Abstract
Carcinoma ex pleomorphic adenoma (CA-ex-PA) may arise with nearly any histologic subtype of carcinoma of the salivary gland. In the absence of recognizable residual pleomorphic adenoma (PA) or a prior history of PA, distinction of CA-ex-PA from morphologically similar de novo carcinomas may be difficult. Oncogenic rearrangement of PLAG1 (pleomorphic adenoma gene 1) has been established in PA; however, it has not yet been proven that PLAG1 alteration persists in carcinomas developed from preceding PA. We evaluated 22 histologically diverse CA-ex-PA by immunohistochemistry for PLAG1, and/or by FISH targeting PLAG1. Of these, 17 cases were immunoreactive (1+ to 3+) and 5 were immunonegative/rare positive for PLAG1. For comparison, 39 various salivary gland neoplasms were immunostained for PLAG1, of which all scored negative/rare positive. Twelve of 19 CA-ex-PA analyzed by PLAG1 FISH (63 %) were positive for gene rearrangement, 2 showed only a trisomy/polysomy profile, and 5 had a normal pattern. One FISH-positive tumor showed amplification of PLAG1. One of 3 cases analyzed for HMGA2 FISH was positive for gene rearrangement. In our series, the majority of CA-ex-PA harbored altered PLAG1 or HMGA2 genes detectable by FISH. While PLAG1 immunostain was specific for CA-ex-PA against other carcinomas, its application as a standalone discriminatory test was limited by variable expression. We conclude that most CA-ex-PA, regardless of morphologic subtype, carry altered PLAG1 or HMGA2 genes, and that FISH for PLAG1, along with immunohistochemistry for PLAG1, may help discriminate CA-ex-PA from its de novo carcinoma counterpart.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The oncogenic effect of recurrent chromosomal translocations in leukemias and sarcomas has been well established over the past few decades. In contrast, in epithelial neoplasms, the role of fusion transcripts has only recently gained recognition. This difference may be attributable to the karyotypic complexity of many carcinomas and the failure of traditional cytogenetic techniques to find putative fusion genes amidst other genetic imbalances accumulated during tumor progression. Recently, recurrent chimeric genes have been described in a number of salivary gland malignancies, including mucoepidermoid, hyalinizing clear cell, and adenoid cystic carcinomas, adding to the small group of translocation-associated carcinomas known to date [1, 2]. Carcinoma ex pleomorphic adenoma (CA-ex-PA), however, is unique among both salivary gland tumors and other epithelial malignancies for its association with a benign preexisting lesion whose genetic characteristics have already been well described.
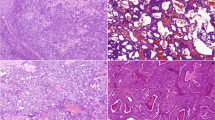
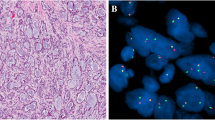
H&E microscopy, immunohistochemistry for PLAG1, and fluorescence in situ hybridization study for PLAG1 in 4 cases of carcinoma ex pleomorphic adenoma (CA-ex-PA) of the salivary gland. Low-grade invasive CA-ex-PA with histologic appearance of adenocarcinoma NOS (1-A), showing variable PLAG1 expression with strong intensity (1-B). FISH for PLAG1 shows one pair of split signals indicative of PLAG1 rearrangement with a balanced translocation pattern (1-C). Intermediate-grade myoepithelial CA-ex-PA with spindle cell morphology (2-A), showing diffuse expression of PLAG1 (2-B), and PLAG1 rearrangement with additional copies of red signal, corresponding to the 5′ end of PLAG1 (2-C). Intermediate-grade epithelial-myoepithelial cell CA-ex-PA (3-A), with a diffuse expression of PLAG1 (3-B). FISH demonstrates one or more copies of rearranged PLAG1 with an unbalanced translocation pattern and a polysomy profile (3-C). Intermediate-grade recurrent adenocarcinoma ex pleomorphic adenoma (4-A), showing strong immunoreactivity for PLAG1 (4-B). FISH for PLAG1 reveals a polysomy pattern with up to 8 copies of the intact gene (4-C)
H&E microscopy, immunohistochemistry for PLAG1, and FISH for PLAG1 in 2 cases of metastatic CA-ex-PA. High-grade metastatic adenocarcinoma ex pleomorphic adenoma to the lung (1-A), with weak nuclear expression of PLAG1 (1-B). FISH demonstrates 1–2 copies of rearranged PLAG1 and a trisomy profile (1-C). High-grade metastatic salivary duct CA-ex-PA to the liver (2-A) with nuclear expression of PLAG1 (2-B). FISH shows a polysomy pattern and multiple copies of rearranged PLAG1 (2-C)
H&E microscopy, immunohistochemistry for PLAG1, and FISH for HMGA2 in a high-grade primary CA-ex-PA comprising various histologic subtypes (adenocarcinoma, myoepithelial and salivary duct carcinomas) (A), with rare PLAG1 positive cells (B). FISH for PLAG1 had a normal pattern (not shown). FISH for HMGA2 demonstrates one pair of split signals reflective of gene alteration (C)
Conventional cytogenetic analysis has detected recurrent translocations in approximately 70 % of pleomorphic adenomas (PA) [3–6]. These translocations target primarily PLAG1 (pleomorphic adenoma gene 1), resulting in upregulation of this gene, located at 8q12, and overexpression of its protein (PLAG1) [7, 8]. The second, much less frequent target gene in PA is HMGA2, mapped to 12q14-15 [9, 10]. The pathogenic role of PLAG1, however, appears to span all PA, including tumors without cytogenetic evidence of PLAG1 alteration and those with HMGA2 rearrangement. This conclusion is drawn from immunohistochemical and Northern blot studies showing PLAG1 overexpression in nearly all PA, regardless of PLAG1 status [11–13].
Few CA-ex-PA have been evaluated for PLAG1 status [11, 12, 14–16]. Martins et al. [15] found PLAG1 rearrangement in 3 cases and trisomy 8 without gene alteration in 1 case among 4 selected CA-ex-PA that were known to have karyotypic deviations of chromosome 8 [15]. On the other hand, Matsuyama et al. [12] conducted RT-PCR assays for the classic fusion transcripts of PA in 4 CA-ex-PA and found none of them to harbor chimeric genes of interest [12]. Resolving this issue is relevant from both biologic and diagnostic standpoints. CA-ex-PA is histologically diverse with malignant components that can resemble distinct types of salivary gland carcinomas or variable admixtures of different tumor types including heterologous mesenchymal elements in some instances. At clinical presentation, the malignant component of CA-ex-PA often overruns the preexisting PA and the presence of a precursor PA may only be suggested by extensive areas of tumor hyalinization. Moreover, the histologic diversity of CA-ex-PA increases the likelihood of misclassification, both as other salivary and non-salivary malignancies, in small biopsy specimens. Therefore, defining a specific immunohistochemical or genetic marker for CA-ex-PA has practical diagnostic implications. With these thoughts, we conducted the present study with the following objectives: first, to determine the status of PLAG1 rearrangement and overexpression of PLAG1 protein in the carcinomatous component of PA, and second, to discern the validity of PLAG1 protein as a potential diagnostic marker for CA-ex-PA versus other carcinoma types.
Materials and Methods
Materials
This study was approved by the respective institutional review boards at both participating institutions. All tumors were defined per WHO criteria for salivary gland neoplasms [17]. Twenty-two histologically confirmed CA-ex-PA with sufficient material for the study were retrieved from the pathology archives at Brigham and Women’s Hospital. Of these, 20 cases were primary CA-ex-PA (including 4 non-invasive and 1 minimally invasive) and 2 cases were metastatic tumors. All study subjects had either a pre-existing PA adjacent to the carcinoma or a history of prior excision for a PA at the same location (see Table 1). Metastatic tumors were confirmed as being histologically similar to the corresponding CA-ex-PA at the presumed primary site.
For comparison, 39 cases of various benign and malignant salivary gland neoplasms among the following histologic categories were randomly selected: adenocarcinoma, NOS (4 cases), mucoepidermoid carcinoma (8 cases), adenoid cystic carcinoma (7 cases), salivary duct carcinoma (4 cases), epithelial-myoepithelial carcinoma (2 cases), polymorphous low-grade adenocarcinoma (1 cases), myoepithelial carcinoma (1 case), acinic cell carcinoma (1 case), basaloid carcinoma (1 case), pleomorphic adenoma (5 cases), basal cell adenoma (4 cases), and low-grade salivary gland neoplasm, NOS (1 case). In addition, tissue microarray sections of a diverse group of carcinomas, comprising ductal carcinoma of breast (46 samples), gastric adenocarcinoma (10), esophageal squamous carcinoma (9), lung adenocarcinoma (10), colorectal adenocarcinoma (10), papillary thyroid carcinoma (10), and renal cell carcinoma (9), were evaluated to assess PLAG1 immunoreactivity in other carcinoma types.
Immunohistochemical Study
Formalin-fixed, paraffin-embedded tissue blocks from 61 study and control cases were cut into 4 μm-thick sections and immunostained with a monoclonal PLAG1 antibody (clone 3B7, 1:50; Novus, Littleton, CO), using heat-induced epitope retrieval (in citrate buffer for 30 min) and the bond polymer refine detection system (Leica Microsystems, Bannockburn, IL) after 2 h of antibody incubation at room temperature. Positive controls were included in each run, using sections of a classic PA or a lipoblastoma. Immunostains were semiquantitatively scored as follows: No positive cells, “negative”; 1–4 % positive cells, “rare”; 5–25 % positive cells, “1+”; 26–50 % positive cells, “2+”; >50 % positive cells, “3+”. The intensity of staining was graded as weak, intermediate, and strong. Only the carcinomatous component was scored and graded for PLAG1 immunoreactivity in CA-ex-PA subjects.
Fluorescence In Situ Hybridization Study
Break-apart, dual-color FISH probes for PLAG1 were developed using bacterial artificial chromosome (BAC) probes including the RP11-22E14 clone, flanking the 3′ end, and the RP11-1130K23 clone, flanking the 5′ end of the PLAG1 gene. FISH assay for HMGA2 was developed using BAC probes to include the RP11-662G15 clone, flanking the 5′ end, and the RP11-1025D9 clone, flanking the 3′ end. BAC DNA was isolated using a modified Qiagen plasmid extraction protocol. The RP11-22E14 sequence was labeled with AlexaFluor 488 (green fluorochrome), and the RP11-1130K23 sequence with Rhodamine (red fluorochrome). The RP11-662G15 sequence was labeled with the green and the RP11-1025D9 sequence with the red fluorochrome. Dual-color FISH was performed on 4 micron-thick FFPE tissue sections, as previously described [18]. For CA-ex-PA cases, a section comprising the largest available carcinomatous component was selected. A normal control produced two joined green and red (yellow) signals on chromosome 8 in each nucleus.
Material was available to perform FISH for PLAG1 in 19 of 22 CA-ex-PA, and in all 5 PA. Subsequently, FISH for HMGA2 was conducted in CA-ex-PA subjects that were found to be negative for PLAG1 rearrangement by FISH (when adequate material was available).
Results
Clinicopathologic Findings
The demographics and pathologic findings of the 22 study cases are presented in Table 1. In summary, CA-ex-PA occurred in 13 males and 9 females, ranging from 24 to 86 years old (median, 58 years old), and involved the following salivary glands/anatomic locations: parotid gland (13 cases), submandibular gland (4 cases), parapharynx (2 cases), oropharynx (1 case), and metastatic sites (2 cases, 1 each in liver and lung). The malignant component comprised one or more of these histologic subtypes: adenocarcinoma, NOS (7 cases), myoepithelial carcinoma (4 cases), salivary duct carcinoma (3 cases), epithelial-myoepithelial carcinoma (2 cases), squamous cell carcinoma (1 case), carcinosarcoma (1 case), and mixed histology with a combination of 2 or more of adenocarcinoma NOS, salivary duct, myoepithelial, mucinous, and squamous cell carcinoma (4 cases). Ten CA-ex-PA were high-grade, 8 were intermediate-grade, and 4 were low-grade.
Immunohistochemical Findings
Of 22 CA-ex-PA examined for PLAG1 expression, 7 cases scored “3+”, 6 cases scored “2+”, 4 cases scored “1+”, 2 cases scored “rare”, and 3 cases scored “negative” (Table 1). Of 5 pleomorphic adenomas, 3 cases scored “3+” and 2 cases scored “2+”. The intensity of staining was variable, and sometimes weak. All other salivary gland neoplasms scored “rare/negative” for PLAG1 immunostain. Tissue microarray sections of carcinomas from various organs were negative in all but 1 infiltrating ductal carcinoma, which was positive for PLAG1 in rare cells.
Fluorescence In Situ Hybridization Study Findings
The details of FISH findings are presented in Table 1. In summary, of 19 CA-ex-PA analyzed by PLAG1 FISH, 12 cases were positive for gene rearrangement with a balanced or an unbalanced translocation pattern, two cases showed a trisomy/polysomy pattern without gene rearrangement (increased copies of an intact PLAG1), and 5 had a normal FISH pattern. In tumors positive for PLAG1 alteration, additional chromosomal abnormalities were found, including a trisomy/polysomy profile (6 cases), and amplification of PLAG1 (1 case). The gene rearrangement could be seen in one or both genes (one to two pairs of split signals in a cell), or in multiple genes (rearrangement in the setting of polysomy). In cases positive for PLAG1 alteration, split signals were seen in 56–95 % of examined cells. Of 3 cases analyzed for HMGA2 FISH, 1 case was positive for gene rearrangement (in 94 % of cells analyzed). Eleven of 12 FISH positive cases expressed PLAG1 by immunohistochemistry (1+ to 3+ ; weak to strong), and 1 case was negative (on one representative section). Of the 5 PA, FISH was positive for gene rearrangement with a balanced translocation pattern in 3 cases, negative in 1 case, and failed in 1 case on multiple attempts due to poor hybridization.
Discussion
Carcinomatous transformation is an uncommon but well-known risk associated with PA (referred to as CA-ex-PA). It accounts for <4 % of all salivary gland neoplasms, and <12 % of malignant salivary gland tumors [17]. CA-ex-PA may develop with a wide-spectrum of histologic subtypes; however, adenocarcinoma NOS, salivary duct carcinoma, or myoepithelial carcinoma [19–21] occurs most commonly.
The diagnosis of CA-ex-PA requires recognition of a co-existing component of PA, or a history of prior excision of a PA at the same site. However, the malignant component frequently takes over the entire tumor; as a result, the underlying PA may no longer be identifiable. According to one account, examination of up to 100 cut sections is sometimes needed to find traces of small residual preexisting PA [22]. While having multiple carcinoma subtypes in a given mass or marked stromal hyalinization strongly suggests the possibility of CA-ex-PA, these features are not sufficiently specific for a definitive diagnosis. A reliable marker for CA-ex-PA, therefore, would be a valuable ancillary tool in cases of diagnostic uncertainty. In the salivary gland, it is important to be able to distinguish CA-ex-PA from metastatic disease. Conversely, at metastatic sites, it is important to be able to distinguish metastatic CA-ex-PA from other potential primary tumors. With the prospect of increasingly targeted cancer therapies, distinction of CA-ex-PA from other clinically aggressive salivary gland malignancies is also likely to take on greater significance.
Based on the current state of knowledge, translocations involving PLAG1 are known to be specific for two pathologic entities, PA and lipoblastoma. In addition, we have recently demonstrated PLAG1 alteration in a subset of mixed tumors of the skin and soft tissue [18]. The PLAG1 gene produces a zinc finger protein with roles during embryogenesis and fetal development. In adult tissues, on the other hand, the protein is either undetectable or present at negligible levels [23, 24]. Translocations involving PLAG1 lead to upregulation of the gene and ectopic production of PLAG1 protein mediated by promoter swapping between PLAG1 and a ubiquitously expressed partner gene [13, 23, 24]. The preferential partner gene in almost half of PA is the gene for ß-catenin (CTNNB1) [23]. The second most common participating gene is the leukemia inhibitory factor receptor (LIFR) gene [25]. The molecular mechanisms of PLAG1 alteration in lipoblastoma are similar to those in PA but involve different partner genes, COL1A2 or HAS2 [26, 27].
PLAG1 protein expression, on the other hand, has been seen in other tumors including a subset of acute myeloid leukemia [28], hepatoblastoma [29], and a variety of mesenchymal tumors [11, 30]. PLAG1 overexpression also encompasses PA without cytogenetic finding of PLAG1 alteration, including those with HMGA2 rearrangement and karyotypically normal tumors [11, 12]. Cryptic intrachromosomal rearrangements, not detectable by routine cytogenetic studies, involving two genes close to PLAG1, namely TCEA1 and CHCHD7, account for a subset of these tumors [11, 31]. Other mechanisms, such as mutational activation or copy number gain of PLAG1, have been postulated in PLAG1 overexpression [11, 32]. Numerical gain of chromosome 8 or increased copy number of intact PLAG1, as an isolated anomaly, was seen in 2 subjects in our series, although the mechanism is unclear through which increased dosage of a putatively inactive gene ultimately leads to protein overexpression.
The findings from our study support that PLAG1 is a specific marker for CA-ex-PA among other carcinomas, both those primary to the salivary gland and a variety of other common carcinomas from other primary sites. Outside the salivary gland, one should also consider the uncommon possibility of malignant mixed tumors of skin/soft tissue. Obviously neither FISH nor immunohistochemistry can help to discriminate between a malignant mixed tumor of soft tissue and a metastatic CA-ex-PA, and clinical history would be the most helpful clue in this situation.
While PLAG1 was not an entirely robust immunostain for CA-ex-PA in our series, we found it a useful adjunctive marker, especially when interpreted in conjunction with PLAG1 FISH findings. The utility of PLAG1 immunostaining was limited in some cases by variable, focal, or weak staining. In our experience, however, a diffuse and unequivocal expression for PLAG1 in a carcinoma is a strong indication for the diagnosis of CA-ex-PA in the proper clinicopathologic setting. One case in our series, despite cytogenetic confirmation of PLAG1 rearrangement, failed to show PLAG1 immunoreactivity, and 4 additional pathologically confirmed CA-ex-PA were also negative. Since some of our samples were retrieved among old archival material, we cannot exclude that the mode of tissue fixation had not contributed to the lack of protein expression in some cases.
The combination of immunohistochemistry and FISH for PLAG1, with or without FISH for HMGA2, appears to be a more practical approach in investigating the possibility of CA-ex-PA compared to RT-PCR, in view of the multiplicity of PLAG1 partner genes. Matsuyama et al. [12] performed RT-PCR on 45 PA, screening for multiple known fusion genes with the following positive results: CTNNB1-PLAG1 (8 cases), LIFR-PLAG1 (2 cases), and one each for CHCHD7-PLAG1 and HMGA2-WIF1. None of the cases had TCEA1-PLAG1, HMGA2-FHIT, and HMGA2-NFIB fusion transcripts. In our series, 63 % of CA-ex-PA had PLAG1 rearrangement, closely matching the frequency of translocations in PA observed by cytogenetic studies (nearly 70 %). Considering that alterations of PLAG1 and HMGA2, and a trisomy/polysomy profile for chromosome 8 (or gain of intact PLAG1) are the relevant genetic alterations in PA, only 4 (21 %) of CA-ex-PA in our series had a normal pattern for PLAG1 or HMGA2 FISH assays. It is notable that cryptic intrachromosomal PLAG1 rearrangements are likely undetectable by FISH; a normal FISH pattern, therefore, does not entirely exclude the possibility of gene rearrangement.
The pathogenesis and genetic events in malignant progression of PA are not entirely understood. The risk is likely increased with the duration of the mass [19, 33]; therefore, it is conceivable that as a tumor grows, acquisition of additional genetic anomalies leads to malignant progression. Marked numerical and structural chromosomal aberrations and DNA aneuploidy are found in CA-ex-PA, but which, if any, of these changes are crucial in malignant transformation is yet to be determined [34, 35]. A study by El-Naggar et al. [35] suggested that LOH in the 12q region identifies a group of PA with a potential for evolution to carcinoma, and that additional changes at 17p may be the preceding events for malignant transformation [36]. Using a genome-wide, high-resolution array-CGH, Persson et al. [37] found amplification of 12q genes (specifically MDM2), deletions of 5q23.2-q31.2, gains of PLAG1 and MYC, and amplification of ERBB2 to be relevant events in malignant change [37]. Tsang et al. [16] reported a case of CA-ex-PA with amplification of multiple genes including PLAG1 [16]. We have also recently demonstrated increased copy number and amplification of rearranged PLAG1 in a CA-ex-PA in a young patient who had a history of PA with simple PLAG1 rearrangement [38]. An additional case in our current series showed amplification of altered PLAG1. In addition, by FISH, six of CA-ex-PA in our study had PLAG1 alteration with a trisomy/polysomy pattern. Our findings, in addition to previous reports, suggest that copy number gain and amplification of PLAG1 may contribute to malignant progression in a subset of tumors. Amplification and overexpression of HMGA2 has also been postulated in transformation to CA-ex-PA [39]. It seems more likely that malignant degeneration may be triggered by a variety of mechanisms, rather than a single genetic event responsible for all tumors. The role of p53 mutation in malignant transformation of PA, however, has already been looked at, and refuted by mutational analysis of 11 cases [40].
In summary, we evaluated 22 cases of CA-ex-PA by immunohistochemistry for PLAG1 and by FISH targeting PLAG1. In our series, the majority of CA-ex-PA carried an altered PLAG1 gene that could be identified by FISH technique. We conclude that PLAG1 rearrangement persists in carcinomas arising from PA, confirming the few previous observations. PLAG1 expression in CA-ex-PA was variable but was specific against other salivary gland carcinomas and carcinomas of other primary sites when staining was robust. In situations where there is a question of malignancy arising in the context of preexisting PA, FISH for PLAG1 and immunohistochemistry for PLAG1, particularly in combination, are helpful ancillary studies to facilitate diagnostic interpretation.
References
Mitani Y, Rao PH, Futreal PA, et al. Novel chromosomal rearrangements and break points at the t(6;9) in salivary adenoid cystic carcinoma: association with MYB-NFIB chimeric fusion, MYB expression, and clinical outcome. Clin Cancer Res. 2011;17:7003–14.
Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–13.
Bullerdiek J, Wobst G, Meyer-Bolte K, et al. Cytogenetic subtyping of 220 salivary gland pleomorphic adenomas: correlation to occurrence, histological subtype, and in vitro cellular behavior. Cancer Genet Cytogenet. 1993;65:27–31.
Mark J, Dahlenfors R. Cytogenetical observations in 100 human benign pleomorphic adenomas: specificity of the chromosomal aberrations and their relationship to sites of localized oncogenes. Anticancer Res. 1986;6:299–308.
Mark J, Sandros J, Wedell B, et al. Significance of the choice of tissue culture technique on the chromosomal patterns in human mixed salivary gland tumors. Cancer Genet Cytogenet. 1988;33:229–44.
Sandros J, Stenman G, Mark J. Cytogenetic and molecular observations in human and experimental salivary gland tumors. Cancer Genet Cytogenet. 1990;44:153–67.
Kas K, Roijer E, Voz M, et al. A 2-Mb YAC contig and physical map covering the chromosome 8q12 breakpoint cluster region in pleomorphic adenomas of the salivary glands. Genomics. 1997;43:349–58.
Kas K, Voz ML, Hensen K, et al. Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J Biol Chem. 1998;273:23026–32.
Geurts JM, Schoenmakers EF, Roijer E, et al. Expression of reciprocal hybrid transcripts of HMGIC and FHIT in a pleomorphic adenoma of the parotid gland. Cancer Res. 1997;57:13–7.
Geurts JM, Schoenmakers EF, Roijer E, et al. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–72.
Astrom AK, Voz ML, Kas K, et al. Conserved mechanism of PLAG1 activation in salivary gland tumors with and without chromosome 8q12 abnormalities: identification of SII as a new fusion partner gene. Cancer Res. 1999;59:918–23.
Matsuyama A, Hisaoka M, Nagao Y, et al. Aberrant PLAG1 expression in pleomorphic adenomas of the salivary gland: a molecular genetic and immunohistochemical study. Virchows Arch. 2011;458:583–92.
Stenman G. Fusion oncogenes and tumor type specificity—insights from salivary gland tumors. Semin Cancer Biol. 2005;15:224–35.
Jin C, Martins C, Jin Y, et al. Characterization of chromosome aberrations in salivary gland tumors by FISH, including multicolor COBRA-FISH. Genes Chromosomes Cancer. 2001;30:161–7.
Martins C, Fonseca I, Roque L, et al. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–55.
Tsang YT, Chang YM, Lu X, et al. Amplification of MGC2177, PLAG1, PSMC6P, and LYN in a malignant mixed tumor of salivary gland detected by cDNA microarray with tyramide signal amplification. Cancer Genet Cytogenet. 2004;152:124–8.
Gnepp DR, Brandwein M, El-Naggar AK, et al. Pleomorphic adenoma. In: Eveson JW, Reichart P, Sidransky D, Barnes L, editors. World health organization classification of tumours. Pathology and genetics of head and neck tumours. France: IARC Press; 2005. p. 242–4.
Bahrami A, Dalton JD, Krane JF, et al. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–8.
Olsen KD, Lewis JE. Carcinoma ex pleomorphic adenoma: a clinicopathologic review. Head Neck. 2001;23:705–12.
Katabi N, Gomez D, Klimstra DS, et al. Prognostic factors of recurrence in salivary carcinoma ex pleomorphic adenoma, with emphasis on the carcinoma histologic subtype: a clinicopathologic study of 43 cases. Hum Pathol. 2010;41:927–34.
Lewis JE, Olsen KD, Sebo TJ. Carcinoma ex pleomorphic adenoma: pathologic analysis of 73 cases. Hum Pathol. 2001;32:596–604.
Foote FW Jr, Frazell EL. Tumors of the major salivary glands. Cancer. 1953;6:1065–133.
Kas K, Voz ML, Roijer E, et al. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–4.
Voz ML, Van d V, Kas K. First insights into the molecular basis of pleomorphic adenomas of the salivary glands. Adv Dent Res. 2000;14:81–3.
Voz ML, Astrom AK, Kas K, et al. The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of PLAG1 gene expression under control of the LIFR promoter. Oncogene. 1998;16:1409–16.
Astrom A, D’Amore ES, Sainati L, et al. Evidence of involvement of the PLAG1 gene in lipoblastomas. Int J Oncol. 2000;16:1107–10.
Hibbard MK, Kozakewich HP, Dal CP, et al. PLAG1 fusion oncogenes in lipoblastoma. Cancer Res. 2000;60:4869–72.
Landrette SF, Kuo YH, Hensen K, et al. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood. 2005;105:2900–7.
Zatkova A, Rouillard JM, Hartmann W, et al. Amplification and overexpression of the IGF2 regulator PLAG1 in hepatoblastoma. Genes Chromosomes Cancer. 2004;39:126–37.
Matsuyama A, Hisaoka M, Hashimoto H. PLAG1 expression in mesenchymal tumors: an immunohistochemical study with special emphasis on the pathogenetical distinction between soft tissue myoepithelioma and pleomorphic adenoma of the salivary gland. Pathol Int. 2012;62:1–7.
Asp J, Persson F, Kost-Alimova M, et al. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer. 2006;45:820–8.
Persson F, Winnes M, Andren Y, et al. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27:3072–80.
Spiro RH, Huvos AG, Strong EW. Malignant mixed tumor of salivary origin: a clinicopathologic study of 146 cases. Cancer. 1977;39:388–96.
Vargas PA, Torres-Rendon A, Speight PM. DNA ploidy analysis in salivary gland tumours by image cytometry. J Oral Pathol Med. 2007;36:371–6.
El-Naggar AK, Lovell M, Callender DL, et al. Concurrent cytogenetic, interphase fluorescence in situ hybridization and DNA flow cytometric analyses of a carcinoma ex-pleomorphic adenoma of parotid gland. Cancer Genet Cytogenet. 1998;107:132–6.
El-Naggar AK, Callender D, Coombes MM, et al. Molecular genetic alterations in carcinoma ex-pleomorphic adenoma: a putative progression model? Genes Chromosomes Cancer. 2000;27:162–8.
Persson F, Andren Y, Winnes M, et al. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82.
Bahrami A, Dalton JD, Bangalore S, et al. Disseminated carcinoma ex pleomorphic adenoma in an adolescent confirmed by application of PLAG1 immunohistochemistry and FISH for PLAG1 rearrangement. Head Neck Pathol. 2012. doi:10.1007/s12105-012-0330-2.
Roijer E, Nordkvist A, Strom AK, et al. Translocation, deletion/amplification, and expression of HMGIC and MDM2 in a carcinoma ex pleomorphic adenoma. Am J Pathol. 2002;160:433–40.
Gedlicka C, Item CB, Wogerbauer M, et al. Transformation of pleomorphic adenoma to carcinoma ex pleomorphic adenoma of the parotid gland is independent of p53 mutations. J Surg Oncol. 2010;101:127–30.
Acknowledgments
This work was supported in part by the American Lebanese Syrian Associated Charities (ALSAC). The study was performed with the approval of the Institutional Review Boards of St. Jude Children’s Research Hospital and Partners Health Care. IRB numbers: NR10-130 and 2010-P-001957/1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bahrami, A., Dalton, J.D., Shivakumar, B. et al. PLAG1 Alteration in Carcinoma Ex Pleomorphic Adenoma: Immunohistochemical and Fluorescence In Situ Hybridization Studies of 22 Cases. Head and Neck Pathol 6, 328–335 (2012). https://doi.org/10.1007/s12105-012-0353-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12105-012-0353-8