanghai) into the reaction mixture. The color of reaction mixture will turn green in the presence of LAMP-amplified products, while it remains orange in reaction mixture with no amplification. The sensitivities of gel electrophoresis analysis and visual observation by addition of SybrGreen I were compared using different LAMP products amplified from serially diluted soybean genomic DNA samples.
Results and Discussion
Design of the LAMP Systems for Detecting GM Soybeans
One ideal LAMP assay relies on the concurrence of the Bst DNA polymerase with strand displacement activity and a set of four specially designed primers capable of recognizing a total of six distinct regions on the target DNA (Notomi et al. 2000). LAMP primers of GTS 40-3-2 event were designed based on the event-specific sequence of 3′ end of exogenous integration (EMBL accession code AJ308515), recognizing a 214 bp target sequence (Windels et al. 2001). The MON89788 LAMP primers were designed based on our revealed event-specific sequence of 3′ end of exogenous integration to recognize a 203 bp target sequence (Liu et al. 2009). LAMP primers for the soybean endogenous reference gene were targeted at the Lectin sequence (EMBL accession code. K00821), targeting to a 217 bp target sequence. The detailed sequences of all LAMP primers used in this study are listed in Table 1.
Optimization of the LAMP Assays
In the present study, LAMP assays were performed employing heat-denatured soybean genomic DNA as the template with two inner primers and two outer primers. Previous study reported that LAMP assay could work well using non-denatured DNA samples as templates (Nagamine et al. 2001). However, we found that the amplified efficiency of LAMP assay using non-denatured soybean DNA as template was obviously lower than that using denatured soybean DNA (data not shown). Accordingly, the concentration of base destabilizer, betaine, was decreased to 0.5 M in each reaction. The optimal reaction temperature was determined by comparing the LAMP-amplified efficiencies under various temperatures ranging from 60 °C to 65 °C. In addition, different primer concentrations and the ratios between inner primers (FIP and BIP) and outer primers (F3 and B3) were also optimized. The LAMP assays for soybean Lectin endogenous reference gene, GTS 40-3-2 and MON89788 soybean events were established based on the optimized reaction conditions described in “Materials and Method” section.
Evaluation of the Reaction Efficiency of LAMP Assays
Previously, it has been reported that the LAMP reaction can be accelerated by using two loop primers (Nagamine et al. 2002). However, in the present study, only two inner primers (FIP and BIP) and two outer primers (F3 and B3) were used for amplification. In order to test the reaction efficiency of the developed LAMP assays, the different reaction times (10, 20, 30, 40, 50, and 60 min) were selected in GTS 40-3-2 and MON89788 LAMP assays. As shown in Fig. 2, the GTS 40-3-2 event specific LAMP products could be observed after 30-min amplification with 10 ng DNA template or 40-min amplification with 1 ng DNA template. Also, similar results were obtained in MON89788 event-specific LAMP assay (data not shown). These results indicated that the established LAMP assays have high reaction efficiency without the loop primers, suggesting that these assays are sufficient for rapid amplification of target DNAs.
Determination of the amplified efficiency of LAMP assays. LAMP products were detected on 2% agarose gel after 10 min (lanes 1 and 2), 20 min (lanes 3 and 4), 30 min (lanes 5 and 6), 40 min (lanes 7 and 8), 50 min (lanes 9 and 10), 60 min (lanes 11 and 12); lane M DL2000 marker. a The products were detectable after 40 min when the total template was 1 ng. b The products were detectable after 30 min when the total template was 10 ng
Specificity Test
To evaluate the specificity of the developed LAMP assays (GTS 40-3-2, MON89788 and Lectin), several GM events (GTS 40-3-2 and MON89788 soybean, MON810 maize, Huafan No. 1 tomato, GT73 canola, and MON531 cotton) and non-GM plants (pepper, soybean, potato, wheat, and rice) were used. In the specificity test, 10 ng total corresponding plant genomic DNA was used as the template in each LAMP assay. As expected, in the Lectin assay, the typical ladder-like pattern products and green color of the reactions were only obtained in the tests using soybean genomic DNA samples as templates, no amplified products and orange color were observed in other GM and non-GM crops as well as the NTC (Fig. 3a–b). In the GTS 40-3-2 assay, the typical ladder-like pattern products (Fig. 4a) and green color (data not shown) were obtained only in the reaction using GTS 40-3-2 soybean genomic DNA as template. In the MON89788 LAMP assay, the similar result was observed (Fig. 5a). In addition, we found that all the results obtained from gel electrophoresis and visual observation by addition of SybrGreen I was consistent in the three LAMP assays. These data confirmed that the developed LAMP assays have high specificity for amplifying the target DNAs.
Lectin LAMP assay. a, b Specificity test of LAMP assay by agarose gel electrophoresis analysis and visual observation using SybrGreen I. Lane 1 no template control (NTC); lanes 2–11 GTS 40-3-2, MON89788, MON810, Huafan No. 1, GT73, MON531, non-GM soybean, pepper, wheat, and rice; lane M DL2000 marker. c, d Sensitivity test of LAMP products by agarose gel electrophoresis analysis and visual observation using SybrGreen I. Lane 1 NTC; lanes 2–9 correspond to 100, 10, 1, 0.1, 0.01, 0.005, 0.001, and 0.0005 ng; lane M DL2000 marker
GTS 40-3-2 LAMP assay. a Specificity test of the LAMP assay. Lane 1 NTC; lanes 2–11 GTS 40-3-2, MON89788, MON810, Huafan No. 1, GT73, MON531, non-GM soybean, pepper, wheat, and rice; lane M DL2000 marker. b, c Sensitivity test of LAMP products by agarose gel electrophoresis analysis and visual observation using SybrGreen I. Lanes 1–8 mixed GM GTS 40-3-2 soybean samples with GM contents of 10%, 5%, 1%, 0.1%, 0.05%, 0.01%, 0.005%, and 0%; lane 9 NTC; lane M DL2000 marker
MON89788 LAMP assay. a Specificity test of the LAMP assay. Lane 1 NTC; lanes 2–11 MON89788, GTS 40-3-2, MON810, Huafan No. 1, GT73, MON531, non-GM soybean, pepper, wheat, and rice; lane M DL2000 marker. b, c Sensitivity test of LAMP products by agarose gel electrophoresis and visual observation using SybrGreen I. Lane 1 NTC; lanes 2–9 mixed GM MON89788 soybean samples with GM contents of 10%, 5%, 1%, 0.1%, 0.05%, 0.01%, 0.005%, and 0%; lane M DL2000 marker. d Sensitivity test of MON89788 conventional PCR. PCR products were amplified from MON89788 soybean DNAs with different concentrations. Lanes 1–7, 5%, 1%, 0.1%, 0.05%, 0.01%, 0.005%, and 0%; lane 8 NTC; lane M DL2000 marker
Limit of Detection
For GMOs detection, the high sensitivity is important and necessary because the degradation of low quantity DNA derived from GMOs often occurs in practical detection. To test the LOD of Lectin LAMP assay, non-GM soybean genomic DNA was serially diluted with 0.1 × TE buffer to final concentrations of 20, 2, 0.2, 0.02, 0.002, 0.001, 0.0002, and 0.0001 ng/μL. Five microliter diluted DNA sample was used as template in each reaction. As shown in Fig. 3c–d, the amplified LAMP products and green color were observed in all the dilutions except for the levels of 0.0002 and 0.0001 ng/μL. Thus, the LOD of Lectin assay was as low as 5 pg, corresponding to about four copies soybean haploid genomic DNA according to the soybean genome size (Andersen et al. 2006).
For testing the LODs of GTS 40-3-2 and MON89788 LAMP assays, the tested samples were prepared by mixing the relevant GM soybean event with non-GM soybean at various levels, such as 10%, 5%, 1%, 0.1%, 0.05%, 0.01%, 0.005%, and 0% (w/w). In each reaction, a total of 100 ng soybean genomic DNA was amplified. As shown in Figs. 4b–c and 5b–c, the amplified DNA fragments and the green colors were observed from all the tested levels except for the 0% level in both GTS 40-3-2 and MON89788 LAMP assays. These results indicated that the LODs of both GTS 40-3-2 and MON89788 LAMP assays were 0.005%, corresponding to about 4 copies of haploid soybean genomic DNA. In addition, we also found that the analyzed results from visual observation by addition of SybrGreen I and gel electrophoresis analysis were quite consistent, and there were no more difference between the visual observation and gel electrophoresis analysis. This indicated that the visual observation could be used for LAMP analysis instead of the general agarose gel electrophoresis.
In conventional PCRs, the LODs of Lectin and GTS 40-3-2 were reported with the value of 40 copies (Rott et al. 2004), and the LOD of MON89788 was tested as low as eight copies of haploid soybean genomic DNA (Fig. 5d), respectively. Compared with the LODs of conventional PCR assays, we believed that the developed LAMP assay is more sensitive and could be feasibly used for detection of GTS 40-3-2 and MON89788 soybeans.
Conclusion
In this study, we developed the visual and rapid detection methods for GM soybean MON89788 and GTS 40-3-2 events using the optimized LAMP technique. Compared to the classical PCR analysis, the LAMP assay has the advantages such time saving and low cost or simple procedures for result analysis. LAMP assay can be performed under isothermal conditions (60-65 °C) without PCR thermal cyclers within shorter time (40 min in this study). In combination with SybrGreen I dye, the amplified products can be directly detected by naked eye instead of the conventional gel electrophoresis analysis or fluorescent detection...In addition, the LODs of developed MON89788 and GTS 40-3-2 LAMP assays with the values of four copies are more sensitive than conventional PCR methods. High specificity and sensitivity, cost-effectiveness, and low requirement of equipment of the developed visual LAMP assays allow this method to be useful in GM soybean samples analysis, especially on spot detection combined with the fast DNA extraction method.
References
Andersen CB, Holst-Jensen A, Berdal KG, Thorstensen T, Tengs T (2006) Equal performance of TaqMan, MGB, molecular beacon, and SYBR green-based detection assays in detection and quantification of roundup ready soybean. J Agric Food Chem 54:9658–9663
Bellocchi G, Acutis M, Paoletti C, Confalonieri R, Trevisiol P, Grazioli E, Delobe Ch, Savini C, Mazzara M, Van den Eede G (2008) Expanding horizons in the validation of GMO analytical methods: fuzzy-based expert systems. Food Analytical Methods 1:126–135
Berdal KG, Holst-Jensen A (2001) Roundup Ready soybean event specific real-time quantitative PCR assay and estimation of the practical detection and quantification limits in GMO analyses. Eur Food Res Technol 213:432–438
David L, La Mura M, Allnutt TR, Powell W (2009) Detection of genetically modified organisms (GMOs) using isothermal amplification of target DNA sequences. BMC Biotechnology 9:7
Enosawa M, Kageyama S, Sawai K, Watanabe K, Notomi T, Onoe S, Mori Y, Yokomizo Y (2003) Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol 41:4359–4365
Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T (2007) Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol 45:2521–2528
Hernández M, Rodríguez-Lázaro D, Ferrando A (2005) Current methodology for detection, identification and quantification of genetically modified organisms. Current Anal Chem 1:203–221
Holst-Jensen A, Rønning SB, Løvseth A, Berdal KG (2003) PCR technology for screening and quantification of genetically modified organisms (GMOs). Anal Bioanal Chem 375:985–993
Huang C, Pan T (2005) Event-specific real-time detection and quantification of genetically modified roundup ready soybean. J Agric Food Chem 53:3833–3839
Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, Van Tu P, Tien NT, Tashiro M, Odagiri T (2007) Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J Virol Methods 141:173–180
Iwamoto T, Sonobe T, Hayashi K (2003) Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol 41:2616–2622
James C (2008) Global status of Commercialized Biotech/GM Crops: 2008. ISAAA Briefs 39
Liu J, Yang L, Guo J, Zhang D (2009) Development and in-house validation of the event-specific PCR detection methods for genetically modified soybean MON89788 based on the cloned integration flanking sequence. J Agric Food Chem 57:10524–10530
Marmiroli N, Maestri E, Gullì M, Malcevschi A, Peano C, Bordoni R, De Bellis G (2008) Methods for detection of GMOs in food and feed. Anal Bioanal Chem 392:369–384
Misawa Y, Yoshida A, Saito R, Yoshida H, Okuzumi K, Ito N, Okada M, Moriya K, Koike K (2007) Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J Infect Chemother 13:134–140
Mori Y, Nagamine K, Tomita N, Notomi T (2001) Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun 289:150–154
Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T (2001) Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem 47:1742–1743
Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 16:223–229
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:e63
Poon LL, Leung CS, Chan KH, Lee JH, Yuen KY, Guan Y, Peiris JS (2005) Detection of human influenza A viruses by loop-mediated isothermal amplification. J Clin Microbiol 43:427–430
Rodríguez-Lázaro D, Lombard B, Smith H, Rzezutka A, D'Agostino M, Helmuth R, Schroeter A, Malorny B, Miko A, Guerra B, Davison J, Kobilinsky A, Hernández M, Bertheau Y, Cook N (2007) Trends in analytical methodology in food safety and quality: monitoring microorganisms and genetically modified organisms. Trends Food Sci Technol 18:306–319
Rott ME, Lawrence TS, Wall EM, Green MJ (2004) Detection and quantification of roundup ready soy in foods by conventional and real-time polymerase chain reaction. J Agric Food Chem 52:5223–5232
Singh CK, Ojha A, Bhatanagar RK, Kachru DN (2008) Detection and characterization of recombinant DNA expressing vip3A-type insecticidal gene in GMOs–standard single, multiplex and construct-specific PCR assays. Anal Bioanal Chem 390:377–387
Taverniers I, Windels P, Vaïtilingom M, Milcamps A, Van Bockstaele E, Van den Eede G, De Loose M (2005) De Loose, M. Event-specific plasmid standards and real-time PCR methods for transgenic Bt11, Bt176, and GA21 maize and transgenic GT73 canola. J Agric Food Chem 53:3041–3052
Windels P, Taverniers I, Depicker A, Van Bockstaele E, De Loose M (2001) Characterisation of the roundup ready soybean insert. Eur Food Res Technol 213:107–112
Yang L, Guo J, Zhang H, Liu J, Zhang D (2008) Qualitative and quantitative event-specific PCR detection methods for oxy-235 canola based on the 3′ integration flanking sequence. J Agric Food Chem 56:1804–1809
Zel J, Mazzara M, Savini C, Cordeil S, Camloh M, Stebih D, Cankar K, Gruden J, Morisset D, Van den Eede G (2008) Method validation and quality management in the flexible scope of accreditation: an example of laboratories testing for genetically modified organisms. Food Anal Methods 1:61–72
Acknowledgments
This work was supported by the National Key Basic Research Program (2007CB109201, 2007FY230100), the National Transgenic Plant Special Fund (2008ZX08012-002, 003,005, and 2009ZX08012-002B), the National Natural Science Foundation of China (30725022, 30700499), the national high-tech project “863” (2006AA10Z443), and Shanghai Chenguang project (2008CG16).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiaoyan Guan and Jinchao Guo contributed equally to this work.
Rights and permissions
About this article
Cite this article
Guan, X., Guo, J., Shen, P. et al. Visual and Rapid Detection of Two Genetically Modified Soybean Events Using Loop-mediated Isothermal Amplification Method. Food Anal. Methods 3, 313–320 (2010). https://doi.org/10.1007/s12161-010-9132-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-010-9132-x