Abstract
Group-wise diversity of sediment methylotrophs of Chilika lake (Lat. 19°28′–19°54′N; Long. 85°06′–85°35′E) Odisha, India at various identified sites was studied. Both the culturable and unculturable (metagenome) methylotrophs were investigated in the lake sediments employing both mxaF and 16S rRNA genes as markers. ARDRA profiling, 16S rRNA gene sequencing, PAGE profiling of HaeIII, EcoRI restricted mxaF gene and the mxaF gene sequences using culture-dependent approach revealed the relatedness of α-proteobacteria and Methylobacterium, Hyphomicrobium and Ancyclobacter sp. The total viable counts of the culturable aerobic methylotrophs were relatively higher in sediments near the sea mouth (S3; Panaspada), also demonstrated relatively high salinity (0.1 M NaCl) tolerance. Metagenomic DNA from the sediments, amplified using GC clamp mxaF primers and resolved through DGGE, revealed the diversity within the unculturable methylotrophic bacterium Methylobacterium organophilum, Ancyclobacter aquaticus, Burkholderiales and Hyphomicrobium sp. Culture-independent analyses revealed that up to 90 % of the methylotrophs were unculturable. The study enhances the general understandings of the metagenomic methylotrophs from such a special ecological niche.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chilika (Lat. 19°28′–19°54′N; Long. 85°06′–85°35′E), a biodiversity hotspot of the country, is a simple cone-shaped, largest brackish water lake in Asia and second largest in the world, situated in humid tropical climatic coastal zone of Odisha (former Orissa), India, declared as a Ramsar site (listed as a wetland for intensive conservation and management by the ministry of Environment and Forests, Government of India) under the convention on ‘Wetlands of international importance’ [1]. Chilika, slowly but steadily transforming into a lagoon, is an attractive and unparalleled fusion of marine, river, and estuarine habitat that supports unique assemblage of marine, brackish water and freshwater microbes [2], presents a challenge to physiologists and biochemists [3]. Biological effects of siltation and salinity, overall biodiversity loss due to the changes in phytoplankton communities, and degradation of lake ecosystem owing to the freshwater invasive species thereby declining the overall productivity, have been profoundly documented in recent times [2, 3, 5, 7, 8].
Bacterial assimilation of methane, methanol and methylamine is important in the detritus food chain. Methylotrophs are globally distributed and phylogenetically dispersed. These have been isolated from various environments, including soils, sediments, freshwater, marine sediments, seawater, acid peat bogs, hot springs and cold environments such as the Antarctic, and have attracted a great deal of interest due to their considerable commercial potential to produce bulk/fine chemicals and in bioremediation. Due to their specific properties, for instance, haloalkaliphilic methanotrophs could be of use in modern biotechnology [4]. Methylotrophs have been reported from diverse aquatic niches, such as, Washington Lake, USA [5], Mono Lake, USA [6], Lonar Lake, India [7], Weyerhaeuser [8], Warm pool, China [9], Colne Estuary, UK [10], Brackish marsh, Portugal [11], and as a marine symbiont [12]. There is increasing evidence for the striving methanotrophs in various ecogeographical regions including the saline and alkaline aquatic environments. Although the vast biodiversity of the brackish water system Chilika has been appreciated unquestionably [1–3], reports have been feeble.
These are being extensively studied in a range of environments due to their critical role in global methane cycle as well. PCR-based methods facilitate the methanotrophs ecology and diversity studies, viz., 16S ribosomal RNA technology and specific amplification of ‘functional genes’, such as those encoding unique enzymes in the organismal metabolism including methane monooxygenase and methanol dehydrogenase. Methanol-oxidizing bacteria play significant role in biogeochemical carbon cycling by facilitating incorporation of C1 derivatives into biomass [13, 14] using methanol as the sole carbon and energy source. Cyto- and bio-chemical properties, such as, synthesis of osmoprotectants, accumulation of potassium ions, formation of glycoprotein S-layers on the outer cell wall surface, and modification of the chemical composition of their membranes, allow these specialized group (haloalkaliphilic methanotrophs) to adapt to saline and alkaline habitats [4, 15].
Global cycling of C1 compounds affect important environmental phenomena related to climate change. Methylotrophs thriving on C-substrates like methane, methylated sulfur sp., methylated amines, halogenated methanes and methanol might play a crucial role in global warming and groundwater contamination, the two major environmental concerns. Alkalitolerant halophilic and type I alkaliphilic halotolerant methanotrophs utilize methane and methanol, to oxidize ammonium ions, and to transform various organic compounds even at 12 ‰ salinity and 5–11 pH [15]. Recent investigations reported methanol utilizing methylotrophs from salty water-bodies having impact on global warming and bioremediation of pollution by methanol and other C1 compounds [15–18]. Microscopic and enzymatic evidence of the reducing sediments at hypersaline seeps in the abyssal Gulf of Mexico supported the hypothesis that methylotrophs capable of using reduced C1 compounds as their carbon and energy sources occur as intracellular symbionts of the seep mussel and other benthic invertebrates resembling hydrothermal vent community assemblages [12]. These symbioses differed from those reported for bivalves from hydrothermal vents and reducing sediments (23 to −34 ‰ δC13). The microbiology, the bioprocess of CH4, N2O, and CO2 abatement, potential and limitations of the GHG biodegradation processes, technology niches and the knowledge gaps have been reviewed [19]. It is essential to mention that, methane (CH4), nitrous oxide (N2O), and carbon dioxide (CO2) emissions represent approximately 98 % of the global greenhouse gas (GHG) inventory.
The ecophysiological importance of methanotrophs in microbial communities inhabiting haloalkaline aquatic environments is due to their involvement in the global cycles of methane and major bioelements (such as, C, N, and S). The key microbial enzyme, methane monooxygenase (MMO; especially the soluble one), is remarkably substrate nonspecific. This unique capability, i.e., catalyzing reactions of environmental importance, has attracted great attention for applied microbiologists and bioengineers. Prudent biological technologies can become low-cost ecofriendly alternative to physicochemical methods for GHGs abatement. Molecular phylogenetic studies of microbial diversity based on the conserved functional gene sequences have greatly expanded our knowledge [9, 20–23]. The mxaF gene is supposed to be a phylogenetic chronometer for methylotrophs. This is also a conserved functional gene like 16S rRNA, primarily responsible for methylotrohy. With the functional methylotroph group as the target, this gene was used to map the diversity of the unculturable methanol oxidizers in Chilika Lake, which is slowly but steadily transforming into a lagoon. The mxa genes are well conserved among α-, β- and γ-proteobacteria classes as expressed gene sequences, suggesting a methylotrophic origin for methanol oxidation machinery encoded by these genes. The probes targeting the functional mxaF gene coding for MDH and 16S rRNA has been widely used to detect and analyze the methylotroph diversity within proteobacteria [24, 25]. Being a conserved gene, mxaF served dubiously as a genetic marker to detect methylotrophy in the environment [26, 27].
This study of summer 2010 isolated and characterized the aerobic methanol-utilizers from Chilika lake sediments, and analyzed their diversity using polyphasic approaches (ARDRA profiling, 16S rRNA sequencing, mxaF-RFLP, mxaF gene sequencing and real time quantification). Due to geographical vastness, the methylotrophs diversity of entire lake needs bigger comprehensive approach. None-the-less, the present study is a first such attempt.
Materials and Methods
Sampling Site
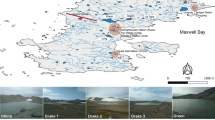
Samples were collected randomly by grab sampling method from the sediment surface, up to 10–20 cm deep, from diverse sites of Chilika lake (Lat. 19°28′–19°54′N; Long. 85°06′–85°35′E) from Balugaon, Bhusandapur, Panaspada, Nalaban island, and Breakfast island (Fig. 1). There was variation in the length of water column due to variation in the sea flow attached to the lake. At each of these identified sampling sites, one composite sample was prepared by pooling five sediment samples covering an area of 5 km2, each sampling point being located at least 10 km distant from each other. In all, five composite samples S1 (Balugaon), S2 (Bhusandapur), S3 (Panaspada), S4 (Nalaban island), and S5 (Breakfast island) were made. Samples were transported to the lab on ice and stored at 4 °C until further analyses. A part of the samples was immediately stored at −80 °C for culture-independent studies. Samples were analyzed for various parameters, such as, pH, salinity, organic carbon, and aerobic heterotrophic (total) microbial counts following standard procedures [28]. The average mean values of these physiochemical and biological attributes are detailed sampling site-wise in Table 1.
Enrichment and Isolation
Nitrate mineral salt (NMS) medium was used for methylotrophs enrichment, a liter of which contained NaCl 5.0 g, Na2HPO4 0.72 g, KH2PO4 0.28 g, MgSO4·7H2O 0.2 g, CaCl2·2H2O 0.02 g, FeSO4·7H2O 5.0 mg, ZnSO4·7H2O 70 µg, MnCl2·4H2O 30 µg, H3BO3 300 µg, CoCl2 200 µg, NiCl2·6H2O 20 µg, Na2MoO4·2H2O 30 µg, CuCl2·2H2O 10 µg, Na4-EDTA 1.0 mg, KNO3 1.0 g, methanol 0.1 % v/v [29]. Antifungal cycloheximide (40 mg/ml) was added at the rate of 500 µl per liter, and pH was adjusted by inorganic (sodium; NaHCO3–Na2CO3) buffer as per pH of the sample at sampling. The prepared sediment samples (10.0 g in 100 ml double distilled water) were placed in 500 ml Erlenmeyer flasks with 100 ml methanol-fortified (as above) NMS medium and shaker incubated (120 rpm; 30 °C) overnight. The samples were enriched following three transfers in the medium over 4–6 days, serially diluted, and plated on fortified-NMS agar for up to 24 h. After incubation colonies were picked and transferred on to the slants, flooded with glycerol and maintained as stocks.
Genomic DNA Extraction and PCR Amplification of 16S rRNA and mxaF Genes
Genomic DNA was isolated from the log phase broth cultures. Pelleted cells from 1.5 ml media were resuspended in 0.5 ml SET buffer (75 mM NaCl, 25 mM EDTA and 20 mM Tris) with 10 µl of lysozyme (10 mg/ml) and the genomic DNA was extracted [30]. The integrity and concentration of purified DNA was determined by ethidium bromide stained agarose gel (0.8 %) electrophoresis by comparing with the commercial DNA ladder concentration (Hyper ladder genetix). DNA from raw sediment samples were extracted using UltraClean Soil DNA isolation kit (MoBio Laboratories, Carlsbad). For PCR amplification, final genomic DNA concentration was adjusted to 50 ng/µl. 16S rRNA gene was partially amplified using primers pA (5′-AGAGTTTGATCCTGGCTCAG3′; E. coli position 8–27) and pH (5′-AAGGAGGTGATCCAGCCGCA3′; E. coli position 1525–1544) [31]. Amplification was carried out on a thermal cycler (Biorad PTC0220) in 100 µl volume by mixing 50–90 ng template DNA with polymerase reaction buffer (10×); 100 µM (each) dATP, dCTP, dTTP and dGTP; primers pA and pH (20 ng each) and 1.0 U Taq polymerase using following conditions: initial denaturation at 94 °C for 1.5 min; 35 cycles at 95 °C for 1.0 min, 55 °C for 1.0 min, 72 °C for 1.0 min; and final extension at 72 °C for 5 min.
The mxaF gene was used to identify/authenticate methanol-oxidizing population downstream [6]. The mxaF gene in the isolates was partially amplified using specific primers, mxaF-1003 (5′GCGGCACCAACTGGGGCTGGT3′; forward) and mxaR-1561 (5′GGGCAGCATGAAAGGGCTCCC3′; reverse) [24]. For environmental DNA, GC clamp (CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G) attached at 5′ end with one of the primer was used to increase the bands separation in DGGE gel [32]. The PCR condition was similar to 16S rDNA amplification. PCR products were separated on ethidium bromide stained 1.5 % agarose gel, and documented in Alpha Imager TM1200 (Alpha InfoTech).
ARDRA and RFLP
Products were digested with selected restriction enzymes with different restriction sites after 16S amplifying rDNA. About 1 µg of PCR-amplified 16S rDNA fragment was restricted with three different endonucleases HaeIII, MspI and EcoRI (Fermentas) separately, incubated at 37 °C overnight and resolved on 2 % agarose gel. The mxaF gene PCR products were digested with HaeIII and EcoRI and separated through PAGE (PolyAcrylamide Gel Electrophoresis). Banding patterns were visualized by ethidium bromide staining and analyzed in Alpha Imager TM1200 (Alpha InfoTech). Thus, amplified rDNA restriction analysis and restricted fragment length polymorphism were performed. Different phylotypes or operational taxonomic units were obtained by similarity and clustering analysis using NTSYSpc-2.02e. Similarities among isolates were calculated by Jaccard’s coefficient [33], and dendrogram constructed using UPGMA method [34]. Since ARDRA is restriction digestion for (16S) ribosomal gene while RFLP is restriction digestion of mxaF gene, the aim and importance of ARDRA and RFLP in the investigation was to group the isolates based on structural and functional genes.
DGGE (Denaturing Gradient Gel Electrophoresis) Profiling
The mxaF PCR products were purified using SV-PCR purification kit (Promega). PCR products were separated on a 1.0 mm thick, vertical polyacrylamide gel (6.5 % w/v acrylamide:bisacrylamide::37.5:1, Bio-Rad) prepared with and electrophoresed in 1.0 × TAE, pH 7.4 (0.04 M Tris-base, 0.02 M sodium acetate, 1 mM EDTA) at 60 °C and a constant voltage of 150 V for 16 h [35]. A denaturing gradient of 100 % denaturant corresponded to 7 M urea plus 40 % v/v formamide. Gels were loaded with 30 µl of PCR product depending on the band intensity and electrophoresed on 1.5 % agarose gels. DGGE gels were stained for 20 min in water containing 0.5 µg/ml ethidium bromide, and the images were recorded in an Alpha Imager TM1200 (Alpha InfoTech) documentation analysis system. DNA bands migrating to the same position on the gel were assumed to be identical amplicons [32].
16S rRNA and mxaF Gene Sequencing
PCR amplified 16S rDNA products were purified with a Quaquick purification kit (Qiagen). DNA sequences were double checked by sequencing both strands using primers pA and pH for forward and reverse reactions, respectively. Nucleotide sequences were dideoxy-cycle sequenced with fluorescent terminators (Big Dye, Applied Biosystems) and run in a 3130×l ABI prism automated DNA sequencer (Applied Biosystems). DGGE bands were excised from the gel using a sterile scalpel and incubated in 60 µl sterile Milli-Q purified water at 4 °C for 24 h. DNA was diffused out of the gel and the solution used as the template in a reamplification PCR, performed using the original primers but modified PCR regime (Table 2) and run on DGGE to confirm its identity. Only pure bands were used for sequencing by amplifying with primers without a GC clamp. PCR products for sequencing were purified and sequenced using ABI prism sequencer. Representatives of the dendrogram constructed from mxaF-RFLP PAGE patterns were also sequenced with fluorescent terminators (Big dye, Applied Biosystems) and run in the same DNA sequencer.
The sequences of 24 representative isolates are submitted to GenBank with accession numbers GQ281064-GQ281070, GQ281072-GQ281076, GQ354269-GQ354270, GQ411497-GQ411503, GQ411505, GQ227415 and GQ332407. Partial mxaF gene fragments sequences (HM765479–HM765503) and the excised DGGE bands sequences (HM628891–HM628901) have also been submitted to the GenBank.
Quantification Through RT PCR
The mxaF gene quantification was done in copies based on ‘second derivative maximum method’ (using LightCycler software 3.5; Roche diagnostics), wherein exponential phase of amplification curve was linearly related to a starting concentration of template DNA molecules. Each QC-PCR standard curve was generated by using six dilutions of standard DNA template from 102 to 106 copies of mxaF gene, and the standard DNA template was tenfold serial-diluted. Quantitative PCR using SYBR Green I technology [36] with the primers mxaF and mxaR was carried out amplifying five metagenome samples, negative control and five plasmid DNA standards. Mastermix [14 µl sterile water, 2 µl MgCl2 (25 mM), 1 µl of each primer (20 pmol), 2 µl of SYBR Green master mix (20 pmol; Roche diagnostics) and 50 ng DNA] was prepared. Amplification started with a 10 min denaturation at 95 °C followed by a 40-cycles 4-segment amplification (denaturation for 15 s at 95 °C, annealing for 10 s at 55 °C, elongation for 20 s at 72 °C and appended for 5 s at 83 °C for possible primer-dimers through a single fluorescence measurement). The last step ensured the elimination of nonspecific fluorescence signal and ensured accurate quantification of the desired product. Finally, a systematic melting step (10 s at 95 °C, 10 s at 60 °C and slow heating at a rate of 0.1 °C per second up to 99 °C) with continuous fluorescence measurement was performed.
BLAST Search and Phylogeny Analysis
The partial 16S rDNA sequences, mxaF gene sequences of isolated strains and mxaF gene sequences from environmental DNA were compared with those available in the databases. Identification was based on sequence similarity of ≥97 % with that of public database sequences, by BLAST homology. The sequence alignment and comparison were performed using multiple sequence alignment program ClustalW2 [37] with default parameters, and data converted to PHYLIP format. Minor modifications were done manually on the basis of conserved domains, and columns containing more than 50 % gaps were removed. The phylogenetic trees were constructed on the aligned datasets using MEGA 4.0.2 [38] using the neighbor-joining method [39]. Bootstrap analysis [40] was performed on 1,000 random samples taken from the multiple alignments.
Results and Discussion
Methylotrophs are distributed in diverse environments from freshwater Lake [5], deep-sea sediments [9], hypersaline lake [24], chlorinated environments [41], plant phyllosphere [42] to hot water effluent [43], suggesting their ubiquity. Present study focused on the aerobic methylotrophs covering 1,100 km2 of the Chilika Lake sediments through established molecular markers (such as the functional gene mxaF and phylogenetic 16S rRNA gene probes) which demonstrated the presence of phylogenetically diverse aerobic methylotrophs. The total viable counts were relatively higher in sediments near the sea mouth (S3; Panaspada) as also viewed by Joshi et al. [44].
Enumeration of Methanol Oxidizing Bacteria
The bacterial counts were from 5.4 to 6.4 log CFUs/g sediment. A total of 80 isolates from the samples were selected for further study. Maximum methylotroph population was observed at relatively nearer to the sea mouth (at S3; Panaspada), followed by S5 (Breakfast island) down south, and minimum was in S1 (Balugaon), far away from sea mouth. All isolates were aerobic, catalase, urease positive and weakly oxidase positive. The halotolerance ability of the representative isolates was studied. Methylobacterium aerobically produces carotenoid pigment and bacteriochlorophyll A indicating that their ability to acquire ATP helps them to survive even in carbon deprivation [41]. Two Methylobacterium sp. isolated from the sediment samples from S3 (Panaspada) endured up to 1.0 M NaCl concentration (Table 3). The mixing of the sea water in the lake might be a reason for the relatively high halotolerance. Surface sediment was aerobic, whereas the oxygen typically depleted within millimeters below possibly due to the oxygen diffusional limitation across the column coupled with the active aerobic respirers [36].
Isolates growing on methanol, but not responding to the PCR amplification with mxaF-primers, may have alternate mechanism for primary methanol metabolism [26], or might be growing autotrophically. The lack of mxaF gene amplification in isolates CS3, CS8 and CS15 point to such alternate methylotrophy. Negative results suggest the role of some other alcohol dehydrogenase in methylotrophic metabolism.
The mxaF Gene Amplification
Results of the amplified gene mxaF coding for α-subunit of methanol dehydrogenase indicated that the C1 compounds in the lake sediments may be derived from the organic sedimentation and degradation. mxaF gene quantification gives the real scenario of methylotroph biocoenose [9]. Most isolates gave a fine 550 bp partial gene amplification, which authenticated that all of them were methylotrophically active methanol oxidizers. However, a few (viz., CS3, CS15, CS8; Table 2) did not exhibit such amplification but still could grow on agar plates even after several streaks. The genus Methylobacterium, a pink pigmented facultative methylotroph follows Serine pathway to metabolize formaldehyde [41], also reported from sea water [45], was dominant in the explored diverse methylotroph communities.
Amplified Ribosomal DNA Restriction Analysis and RFLP
Restriction digestion of 16S rDNA gene (using HaeIII, MspI and EcoRI) and restriction digestion of mxaF gene (using HaeIII and EcoRI) yielded distinct restriction patterns, with about two to five restricted fragments of varying sizes in each pattern. Cluster analysis of combined 16S rDNA restriction patterns based on the Jaccard’s similarity index grouped all 80 isolates in different groups (data not given). The majority of the isolates formed two (I—52.5 %; II—37.5 %) major clusters, whereas the remaining ones formed small clusters. A similar cluster analysis of the mxaF gene combined restriction patterns (data not provided) grouped the 77 isolates in two major groups, of which three isolates did not exhibit mxaF gene amplification.
16S rRNA Gene Sequence Analysis
All 24 pigmented and nonpigmented representative isolates with similar ARDRA pattern generated by three restriction enzymes were selected for sequence analysis, to study the species-level diversity of methylotrophs (Fig. 2). Based on 16S rDNA sequencing, the isolates identified were Methylobacterium radiotolerans, M. extorquens, M. hispanicum, M. organophilum, M. lusitanum, M. zatmanii, Hyphomicrobium facile, Methyloversatilis sp., Mycobacterium brisbanense and Pseudomonas sp. The prevalent genera were α-proteobacteria, Methylobacterium and Hyphomicrobium. CS11 showed distinctly high sequence similarity (100 %) with Methylobacterium sp. CS3, reportedly a novel strain from a brackish water environment, showed a lower similarity (85 %) with Methyloversatilis universalis (Table 3). Some reported gene sequences here matched with the gene sequences of active methylotrophs from Lake Washington [5] and from the deep sea [9].
Phylogenetic tree based on the 16S rRNA gene sequences of methylotrophs using neighbor-joining method. Data of all genera obtained are from GenBank database—sequence accession numbers are in parentheses, the numbers on the tree indicate the percentages of boot-strap sampling derived from 1,000 replicates, and the bar infers nucleotide substitutions
mxaF Gene Sequence Analysis
From Culturable Ones
For sequence analysis, all 25 pigmenting and non-pigmenting representative isolates showing similar mxaF-RFLP pattern generated by restriction enzyme HaeIII and EcoRI (data not presented) were selected. Genes coding for the larger subunit of methanol dehydrogenase were also sequenced and it was observed that the genus Methylobacterium was dominant followed by Methylophilus and Hyphomicrobium sp.
From mxaF Gene Sequences of Unculturable Ones
The mxaF gene amplified metagenome products gave distinct variation on DGGE profiling (Fig. 3). Culture-independent mxaF gene sequences revealed them as methylotrophs Methylobacterium organophilum, Ancyclobacter aquaticus, Burkholderiales and Hyphomicrobium sp. with 73.3 % unculturable methylotrophs, 6.6 % Ancyclobacter aquaticus, 6.6 % Burkholderiales, 6.6 % Hyphomicrobium and 6.6 % Methylobacterium sp. (Fig. 4).
Phylogenetic tree based on the mxaF gene sequences of culturable and unculturable methylotrophs using neighbor-joining method. Data of all genera obtained are from Genbank database. The numbers on the tree indicate the percentages of boot-strap sampling derived from 1,000 replicates, and the bar infers nucleotide substitutions
Real Time PCR Quantification
Sensitivity of Lightcycler RT-PCR was evaluated using different starting amount of DNA and compared with the standard curve. SYBR Green I fluorescence determination at the elevated temperature 83 °C resulted in a reliable and sensitive environmental DNA quantification assay with high linearity (Pearson correlation coefficient 0.99) over five orders of magnitude from 102 to 106 standard mxaF cloned DNA start molecules (Fig. S1). All quantitative PCR reactions were performed in replicates. The corresponding specific mxaF gene fragment from different sediment samples were amplified, indicating the vast distribution of methylotrophs in brackish water lake sediment. The mxaF gene copy number per gram sediment was in the range of 4.9 × 106–1.25 × 107 (Table 1). The mxaF gene copy numbers were compared to the CFUs determined by plate counts, and the calculated coefficient of correlation between the log copy number of mxaF gene and log CFU was 0.979 (Fig. 5). The quantification limit was 1.21 × 106 mxaF gene copies per gram sediment.
Of the total 16S rRNA based identified isolates, 62.5 % were close to Methylobacterium sp., 16.6 % were Hyphomicrobium facile, 4.1 % Methylophilus sp., Mycobacterium brisbanense, Pseudomonas sp. and 8.2 % Methyloversatilis sp., while Methylobacterium was dominant followed by Methylophilus and Hyphomicrobium sp. in mxaF based identification. The unculturable gene sequence analysis of mxaF reported the dominance of unculturable methylotrophic bacteria. PCR amplification of mxaF gene coding for α-subunit of methanol dehydrogenase indicate a methylotrophic metabolism. Burkholderiales was found through culture independent approach with 92 % mxaF gene sequence similarity. Bodrossy et al. [43] reported that Burkholderiales followed some other alcohol dehydrogenase for methanol assimilation, and thus, it might be one of the possible ecological adaptation reasons that Burkholderiales possesses the mxaF gene. Successful species-level identification and validation strategies for Bacillus [21], Clostridium [22], Pseudomonas [23], Streptococcus [48] based on the 16S rDNA (rrs) gene sequence through species-specific phylogenetic frameworks, 30–50 nucleotide long motifs (signature sequences) and (in silico) RE digestion patterns in the rrs have been reported. Kalia et al. [22] identified 84 novel Clostridia and advocated the approach to identify important ‘food and healthcare’ microbes.
High organic matter including lipids imparts toxic effects on the thriving microbial communities. Present study reports a more diversified methylotrophs group from saline sediments, like Methylobacterium, Methyloversatilis, Pseudomonas, Hyphomicrobium and Mycobacterium. Methylophilus methylotrophus could be a crop plants growth promoter under nitrogen stress conditions as it produced significant amount of low-viscosity extracellular polysaccharides from methanol in a chemostat culture under nitrogen limiting condition [46]. The bioengineering applications of methanotrophs are limited such as, the lack of suitable methanotrophic isolate, gas transfer limitation, competitive inhibition of methyl monoxygenase (MMO), regeneration of reducing equivalents for MMO and product toxicity [47]. Hyphomicrobium facile, an aerobic chemoorganotroph, plays an important role in denitrification to remove nitrate at drinking water treatment facilities [48], and sewage treatment plants [49]. Methylobacterium is a facultative methylotroph and cannot use methane, but is capable of utilizing methanol and some other C1 compounds, as well as a wide range of multi-carbon substrates, as their sole carbon and energy source while Hyphomicrobium is a facultative methylotroph. Present study not only describes the methylotrophic diversity of the Chilika Lake but also indicates the richness of the sediments in terms of microbial wealth.
16S rDNA signature sequences for methylotrophs following different metabolic pathways were developed by Brusseau et al. [50]. Specific signature Serine pathway methylotrophy 5′-CCC-TGAGTT-ATT-CCG-AAC-3′ was found in isolate CS7 while isolate CS13 exhibited the signature RuMP pathway methylotrophy 5′-ATG-CAT-CTC-TGC-TTC-GTT-3′. As these were not found in the rest of the strains, they could be tentatively considered for novel species characterization.
Conclusion
Comparing the distribution and quantification of mxaF gene by qRT PCR method revealed higher methylotrophs population from S3 (Panaspada), suggesting a higher C1 utilization rate. Higher mxaF gene copy numbers (about one log unit more) in all samples compared to the CFU counts may suggest that only up to 10 % of methylobacteria were culturable (detectable by the plating method). 16S rRNA, mxaF-RFLP, mxaF gene sequencings, mxaF gene quantification and ARDRA could classify and compare the species level communities. The methylotrophs were found to be predominantly α-proteobacteria. Up to a maximum 85 % similarity of CS3 with Methyloversatilis (GenBank sequences) suggests its novelty attributable to its evolution to adapt well to this unique environment, to confirm by polyphasic taxonomy approach. The phylogeny of the C1-metabolism genes gave a better understanding of the origin of related enzymes and interactions between methylotrophs and non-methylotrophs. The present scientific reporting is one-of-its-kind of the specialized methylotrophs comparing their culturable and metagenomic populations in ‘sediment’ niche covering a wider landscape of the Chilika Lake.
References
Nayak BK, Acharya BC, Panda UC, Nayak BB, Acharya SK (2004) Variation of water quality in Chilika Lake, Orissa. Indian J Mar Sci 33:164–169
Rath J, Adhikary SP (2008) Biodiversity assessment of algae in Chilika Lake, East Coast of India. Monitoring and modelling lakes and coastal environments. Springer, Netherlands, pp 22–33
Behera PK (1999) Applied botany: biodiversity and biotechnology. UGC refresher course. Botany Department, Berhampur University, India, pp 1–92
Murrell JC, McDonald IR, Bourne DG (1998) Molecular methods for the study of methanotroph ecology. FEMS Microbiol Ecol 27:103–114. doi:10.1111/j.1574-6941.1998.tb00528.x
Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L (2005) Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol 71:6885–6899. doi:10.1128/AEM.71.11.6885-6899.2005
Nercessian O, Kalyuzhnaya MG, Joye SB, Lidstrom ME, Chistoserdova L (2005) Analysis of fae and fhcD Genes in Mono Lake, California. Appl Environ Microbiol 71:8949–8953. doi:10.1128/AEM.71.12.8949-8953.2005
Antony CP, Kumaresan D, Ferrando L et al (2010) Active methylotrophs in the sediments of Lonar Lake, a saline and alkaline ecosystem formed by meteor impact. ISME J 4:1470–1480. doi:10.1038/ismej.2010.70
Yu Z, Mohn WW (2001) Bacterial diversity and community structure in an aerated lagoon revealed by ribosomal intergenic spacer analyses and 16S ribosomal DNA sequencing. Appl Environ Microbiol 67:1565–1574. doi:10.1128/AEM.67.4.1565-1574.2001
Wang P, Wang F, Xu M, Xiao X (2004) Molecular phylogeny of methylotrophs in a deep-sea sediment from a tropical west Pacific Warm Pool. FEMS Microbiol Ecol 47:77–84. doi:10.1016/S0168-6496(03)00252-6
Moussard N, Stralis-Pavese N, Bodrossy L, Neufeld JD, Murrell JC (2009) Identification of active methylotrophic bacteria inhabiting surface sediment of a marine estuary. Environ Microbiol Rep 1:424–433. doi:10.1111/j.1758-2229.2009.00063.x
De Marco P, Pacheco CC, Figueiredo AR, Moradas-Ferreira P (2004) Novel pollutant-resistant methylotrophic bacteria for use in bioremediation. FEMS Microbiol Lett 234:75–80. doi:10.1111/j.1574-6968.2004.tb09515.x
Cavanaugh CM, Levering PR, Maki JS, Mitchell R, Lidstrom ME (1987) Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325:346–348. doi:10.1038/325346a0
Chistoserdova L, Lidstrom ME (2013) Aerobic methylotrophic prokaryotes. In: Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E (eds) The prokaryotes, 4th edn. Springer, pp 267–285
Vuilleumier S, Chistoserdova L, Lee M-C et al (2009) Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4:e5584. doi:10.1371/journal.pone.0005584
Trotsenko YA, Khmelenina VN (2002) The biology and osmoadaptation of haloalkaliphilic methanotrophs. Microbiology 71:123–132. doi:10.1023/A:1015183832622
Sanseverino AM, Bastviken D, Sundh I, Pickova J, Enrich-Prast A (2012) Methane carbon supports aquatic food webs to the fish level. PLoS One 7:e42723. doi:10.1371/journal.pone.0042723
Yun J, Zhuang G, Ma A, Guo H, Wang Y, Zhang H (2012) Community structure, abundance, and activity of methanotrophs in the Zoige Wetland of the Tibetan Plateau. Microb Ecol 63:835–843. doi:10.1007/s00248-011-9981-x
Meena KK, Kumar M, Kalyuzhnaya MG, Yandigeri MS, Singh DP, Saxena AK, Arora DK (2012) Epiphytic pink-pigmented methylotrophic bacteria enhance germination and seedling growth of wheat (Triticum aestivum) by producing phytohormone. Antonie Van Leeuwenhoek 101:777–786. doi:10.1007/s10482-011-9692-9
López JC, Quijano G, Souza TS, Estrada JM, Lebrero R, Muñoz R (2013) Biotechnologies for greenhouse gases (CH4, N2O, and CO2) abatement: state of the art and challenges. Appl Microbiol Biotechnol 97:2277–2303. doi:10.1007/s00253-013-4734-z
Rani A, Porwal S, Sharma R, Kapley A, Purohit HJ, Kalia VC (2008) Assessment of microbial diversity in effluent treatment plants by culture dependent and culture independent approaches. Bioresource Technol 99:7098–7107. doi:10.1016/j.biortech.2008.01.003
Porwal S, Lal S, Cheema S, Kalia VC (2009) Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus. PLoS One 4:e4438. doi:10.1371/journal.pone.0004438
Kalia VC, Mukherjee T, Bhushan A, Joshi J, Shankar P, Huma N (2011) Analysis of the unexplored features of rrs (16S rDNA) of the genus Clostridium. BMC Genom 12:18. doi:10.1186/1471-2164-12-18
Bhushan A, Joshi J, Shankar P, Kushwah J, Raju SC, Purohit HJ, Kalia VC (2013) Development of genomic tools for the identification of certain Pseudomonas up to species level. Indian J Microbiol 53:253–263. doi:10.1007/s12088-013-0412-1
McDonald IR, Murrell JC (1997) The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol 63:3218–3224
McDonald IR, Kenna EM, Murrell JC (1995) Detection of methanotrophic bacteria in environmental samples with PCR. Appl Environ Microbiol 61:116–121
Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L (2008) Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol 190:3817–3823. doi:10.1128/JB.00180-08
Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME (2009) The expanding world of methylotrophic metabolism. Annu Rev Microbiol 63:477–499. doi:10.1146/annurev.micro.091208.073600
APHA-AWWA-WPCF (2005) Standard methods for the examination of water and waste water, 20th edn. American Public Health Association, Washington
Patt TE, Cole GC, Bland JA, Hanson RS (1974) Isolation and characterization of bacteria that grow on methane and organic compounds as sole sources of carbon and energy. J Bacteriol 120:955–964
Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from gram positive bacteria. Trends Genet 11:217–218
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11:37–50. doi:10.1111/j.1469-8137.1912.tb05611.x
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Henckel T, Friedrich M, Conrad R (1999) Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase and methanol dehydrogenase. Appl Environ Microbiol 65:1980–1990
Pfaffl M (2001) Development and validation of an externally standardised quantitative insulin-like growth factor-1 RT-PCR using LightCycler SYBR Green I technology. Rapid Cycle Real-Time PCR. Springer, Berlin-Heidelberg, pp 281–291. doi:10.1007/978-3-642-59524-0_30
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving sensitivity of progressive multiple sequence alignments through sequence weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Saitou N, Nei M (1987) The neighbor-joining method a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evol 39:783–791
Hiraishi A, Shimada K (2001) Aerobic anoxygenic photosynthetic bacteria with zinc-bacteriochlorophyll. J Gen Appl Microbiol 47:161–180. doi:10.2323/jgam.47.161
Raja P, Balachandar D, Sundaram SP (2008) Genetic diversity and phylogeny of pink pigmented facultative Methylotrophic bacteria isolated from the phyllosphere of tropical crop plants. Biol Fertil Soils 45:45–53. doi:10.1007/s00374-008-0306-2
Bodrossy L, Murrell JC, Dalton H, Kalman M, Puskas LG, Kovacs KL (1995) Heat-tolerant methanotrophic bacteria from the hot water effluent of a natural gas field. Appl Environ Microbiol 61:3549–3555
Joshi AA, Kanekar PP, Kelkar AS, Shouche YS, Vani AA, Borgave SB, Sarnaik SS (2008) Cultivable bacterial diversity of alkaline Lonar Lake, India. Microbial Ecol 55:163–172
Wang X, Sahr F, Xue T, Sun B (2007) Methylobacterium salsuginis sp. nov. isolated from sea water. Int J Syst Evol Microbiol 57:1699–1703. doi:10.1099/ijs.0.64877-0
Jenkins O, Bryom D, Jones D (1987) Methylophilus: a new genus of methanol utilizing bacteria. Int J Syst Bacteriol 37:446–448. doi:10.1099/00207713-37-4-446
Jiang H, Chen Y, Jiang P, Zhang C, Smith TJ, Murrell JC, Xing X-H (2010) Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J 49:277–288. doi:10.1016/j.bej.2010.01.003
Liessens J, Germonpre R, Kersters I, Beernaert S, Verstraete W (1993) Removing nitrate with a methylotrophic fluidized bed: microbiological water quality. J Am Water Works Assoc 85:155–161
Schmider F, Ottow JCG (1986) Characterization of denitrifying bacteria in the various compartments of a biological sewage plant. Arch Hydrobiol 106:497–512
Brusseau GA, Bulygina ES, Hanson RS (1994) Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol 60:626–636
Acknowledgments
The authors are thankful to the ICAR (Indian Council of Agricultural Research), India for providing the financial and research support. The authors also would like to acknowledge their respective institutional Directors.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors Kamlesh Kumar Meena, Snehasish Mishra and Manish Kumar should be regarded as the joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meena, K.K., Kumar, M., Mishra, S. et al. Phylogenetic Study of Methanol Oxidizers from Chilika-Lake Sediments Using Genomic and Metagenomic Approaches. Indian J Microbiol 55, 151–162 (2015). https://doi.org/10.1007/s12088-015-0510-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-015-0510-3