Abstract
Objective
6-[18F]Fluoro-l-DOPA (FDOPA) is a radiopharmaceutical valuable for assessing the presynaptic dopaminergic function when used with positron emission tomography (PET). More specifically, the striatal-to-occipital ratio (SOR) of FDOPA uptake images has been extensively used as a quantitative parameter in these PET studies. Our aim was to develop an easy, automated method capable of performing objective analysis of SOR in FDOPA PET images of Parkinson’s disease (PD) patients.
Methods
Brain images from FDOPA PET studies of 21 patients with PD and 6 healthy subjects were included in our automated striatal analyses. Images of each individual were spatially normalized into an FDOPA template. Subsequently, the image slice with the highest level of basal ganglia activity was chosen among the series of normalized images. Also, the immediate preceding and following slices of the chosen image were then selected. Finally, the summation of these three images was used to quantify and calculate the SOR values. The results obtained by automated analysis were compared with manual analysis by a trained and experienced image processing technologist.
Results
The SOR values obtained from the automated analysis had a good agreement and high correlation with manual analysis. The differences in caudate, putamen, and striatum were −0.023, −0.029, and −0.025, respectively; correlation coefficients 0.961, 0.957, and 0.972, respectively.
Conclusions
We have successfully developed a method for automated striatal uptake analysis of FDOPA PET images. There was no significant difference between the SOR values obtained from this method and using manual analysis. Yet it is an unbiased time-saving and cost-effective program and easy to implement on a personal computer.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disorder characterized by presynaptic nigrostriatal dopamine dysfunction. Clinical symptoms include rigidity, bradykinesia and resting tremors, and losses of dopamine in the striatum in PD have been shown by postmortem and imaging studies [1, 2]. For noninvasive imaging assessment of the presynaptic dopaminergic function, 6-[18F]Fluoro-l-DOPA (FDOPA) is a valuable radiopharmaceutical when used with positron emission tomography (PET). The fall in striatal uptake of FDOPA had been visualized in PD patients undergoing PET scans [2–5]. For this purpose, a variety of methods have been developed to analyze FDOPA PET images aiming at correlating the FDOPA uptake with the severity of PD.
In most of the methods, the striatal-to-occipital ratio (SOR) is extensively used as the quantitative parameter [6–12]. SOR offers a practical advantage because it is determined by the static data acquisition making it the easiest to apply in clinical settings, quantifying nigrostriatal dopamine function in PD and related disorders without the need of blood sampling [6, 9, 12]. SOR is mostly computed using regions of interest (ROI) manually drawn over these structures on individual images obtained from PET study or on a coregistered image of magnetic resonance imaging (MRI) for better positioning [8, 12, 13]. However, manual ROI delineation is a tedious, time-consuming, and highly operator-dependent procedure. In addition, the inter- and intra-operator variability may hamper the reproducibility and accuracy of results.
In this study, to avoid the shortcomings caused by the manual ROI delineation, we presented an easy, objective and automated method based on the concept of spatial normalization [14–16]. This efficient method analyzed the striatal uptake of FDOPA in PD patients and calculated the SOR. In addition, the results were compared with those of conventional manual analysis performed by a trained, experienced image processing technologist.
Materials and methods
Subjects
Twenty-one patients (15 men, 6 women) diagnosed clinically with PD and 6 healthy control subjects (3 men, 3 women) free of neurologic diseases were included in this retrospective study analysis. All PD patients were scored with the Hoehn and Yahr (H&Y) scale. One patient was at H&Y stage 1, three at stage 2, eight at stage 2.5, seven at stage 3, one at stage 4, and one at stage 5. The mean H&Y scale was 2.7 ± 0.8. Each subject underwent an FDOPA PET and an MRI brain scan. Informed consent was obtained from both the healthy control subjects and patients prior to the examination.
FDOPA PET and MRI brain scan
All subjects fasted for at least 6 h prior to FDOPA administration. All antiparkinsonian medications were stopped at least 12 h before the PET studies and subjects were given 100 mg of carbidopa 1 h before FDOPA administration. FDOPA was produced in our GMP facility as described elsewhere [17]. Static three-dimensional acquisition was performed 2 h after the intravenous injection of 185 MBq FDOPA with a GE Discovery ST PET/CT scanner (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK) for 30 min. Computed tomography (CT) scan was performed immediately prior to the PET scan with a multi-detector (sixteen-slice) spiral CT scanner for attenuation correction. PET image data sets were reconstructed using ordered-subset expectation maximization (OSEM) with a matrix size of 128 × 128, a pixel size of 1.95 mm × 1.95 mm and a slice thickness of 3.27 mm.
MRI brain scan was performed on a GE Signa Excite 1.5-Tesla MRI scanner, using a 3D-FSPGR T1 sequence (repetition time = 12.14 ms, echo time = 5.31 ms) with a matrix size of 512 × 512, pixel size of 0.39 mm × 0.39 mm and slice thickness of 2 mm.
Creation of FDOPA template
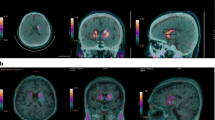
FDOPA template was built by averaging 6 PET scans from healthy control subjects [18, 19]. First, PET images of each control subject were registered to their corresponding MRI using the normalized mutual information algorithm [20, 21]. Thereafter, the MR images of the control subjects were spatially normalized to the T1-weighted MRI template provided within the statistical parametric mapping (SPM) software package (Wellcome Department of Cognitive Neurology, London, UK), and the resulting transformation parameters were applied to the PET scans. Finally, the FDOPA template was built by averaging these normalized PET images and applying a smoothing Gaussian filter with 8 mm FWHM (Fig. 1). All processing methods were performed using SPM2 under MATLAB (The Mathworks, Inc., Natick, Massachusetts, USA). The spatial normalization algorithm provided by the SPM2 software package using its default options: 7 × 9 × 7 discrete cosine transform basis functions, 16 nonlinear iterations and medium nonlinear regularization. Trilinear interpolation was used in all normalizations. The generated FDOPA template matched the standard MNI (Montreal Neurological Institute) space [22] and was capable of being a recognized neuroanatomical model.
FDOPA template (a) created from PET images of 6 healthy subjects and to which a smoothing filter was applied (b). SSmask used for finding the image with the highest level of basal ganglia activity among a series of PET images (c). ROImask on the caudate, putamen, and occipital regions (d). Displaying the un-smooth FDOPA template with SSmask (e) and ROImask (f) superimposed
Automated striatal analysis
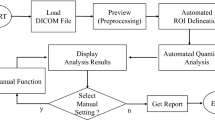
The proposed striatal uptake analysis program was designed with MATLAB programming language and ran on a computer with Windows operating system. The program spatially normalized the subject’s FDOPA PET images into a standard space defined by template images. Subsequently, the images normalized in this way were quantified based on a select-slice mask (SSmask) and ROI mask (ROImask) automatically. The SSmask and ROImask (Fig. 1) were pre-defined according to FDOPA template images. Figure 2 shows an example of our fully automated analysis. The complete execution steps are described as follows:
Step 1
FDOPA PET brain images of each individual subject were spatially normalized into the FDOPA template made earlier, allowing them to have a common coordinate. Following an algorithm developed by others [14–16], this spatial normalization used 6 × 8 × 6 discrete cosine transform basis functions, 16 nonlinear iterations and medium nonlinear regularization. Transformed images were written with a voxel size of 2 mm × 2 mm × 2 mm using trilinear interpolation.
Step 2
Among the series of FDOPA PET brain images that had been spatially normalized, we calculated counts within SSmask of all image slices and identified the one with the highest level of basal ganglia activity. Next, we selected the preceding and following slices (e.g., selecting one that is close to the skull base and one that is close to the vertex) of the above-found image. The summation of these three images was used to quantify and calculate the SOR values.
Step 3
After summing these three images, the SOR values were calculated via ROImask. The ROImask included the caudate (79 pixels), putamen (152 pixels) and occipital cortex (128 pixels) on both sides of the brain, among which the occipital cortices served as the background. The SOR values in the caudate and putamen were calculated by dividing the mean counts per pixel in the caudate or putamen with the mean counts per pixel in the occipital cortex. The average values of the caudate and putamen were then used to calculate the mean value of the whole striatal binding. The SOR values in the caudate, putamen, and striatum mentioned here referred to the average of both sides of the brain.
Manual striatal analysis
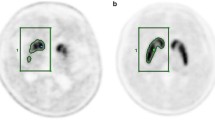
To validate this automated technique, each subject was also evaluated using a conventional manual method as established elsewhere [12] on a GE Xeleris 2 Workstation by a trained, experienced image processing technologist. The technologist was blinded to the results of the automated analysis while performing this manual operation. The MRI scans were registered to the FDOPA PET images and resliced according to the PET images. The ROI were drawn on the MR images and then copied to the ones from PET. The fusion of PET and MR images was further used to verify the correctness of ROI placement (Fig. 3). The ROI was located bilaterally at the caudate and putamen in two slices showing the highest basal ganglia activity. The occipital cortices were also drawn in the same way and served as background. In each slice, the SOR values were calculated using the same way as the automated striatal analytic method. Finally, the average values in two planes were calculated.
Statistical analysis
All data were expressed as mean ± standard deviation (SD) in this study. The Shapiro–Wilk test was used to verify the normality of the data sets in the studied group. The paired t test was used to compare the difference in the SOR values between the manual and the currently being developed automated analysis. If data sets were not normally distributed, the Wilcoxon signed ranks test was used. The Pearson correlation coefficient (r) was calculated to identify the relationship between the SOR values from the manual and the automated analysis. Similarly, if data sets were not normally distributed, the Spearman’s rho correlation (ρ) test would be used. Finally, the statistical method proposed by Bland and Altman [23] was employed to compare these two techniques of measurement. In this graphical method the differences between measurements were plotted against their average derived from the two techniques. Horizontal lines were drawn at 0 (solid line), mean difference (dashed middle line), and the 95% limits of agreement (dashed lines), which are defined as within 1.96 times the standard deviation of the differences (Fig. 6). p values less than 0.05 were considered statistically significant. All statistical analyses were performed using SPSS software (SPSS Inc., Chicago, Illinois, USA).
Results
Table 1 summarizes the SOR values obtained using automated and manual striatal analyses in different subject groups. Figure 4 shows results comparing SOR values in all subjects between the two methods. No significant differences were found in the group of PD subjects (p = 0.147 for caudate, 0.131 for putamen, and 0.067 for striatum) or in all subjects (p = 0.257 for caudate, 0.084 for putamen, and 0.161 for striatum).
From these same SOR values the correlations between automated and manual analyses were calculated and showed good results (Fig. 5). In PD patients, the correlation coefficients were r = 0.943 for caudate, 0.940 for putamen, and 0.960 for striatum; and in all subjects, r = 0.961 for caudate, ρ = 0.957 for putamen and ρ = 0.972 for striatum. All these were statistically significant with p values all less than 0.001.
Figure 6 shows the Bland–Altman plot between the SOR values determined by the manual and the automated striatal analyses for caudate, putamen and striatum in all subjects. The differences (automated minus manual) of the three variables are −0.023, −0.029 and −0.025, respectively. These plots show good agreement among SOR values computed from the manual and the automated analyses. The 95% limits of agreement of the three plots for the manual analysis and the automated analysis measurements were not significantly different.
Discussion
In this study, we successfully developed a software program based on MATLAB programming language for automated striatal uptake analysis of SOR in FDOPA PET brain images. Based on spatial normalization and mask technology, this simple, time-saving technique positioned striatum of the brain automatically and avoided operator-dependent region-of-interest (ROI) drawings. The SOR values of 27 subjects obtained by this fully automated analysis highly correlated with the conventional manual analysis results performed by a trained, experienced image processing technologist.
Conventionally, the SOR is computed in the first step by manual positioning of ROI over certain regions where PET brain image reveals radiopharmaceutical accumulation. Tedious and time-consuming as it is, the manual ROI delineation often results in intra- and inter-operator variations. The automated method developed by us determined ROI automatically and, thus, is operator-independent and has no people-related drawbacks and limitations. This program can easily be run on a personal computer. In our case, it ran on Microsoft Windows XP operation system with Pentium M Centrino 1.86 GHz CPU and 1 GB RAM and it took only about 30 s for the analysis of each patient. In addition, though manual ROI outlining based on a coregistered MRI image could be used to better define the striatum regions, it is not practical to conduct MRI for all patients. On the contrary, our fully automated method requires only MRI images of normal control subjects to create a one-time FDOPA template beforehand and use it for all patients. Consequently, this method for striatal uptake analysis of the FDOPA PET brain images is very efficient and widely applicable.
In some SPECT studies such as the ones using 123I-IBZM to visualize D2 receptor, investigations were based on only SPECT images for template creation and the images of each individual subject were registered to that template [24–27]. Here, we used MRI-aided method to create FDOPA template in a PET study. The MRI belongs to structural images that have much higher spatial resolution, less noise and better anatomical information allowing a more accurate registration than the one performed using only functional images [16, 19]. Even with similar spatial normalization technique as ours, their study did not further differentiate the head of caudate from putamen [26]. Perhaps their SPECT-based template offered insufficient resolution to achieve that task. In our study, we had divided ROI of the striatum into caudate and putamen to derive information of the SOR in greater detail. Another difference from other automated approaches is that our new method located the maximum activity of basal ganglia within the brain volume based on SSmask. In contrast to other techniques [27, 28], this was rather simple to implement.
To further evaluate the reliability of the automated technique, the results obtained from it were subjected to a comparison with a trustworthy manual analysis. For this purpose, we manually drew ROI based on the coregistered MRI images of each patient. Subsequently, the two consecutive slices (total thickness of 6.54 mm) with the highest basal ganglia activity were selected. As a note, the original FDOPA PET brain images were spatially normalized resulting in a slice thickness of 2 mm, and therefore the three slices selected for automated analysis had a total thickness of 6 mm. Therefore, the summation of these three slices best represented the striatal activity as in the two slices selected in the manual analysis. Regardless, labor-intensive as it might be, we believed this manual operation provided objective yet very accurate results for the purpose of evaluating the new automated technique.
Worth mentioning also includes the number of subjects necessary for template creation. In our study, FDOPA template was created by averaging PET scans of six healthy subjects and the result was satisfactory. Undoubtedly, a better template could be achieved using the average of a greater number of healthy subjects. Nonetheless, when considering many practical issues including not least the cost, one would rather use fewer healthy subjects to create an acceptable template. For that reason the most suitable number of scans to create a template deserves further investigation. At the moment, there is no consensus on the minimum number of scans for the creation of a template though there is no argument about its importance.
There is also a potential in other imaging studies generally employing SOR for presynaptic or postsynaptic dopaminergic neuronal functions that a respective radiopharmaceutical-matched template for spatial normalization could be created with our automated method. These might include 123I-FP-CIT, 99mTc-TRODAT, 11C-raclopride, and 123I-IBZM, in PET or SPECT brain studies [29] for the assessment of various striatal statuses. Actually, the image contrast in these various dopaminergic PET or SPECT studies of different radiotracers and our FDOPA template were reasonably similar. One can imagine the possibility that our FDOPA template might directly apply to those scans.
As a general consideration, there are cautions to be taken when SOR is engaged in the measurement of dopaminergic functions. SOR is a semiquantitative measure of radioactivity concentration and, thus, its value may not stay constant in changed biologic conditions or under different technical factors. Although there is so far no systematic study on which factor and in what degree might influence SOR values, one should keep this situation in mind when comparing results between FDOPA PET studies from different imaging centers. In fact, further exploration into the influence of these factors on our presented automated analysis is warranted. One familiar example regarding semiquantitative measurement is the popularly utilized “standardized uptake value” (SUV) method in 18F-fludeoxyglucose (FDG) PET studies where SUV is mainly for the evaluation of the early response to cancer treatment. Both SOR and SUV are based essentially on the radioactivity concentration within selected ROI. Studies have confirmed that many factors affect SUV measurements, including biologic factors, such as patient size, uptake time of the radiotracer, and technologic factors, such as scanner variability, dose of radiopharmaceuticals, image acquisition, and reconstruction parameters [30].
Finally, the correlation of the analysis results and the clinical severity of the disease also need to be established in the future for the differential diagnosis in a larger patient population. This is especially important for the method’s applicability in very severe PD patients. Most PD patients referred to nuclear medicine for FDOPA PET scans and thus included in the present study were at H&Y stage 2–3. Consequently, the method’s reliability might not be as great in very severe PD subjects whose FDOPA uptakes in the relevant regions were rather low.
Conclusion
In this study, a fully automated striatal uptake analysis was developed for FDOPA PET images. The SOR values obtained from the automated method showed a good agreement (the differences between the two methods of caudate, putamen, and striatum were −0.023, −0.029, and −0.025, respectively) and high correlation with manual analysis (correlation coefficients of caudate, putamen, and striatum were 0.961, 0.957, and 0.972, respectively) by a trained and experienced image processing technologist. It is a time-saving and cost-effective program yet remains objective and is easy to implement on a personal computer. We expect this automated striatal uptake analytic program will help nuclear medicine physicians in the interpretation of FDOPA PET brain images for the diagnosis of movement disorder patients.
References
Kish SJ, Shannak K, Hornykiewicz O. Uneven pattern of dopamine loss in the striatum of patients with idiopathic Parkinson’s disease: pathophysiologic and clinical implications. N Engl J Med. 1988;318:876–80.
Garnett ES, Firnau G, Nahmias C. Dopamine visualized in the basal ganglia of living man. Nature. 1983;305:137–8.
Nahmias C, Garnett ES, Firnau G, Lang A. Striatal dopamine distribution in parkinsonian patients during life. J Neurol Sci. 1985;69:223–30.
Leenders KL, Palmer AJ, Quinn N, Clark JC, Firnau G, Garnett ES, et al. Brain dopamine metabolism in patients with Parkinson’s disease measured with positron emission tomography. J Neurol Neurosurg Psychiatry. 1986;49:853–60.
Morrish PK, Sawle GV, Brooks DJ. An [18F]dopa-PET and clinical study of the rate of progression in Parkinson’s disease. Brain. 1996;119:585–91.
Takikawa S, Dhawan V, Chaly T, Robeson W, Dahl R, Zanzi I, et al. Input functions for 6-[fluorine-18]fluorodopa quantitation in parkinsonism: comparative studies and clinical correlations. J Nucl Med. 1994;35:955–63.
Ishikawa T, Dhawan V, Chaly T, Margouleff C, Robeson W, Dahl JR, et al. Clinical significance of striatal DOPA decarboxylase activity in Parkinson’s disease. J Nucl Med. 1996;37:216–22.
Ishikawa T, Dhawan V, Kazumata K, Chaly T, Mandel F, Neumeyer J, et al. Comparative nigrostriatal dopaminergic imaging with iodine-123-βCIT-FP/SPECT and fluorine-18-FDOPA/PET. J Nucl Med. 1996;37:1760–5.
Dhawan V, Ma Y, Pillai V, Spetsieris P, Chaly T, Belakhlef A, et al. Comparative analysis of striatal FDOPA uptake in Parkinson’s disease: ratio method versus graphical approach. J Nucl Med. 2002;43:1324–30.
Eshuis SA, Maguire RP, Leenders KL, Jonkman S, Jager PL. Comparison of FP-CIT SPECT with F-DOPA PET in patients with de novo and advanced Parkinson’s disease. Eur J Nucl Med Mol Imaging. 2006;33:200–9.
Eshuis SA, Jager PL, Maguire RP, Jonkman S, Dierckx RA, Leenders KL. Direct comparison of FP-CIT SPECT and F-DOPA PET in patients with Parkinson’s disease and healthy controls. Eur J Nucl Med Mol Imaging. 2009;36:454–62.
Jokinen P, Helenius H, Rauhala E, Brück A, Eskola O, Rinne JO. Simple ratio analysis of 18F-fluorodopa uptake in striatal subregions separates patients with early Parkinson disease from healthy controls. J Nucl Med. 2009;50:893–9.
Huang WS, Chiang YH, Lin JC, Chou YH, Cheng CY, Liu RS. Crossover study of 99mTc-TRODAT-1 SPECT and 18F-FDOPA PET in Parkinson’s disease. J Nucl Med. 2003;44:999–1005.
Friston KJ, Ashburner J, Frith CD, Poline J-B, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Hum Brain Mapp. 1995;2:165–89.
Ashburner J, Neelin P, Collins DL, Evans A, Friston KJ. Incorporating prior knowledge into image registration. Neuroimage. 1997;6:344–52.
Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–66.
Kao CH, Hsu WL, Xie HL, Lin MC, Lan WC, Chao HY. GMP production of [18F]FDOPA and issues concerning its quality analyses as in USP “Fluorodopa F 18 injection”. Ann Nucl Med. 2011;25:309–16.
Meyer JH, Gunn RN, Myers R, Grasby PM. Assessment of spatial normalization of PET ligand images using ligand-specific templates. Neuroimage. 1999;9:545–53.
Gispert JD, Pascau J, Reig S, Martínez-Lázaro R, Molina V, García-Barreno P, et al. Influence of the normalization template on the outcome of statistical parametric mapping of PET scans. Neuroimage. 2003;19:601–12.
Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Automated multi-modality image registration based on information theory. In: Bizais Y, Barillot C, Di Paolo R, editors. Information processing in medical imaging. Dordrecht: Kluwer Academic Publishers; 1995. p. 263–74.
Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognit. 1999;32:71–86.
Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3D statistical neuroanatomical models from 305 MRI volumes. In: Proceedings of the IEEE Nuclear Science Symposium and Medical Imaging Conference; 1993. p. 1813–7.
Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10.
Habraken JB, Booij J, Slomka P, Sokole EB, van Royen EA. Quantification and visualization of defects of the functional dopaminergic system using an automatic algorithm. J Nucl Med. 1999;40:1091–7.
Radau PE, Linke R, Slomka PJ, Tatsch K. Optimization of automated quantification of 123I-IBZM uptake in the striatum applied to parkinsonism. J Nucl Med. 2000;41:220–7.
Buchert R, Berding G, Wilke F, Martin B, von Borczyskowski D, Mester J, et al. IBZM tool: a fully automated expert system for the evaluation of IBZM SPECT studies. Eur J Nucl Med Mol Imaging. 2006;33:1073–83.
Zubal IG, Early M, Yuan O, Jennings D, Marek K, Seibyl JP. Optimized, automated striatal uptake analysis applied to SPECT brain scans of Parkinson’s disease patients. J Nucl Med. 2007;48:857–64.
Mirzaei S, Zakavi R, Rodrigues M, Schwarzgruber T, Brücke T, Bakala J, et al. Fully automated 3D basal ganglia activity measurement in dopamine transporter scintigraphy (Spectalyzer). Ann Nucl Med. 2010;24:295–300.
Seibyl JP. Single-photon emission computed tomography and positron emission tomography evaluations of patients with central motor disorders. Semin Nucl Med. 2008;38:274–86.
Adams MC, Turkington TG, Wilson JM, Wong TZ. A systematic review of the factors affecting accuracy of SUV measurements. AJR Am J Roentgenol. 2010;195:310–20.
Acknowledgments
The authors thank Dr. Shin-Yuan Chen and Ms. Li-Chuan Huang for providing the clinical information of PD patients and the MR imaging related parameters, respectively. This study was supported by a grant TCRD 99-27 from the Buddhist Tzu Chi General Hospital.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chang, IC., Lue, KH., Hsieh, HJ. et al. Automated striatal uptake analysis of 18F-FDOPA PET images applied to Parkinson’s disease patients. Ann Nucl Med 25, 796–803 (2011). https://doi.org/10.1007/s12149-011-0533-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12149-011-0533-8