Abstract
Nowadays, breast cancer is the most prevalent and jeopardous disease in women after lung cancer. During the past few decades, a substantial amount of cancer cases have been reported throughout the world. Breast cancer has been a widely acknowledged category of cancer disease in women due to the lack of awareness. According to the world cancer survey report 2020, about 2.3 million cases and 685,000 deaths have been reported worldwide. As, the patient-doctor ratio (PDR) is very high; consequently, there is an utmost need for a machine-based intelligent breast cancer diagnosis system that can detect cancer at its early stage and cure it more efficiently. The plan is to assemble scientists in both the restorative and the machine learning fields to progress toward this clinical application. This paper presents SELF, a stacked-based ensemble learning framework, to classify breast cancer at an early stage from the histopathological images of tumor cells with computer-aided diagnosis tools. In this work, we use the BreakHis dataset with 7909 histopathological images and Wisconsin Breast Cancer Database (WBCD) with 569 instances for the performance evaluation of our proposed framework. We have trained several distinct classifiers on both datasets and selected the best five classifiers on the basis of their accuracy measure to create our ensemble model. We use the stacking ensemble technique and consider the Extra tree, Random Forest, AdaBoost, Gradient Boosting, and KNN9 classifiers as the base learners, and the logistic regression model as a final estimator. We have evaluated the performance of SELF on the BreakHis dataset and WBCD datasets and achieved testing accuracy of approximately 95% and 99% respectively. The result of the other performance parameters on the BreakHis and the WBCD datasets also showed that our proposed framework outperforms with F1-Score, ROC, and MCC scores.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
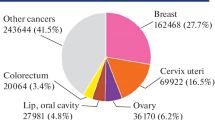
The proper and effective disease treatment with certain information is demanding and challenging in the area of bioinformatics and medical research [33]. As far as dangerous diseases are concerned, cancer is one of the most precarious diseases that occur due to the uncontrolled growth of abnormal cells in the human body. In cancer, the old cells do not decay and their growth gets uncontrolled which turned into the second most deadly disease in the world [11, 41]. Breast cancer is a kind of cancer that forms a mass of cells such as tissues on the breast and the continuous growth of cancer tissues start integrating into other normal human body parts [41]. Breast Cancer is the most prominent type of cancer among others as shown in Fig. 1 from the World Health Organization (WHO) in the year 2020. Most treatments that are offered to cancer patients depend on early diagnosis of cancer symptoms which affects the survival of the cancer patients. When a patient is diagnosed with a tumor, the very first step considered by doctors is identifying whether it is malignant or benign. The malignant type of tumor is cancerous whereas benign is non-cancerous. Therefore, differentiating tumor type is very important for a better and more effective cure and it is also required in the plan of treatment. Though, doctors require a reliable mechanism to differentiate these two types of tumors. In almost all countries, cancer is the root cause of death around eight million people annually. Every possible treatment plan for cancer patients requires a deep study of behavioral changes in cells; approximately 85% of breast cancer cases come from women [8, 10]. Moreover, most nations are in a developing phase where resources are limited and population growth is rapid and the doctors/experts and patients ratio is completely unbalanced. Therefore, it is very important to understand the disease’s behavior through automation so that effective and early treatment can be offered to the patients by accurate classification using machine learning techniques.
Recently, numerous diagnostic institutions, research centers, hospitals, as well as many websites provide a huge amount of medical diagnostic data. So, it is essential to classify and automate them to speed up disease diagnosis. Nowadays, machine learning techniques are extremely admired in most fields of data analysis, classification, and prediction [11]. These techniques are used to facilitate the analysis of these data and produce revolutionary information for medical society and play a vital role in the serious and sophisticated evaluation of various kinds of medical data. Several machine learning and data mining algorithms are widely used for the prediction of breast cancer and it is very important to select the most appropriate algorithms for the classification of breast cancer [24]. These learning techniques require a huge amount of historical data for learning and their prediction results depend on the learning that leads to enormous computing power [36]. Healthcare also believes in large amounts of data from diagnosis and pathology to drug discovery and epidemiology.
Machine learning algorithms can be more reliable, effective, and easier to provide a better solution for automatic cancer disease detection. The formulation of better learning algorithms has the ability to adopt new data independently and iteratively. The machine learning technique prerequisite is continuous learning from historical data. This learning helps machines to identify patterns available in data and produce reliable and informed results for decision-making tasks. The main goal of machine learning is to make a system learn without human intervention which helps to design an automated system for decision making. The tumor type identification is the essential phase to suggest a better treatment plan to a patient that increases the probability of their survival. The main cause of breast cancer disease is the presence of either a benign tumor or a malignant tumor.
A number of algorithms have been utilized in the literature to address breast cancer classification problems by considering histopathological images and may be symptoms like fatigue, headaches, pain, and numbness. Breast cancer can also be categorized by using ensemble-based classification techniques with an improved prediction rate. In this work, we considered Extra tree, Random Forest, AdaBoost, Gradient Boosting, and KNN9 (9-nearest neighbour) as base learners for the proposed stacking ensemble classification framework. Further, we evaluate the performance of the SELF using BreakHis and WBCD datasets [36, 42]. The main objective of this research work is to detect and categorize malignant and benign patients in a faster way and improve prediction accuracy along with other performance parameters. Figure 2 exhibits the structure of a Stacked-based ensemble model for the classification of cancer. Our contributions to this paper are summarized as:
-
1.
Studying the existing literature to identify the research gaps.
-
2.
Identifying appropriate dataset(s) on breast cancer because most of the existing works have considered the datasets with a limited number of images/instances. We have considered the BreakHis dataset with 7909 histopathological images and the WBCD dataset with 569 instances.
-
3.
Identifying the best-performing machine learning models for both datasets to create a stacked-based ensemble learning framework.
-
4.
Proposing SELF, a stacked-based ensemble learning framework, to classify breast cancer with better accuracy and evaluate the performance of the proposed framework on the testing dataset.
The rest of the paper is organized as follows: Sect. 2 discusses the related work in the area of breast cancer prediction and Sect. 3 discusses different machine learning algorithms used in this work. Section 4 provides the details of our proposed ensemble framework for breast cancer classification, and a detailed discussion on performance evaluation of the SELF is presented in Sect. 5. Finally, we concluded our work in Sect. 6 with further possible improvements.
2 Related works
This section presents the existing works related to breast cancer classification/prediction using the image and numerical datasets. The categorization of breast cancer is a kind of classification problem that needs the extraction of relevant features for classification. The amalgamation of medical science and artificial intelligence has great significance and several researchers have been working in the same domain and coming up with extraordinary outputs. Recently, numerous automatic models have been proposed in the literature for breast cancer classification using different machine learning (ML)/deep learning (DL)/ensemble learning (EL) approaches. In the literature, researchers have adopted a number of the existing ML/DL approaches such as K-nearest neighbor (K-NN), Support Vector Machines (SVM), Decision Tree(DT), Random forests (RF), Extra Tree (ET), ResNet 50, VGG16, and VGG19 and a number of ensemble learning approaches to create a better ensemble based classifier using existing machine learning and deep deep learning approaches for breast cancer classification. They have put their good efforts to improve the classification accuracy of their algorithms on different breast cancer datasets. We summarize some of the recent machine learning, deep learning, and ensemble learning techniques based on the breast cancer classification in Tables 1, 2, and 3 respectively.
From Tables 1, 2, and 3, we have made the following observations on the used datasets, learning methodologies/techniques and learning algorithms to address the breast cancer classification problem:
-
Most people in the world are commonly working on either publicly available benchmark datasets such as BreakHis, WBCD, BCW, CBCD, FCNC, BACH, and CBIS-DDSM ROI or hypothetical datasets created by themselves by collecting a smaller number of ultrasound images of the patient to address the problem of breast cancer classification/detection/prediction; the available datasets are either image datasets [2,3,4, 6, 9, 12, 14, 17, 18, 21, 22, 25,26,27,28,29,30,31, 34, 38, 43, 44] or numerical datasets [1, 5, 10, 16, 19, 32, 39, 40], and the researchers are working on them. The existing works on classification are using datasets with fewer sample images [1, 3, 5, 6, 16, 17, 19, 22, 26,27,28, 31, 32, 34, 39, 40, 43] that may not sufficient to train deep learning algorithms because the training process of a deep learning model requires a large amount of image data. The most frequently used datasets are the BreakHis [2, 4, 6, 9, 12, 14, 21, 25, 29, 30, 38, 44] and the WBCD [1, 5, 10, 16, 19, 32, 39, 40]. However, the WBCD dataset consists of only 569 or 699 instances with 32 features, while, the BreakHis dataset consists of 7909 images. Therefore, the BreakHis dataset and WBCD dataset can be the most suitable dataset with a sufficient number of breast cancer instances.
-
The researchers have adopted machine learning [1, 6, 10, 22, 27] or deep learning [2, 4, 12, 14, 17, 26, 29, 38, 43, 44] or ensemble learning [3, 5, 16, 18, 19, 21, 25, 28, 30,31,32, 34, 39, 40] techniques to address the breast cancer classification problem, and put their best efforts to improve the performance of their proposed approach(s). From Tables 1, 2, and 3, we observed that most of the existing works have mainly borrowed deep learning techniques to address the breast cancer classification problem because of image datasets [2, 4, 9, 12, 14, 17, 18, 21, 26, 29, 30, 38, 43, 43, 44]. The Convolutional Neural Network (CNN), a deep learning model, performs well on image datasets because it handles the entire feature engineering phase and extracts features from an image in an efficient way. However, CNN still faces obstacles because CNN training requires a large amount of training data with expensive computational resources which is time-consuming [35]. Other researchers found several machine-learning techniques to work with image datasets and adopted these techniques to address the breast cancer classification problem [6]. A machine learning model can be trained on a small dataset with less computation cost. On the other hand, an ensemble learning technique takes advantage of different machine learning/deep learning algorithms to build a powerful classifier; these techniques are mainly classified as bagging, boosting, and stacking. Several researchers have adopted different ensemble learning techniques to build an efficient breast cancer classifier. However, the existing ensemble-based breast cancer classifiers have been evaluated on either the WBCD dataset or the BreakHis dataset. The ensemble classifier with machine learning algorithms performs well on the WBCD dataset, while, the ensemble classifier with deep learning algorithms performs well on the BreakHis dataset. Therefore, we have an opportunity to take advantage of both ensemble learning techniques and machine learning algorithms to build efficient breast cancer classifiers with reduced computational cost on the BreakHis dataset.
-
The researchers have used different deep learning or machine learning algorithms to build automated breast cancer classifiers on publicly available benchmark datasets which are image datasets. The performance of CNN-based deep learning algorithms is good on image datasets because these algorithms are specially designed for them. On the other hand, some of the machine learning algorithms such as SVM, ANN, random forest, extra tree, and many more are efficiently working on image datasets and can be an alternative for CNN [6]. SVM is the most popular classification algorithm to separate the given data objects into multiple classes using an optimal hyperplane. On the other hand, the RF classifier utilizes the decision tree predictors by combining them into one and produces good results even without hyperparameter tuning [20]. The advantages of the random forest method are: efficiently works on unbalanced datasets and it is very fast. The other machine learning algorithms are also taken into account for breast cancer diagnosis using image classification [15, 37]. Therefore, machine learning techniques can be a good alternative to solve the breast cancer classification problem with limited computational resources.
In this work, we have compiled the findings of the recent approaches for breast cancer classification, and to the best of our knowledge, we found that there is still a scope for improvement on the existing breast cancer classification approaches. Therefore, we opt for an ensemble learning-based classifier with more effective machine learning algorithms to classify breast cancer with improved accuracy and reduced the false negative rate. We also analyzed that the performance of the proposed framework is slightly inferior when compared to the existing deep-learning-based ensemble classifiers on the BreakHis dataset, while, SELF is performing well with the WBCD dataset. Overall, the predicted outcome of the proposed work is at an acceptable level with minimal computational power.
3 Classification algorithms
This section discusses the major classification algorithms used in the proposed ensemble framework. Initially, we attempted different classification algorithms on the breast cancer data to achieve better accuracy, and after that, we created our final ensemble model by using the top-performing classification models. We have trained several distinct classifiers on the training dataset and selected the best five classification models among them on the basis of their accuracy measures to create our resultant ensemble model. A detailed description of the trained classification models is given in the following subsections.
3.1 Random forest (RF)
The Random Forest (RF) algorithm [20] is a well-known supervised learning algorithm that addresses both classification and regression problems in machine learning. This algorithm uses an ensemble learning technique, i.e., bagging, which uses multiple decision trees on different subsets of the given dataset. The resultant accuracy is calculated by taking the average accuracy of all decision trees which gives an improved predictive accuracy. The RF classifier is extensively used to generate a large number of trees as a forest and the quantity of trees introduced in the forest affects its accuracy. As a result, the number of trees created in the forest impact RF accuracy, because the more trees in the forest, the higher the accuracy, and vice versa. Furthermore, while generating a forest of trees, RF employs batching and randomness in the creation of each tree. The nodes in the decision tree branch are decided by an entropy measure which is described by the following equation:
where \(p_i\) is the relative frequency of the ith class and C is total number of classes.
3.2 K-nearest neighbor (kNN)
The K-Nearest Neighbor (kNN) algorithm [23] is a simple and easy-to-implement algorithm that can be used for both classification and regression problems. As the name suggests, this algorithm considers k-nearest neighbors into account during classification and regression. This algorithm is a distance-based algorithm that calculates the distance between the new data point with all the existing data points in the training set and then chooses the k closest data points to the new instance from a set. Finally, the data is classified using the majority class of the k data points chosen. In this work, we have chosen the value of k as 9 by using cross-validation and Euclidean distance measure to get better results. The distance (d) between the two data points is calculated by:
where (\(x_1\),\(y_1\)) and (\(x_2\),\(y_2\)) are any two data points in the given data set.
3.3 Extra tree (ET)
The Extra Tree (ET) algorithm [1] is used to solve both classification and regression problems. Like the RF algorithm, ET algorithm randomly selects subsets of features from the dataset and trains a decision tree on them. This algorithm generates multiple trees and combines the predictions from all decision trees to get the resultant accuracy of the classification algorithm. This algorithm differs from the random forest in two ways: (1) it does not support bootstrap observations and selects samples without replacement. (2) It uses random splits, i.e., randomness that is generated through random splits of all observations rather than bootstrapping sampling. Here, we have also used the entropy measure that can be calculated by Eq. (1).
3.4 Adaptive boosting (AdaBoost)
The Adaptive Boosting (AdaBoost) [7] classifier is a meta-estimator classifier that begins by fitting a classifier on the initial dataset. It fits multiple copies of the classifier on the same dataset while modifying a large number of ineffectively ordered samples so that subsequent classifiers focus more on difficult situations. The idea behind the boosting technique is to associate the multiple weak classifiers in series to build a strong classifier. We build the first classifier on the training dataset and then build the second classifier to rectify the errors made by the first model. This process will continue until the error gets minimized. A boosted classifier is defined as:
where each \({\displaystyle f_{t}}\) is a weak learner that takes an object \({\displaystyle x}\) as input and returns a value indicating the class of the object.
3.5 Gradient boosting (GB)
The Gradient Boosting algorithm [13] is one of the most powerful and widely used techniques in machine learning. Unlike AdaBoost, the base estimators in gradient boosting are fixed, i.e., the Decision Stump. This algorithm is also used to solve both classification and regression problems. It is a numerical optimization approach for determining the best additive model for minimizing the loss function. As a consequence, the GB approach constructs a new decision tree that minimizes the loss function ideally at each step. In regression, the process starts with the first estimate, which is usually a decision tree that minimizes the loss function, and then a new decision tree is fitted to the current residual and added to the previous model to update the residual at each step. This is a step-by-step procedure, which implies that the decision trees used to create the model in previous phases are not changed in subsequent ones. By fitting decision trees to the residuals, the model is enhanced in cases where it does not perform well. Predicted residuals at each leaf can be calculated as follows:
4 SELF: stacked-based ensemble learning framework
This section presents the SELF for breast cancer classification based on the stacking technique. Stacking is an ensemble machine-learning technique that learns how to combine the predictions of well-performing machine-learning models. The following subsections present a detailed description of the dataset, model selection, hyperparameter tuning, phases, and complexity analysis of the SELF.
4.1 Datasets description
In this work, we considered the publicly available BreakHis dataset with a total of 7909 images collected from 82 patients [36] and the WBCD dataset with 569 instances collect from 8 different groups [42]. BreakHis is a Breast Cancer Histopathological database that composes microscopic images of breast tumor tissue with four magnifying factors: 40×, 100×, 200×, and 400×. This dataset contains 2480 benign and 5429 malignant samples (700 × 460 pixels, 3-channel RGB, 8-bit depth in each channel, PNG format). Table 4 exhibits the Images distribution in the BreaKHis dataset, and it can be observed that there is a huge class imbalance in the dataset. The number of malignant images is almost double of benign images. Table 5 exhibits the classification of Malignant images, divided into Ductal Carcinoma (DC), Mucinous Carcinoma (MC), Lobular Carcinoma (LC), and Papillary Carcinoma (PC). Table 6 exhibits the classification of the Benign images, divided into Tubular Adenoma (TA), Adenosis (A), Fibroadenoma (F), and Phyllodes Tumor (PT). We applied different data augmentation techniques, scaling, rotation, flipping, shuffling, zooming, and shearing, to deal with the class imbalance problem. In this work, we have classified the images into two classes, i.e., benign and malignant, which is a supervised learning problem. On the other hand, the WBCD dataset consists of a total of 569 instances with 32 features calculated from a digitized image of a Fine Needle Aspirate (FNA) of a breast mass; the distribution of the malignant and benign tumors is approximately 37% and 63% respectively. Table 7 exhibits the description of the WBCD dataset.
4.2 Model selection
Figure 3 exhibits the proposed framework of the ensemble model for breast cancer prediction. We have taken the breast cancer datasets from Kaggle and preprocessed them to handle the class imbalanced problem. After that, we split the processed dataset into 80% for training and 20% for testing. Our proposed framework is based on stacking ensembling techniques which are deferred from bagging and boosting. In this work, we have trained a total of 9 base learners on a training dataset; these base learners are Random Forest, Support Vector Machine, K-Nearest Neighbor, Extra Tree classifier, AdaBoost, Stochastic Gradient Descent, Gradient Boosting, Multilayer Perceptron, and Classification and Regression Trees (CART). Table 8 exhibits the training and testing accuracy of different base-learners on the BreakHis Dataset. Tables 9 and 10 exhibit the performance of base-learners on the BreakHis and the WBCD testing datasets respectively, out of these, we have selected the top-5 best performing common base-learners to create our final meta-model based on the stacked ensemble. The selected base learners are the Extra Tree classifier, Random Forest, AdaBoost, Gradient Boosting, and 9-Nearest Neighbor (KNN9).
4.3 Hyperparameter tuning
In order to improve the performance of our adopted classifiers, we do the hyperparameter tuning for different classifiers that results in improved performance of the proposed framework. For instance, we have evaluated numerous values of ‘n’ estimators, i.e., 50, 100, 500, 1000, 1500, and 2000 for Random Forest Classifier, and achieved better accuracy for the value of ‘n’ estimator as 500, and employed entropy instead of Gini criterion. Further, we ran with several choices of ‘k’ for our kNN model and achieved better accuracy at k = 9. For the extra tree classifier, we investigate the numerous values of the ’n’ estimators, i.e., 50, 100, 500, 1000, 1500, and 2000, and achieve better accuracy for the value of ‘n’ estimator as 500. The selected hyperparameters with there best values and accuracy for both dataset are given in Table 11.
4.4 SELF
Stacking, also known as Super Learning, is an ensemble strategy that includes training a “meta-learner” using a variety of classification models. The goal of stacking is to bring together diverse groups of strong learners to improve the accuracy of the resultant model. Our proposed model works in three phases: data preprocessing, feature extraction, and model creation and evaluation. Algorithm 1 exhibits the overall working of the proposed stacked classifier for breast cancer prediction.

-
Data preprocessing phase In this phase, we handle the imbalance of images on the BreakHis dataset by applying the resizing of given images and image augmentation techniques. The techniques used for data augmentation are scaling, rotation, flipping, shuffling, zooming, and shearing. At the last, we create an array of training images.
-
Feature extraction phase After data preprocessing, we extracted 10 features such as pixel features, gobor features, and edge features, and remove noise from the images in this phase.
-
Model building and evaluation phase Figure 3 exhibits the working of the proposed framework. After feature extraction, we prepared our final ensemble model for breast cancer prediction. In this phase, we first train the different machine learning models on our training dataset using different features, then, we evaluated all trained models on the testing dataset. We utilized the power of multiple base learners to create a meta-learner classifier using the stacking technique. The architecture of a stacking model involves two or more base learners, often referred to as level-0 learners, and a meta-learner referred to as a level-1 learner that combines the predictions of the base learners. We train the level-0 learners on the training data whose predictions are compiled; the level-1 learner learns how to best combine the predictions of the trained base-learners. We trained multiple base-learners using 10-fold cross-validation on our training data at level-0 and the output of base-learners is used as input to the meta-learner at level-1. The meta-learner trained on the predictions made by the different base-learners on out-of-sample data. Finally, we build the resultant meta-learner that interprets the prediction of base-learners in a very smooth manner, and then uses a linear model, i.e., logistic regression, to handle the classification problem. The resultant meta-learner is validated on test data and its performance has been evaluated on the basis of several performance metrics.
4.5 Complexity analysis of SELF
Table 12 exhibits the training complexity of the different machine-learning models used in our proposed ensemble model. Let the training dataset have n number of training examples, m number of features, k number of neighbors, p number of decision trees (or stumps), and d is the maximum depth of a decision tree. Figure 4 exhibits the inferential flowchart of the proposed SELF classifier in which we have used the kNN, Random Forest, Extra Tree, Gradient Boosting, and Adaptive Boosting classifiers as base-learners at level-0 and the Logistic Regression classifier is used as a meta-learner at level-1 and it produces the final prediction. The time complexity of stacked classifiers actually depends on the training pattern of base learners. The base learners can be trained in either a serial or parallel manner that influences the complexity of the resultant stacked classifier. Suppose, we have the total N base-learners, the complexity of the ith base-learner is \(C_i\), and the complexity of the meta-learner is \(C_{meta}\) .
-
If we train the base-learner in a serial manner then the complexity of the stacked classifier is given as:
$$\begin{aligned} Complexity\_serial = \sum _{i=1}^{N} (C_i) + C_{meta} \end{aligned}$$(5) -
If we train the base-learner in a parallel manner then the complexity of the stacked classifier is given as:
$$\begin{aligned} Complexity\_parallel = \max _{i=1}^{N} (C_i) + C_{meta} \end{aligned}$$(6)
Therefore, we can compute the training complexity of the proposed model by using the equation either (5) or (6).
5 Results and discussion
This section presents the evaluation of the proposed framework along with the detailed findings. We have evaluated the performance of the SELF using several performance metrics on BreakHis and WBCD datasets and compared them with the existing works based on the accuracy, sensitivity, precision, F1-Score, ROC, and MCC. Now, we first present a detailed description of the various performance metrics, then we compare the SELF with the existing works.
5.1 Performance metrics
Figure 5 exhibits a generalized confusion matrix that helps to define the different performance parameters. The performance metrics accuracy, sensitivity, precision, F1-Score, ROC, and MCC are defined as follows:
-
Accuracy: It is one of the most often used measures for assessing the performance of a classifier. It is expressed as a proportion of properly classified samples and defined as:
$$\begin{aligned} Accuracy = \frac{TP + TN}{TP + TN + FP + FN} \end{aligned}$$(7) -
Precision: It describes the ratio of actual positive instances out of predicted positive instances by a potential classifier, and is defined as:
$$\begin{aligned} Precision = \frac{TP}{TP + FP} \end{aligned}$$(8) -
Sensitivity or true positive rate (TPR) or recall: It describes the potential for a classifier to properly predict a favorable outcome in the presence of disease, and is defined as:
$$\begin{aligned} Sensitivity = \frac{TP}{TP + FN} \end{aligned}$$(9) -
Specificity or true negative rate (TNR): It is a classifier’s likelihood of predicting a negative outcome when there is no sickness, and is computed as:
$$\begin{aligned} Specificity = \frac{TN}{ TN + FP} \end{aligned}$$(10) -
F1-score: It is regarded as the weighted average of precision and recall (or harmonic mean). The score of 1 is considered the best model, while, 0 is considered the worst model. The TNs are not taken into account in F-measures. The F1-score is computed as:
$$\begin{aligned} F1{-}Score = 2 * \frac{Precision * Recall}{Precision + Recall} \end{aligned}$$(11) -
Mathews correlation coefficient (MCC) MCC, a correlation coefficient between predicted classes and actual classes, is used for binary classification. The value of MCC ranges between \(-\)1 to +1, where +1 indicates the best prediction result, 0 indicates no better than the random prediction, and \(-\)1 indicates a complete disagreement between predicted and actual results. MCC is defined as:
$$\begin{aligned} MCC = \frac{TP * TN - FP * FN}{\sqrt{(TP + FP)*(TP + FN)*(TN + FP)*(TN + FN)}} \end{aligned}$$(12) -
Area under the curve-receiver operating characteristics (AU-ROC) AU-ROC is one of the most important and extensively used performance metrics for classification problems at various threshold settings. ROC represents a probability curve while AUC represents the degree or measure of separability. The higher value of AUC represents a better predictive model, and the ROC curve is plotted against the True Positive Rate (TPR) versus False Positive Rate (FPR), where TPR is represented in Y-axis and FPR is represented in X-axis. The AUC-ROC is defined as:
$$\begin{aligned} AU{-}ROC = \frac{1}{2}*\left( \frac{TP}{TP + FN} + \frac{TN}{TN + FP}\right) \end{aligned}$$(13)
where TP, FP, TN, and FN denote True Positive, False Positive, True Negative, and False Negative respectively.
5.2 Comparison of SELF with different base-learners
We compared our proposed classifier with different baseline models on BreakHis and WBCD datasets. Figures 6 and 7 exhibit the comparison of our proposed model with the different baseline models for breast cancer prediction on BreakHis and WBCD datasets respectively. From the Figures, we observed that our proposed model performs better in comparison to the baseline machine learning models and other existing models, and gives the approximate 95% and 99% of accuracy on BreakHis and WBCD testing datasets respectively. Tables 13 and 14 exhibit performance comparisons of the SELF with respect to the base learners on BreakHis and WBCD datasets respectively. From Table 13, we can also observe that with respect to other performance parameters, our proposed classifier has the highest F1-Score, ROC, and MCC scores with values of 94.17%, 89.41%, and 80.81% respectively. With respect to sensitivity, our proposed classifier has achieved the second-highest score of 95.96% whereas the random forest and extra tree classifiers have the highest sensitivity score of 97.11%. On the other hand, from Table 14, we can also observe that with respect to other performance parameters, our proposed classifier has the highest sensitivity, F1-Score, and MCC scores with values of 99.09%, 99.09%, and 97.45% respectively. With respect to precision score, our proposed classifier has achieved the second-highest score of 99.09% whereas the random forest has the highest precision score of 100.00%. Figure 8 exhibits the ROC curves for the best-performing models on BreakHis to analyze the developed models; this curve displays the classifier’s diagnostic skills. The closer the area value of the ROC curve is to one, the greater the model’s diagnostic capabilities. From the Figure, we observed that the area covered under our proposed model is the highest, i.e., 0.984. Similarly, Fig. 9 exhibits the comparison of precision, sensitivity, specificity, and ROC of the proposed classifier with different machine learning models. From these figures, we observe that our proposed ensemble classifier outperforms in most of the cases in comparison to other ML models.
5.3 Comparison of SELF with the existing breast cancer classifiers
We have also compared the proposed SELF with the other existing classifiers for breast cancer classification on BreakHis and WBCD datasets. Figure 10 exhibits the comparison of the SELF model with the existing classifiers on the BreakHis dataset. We observed that our ensemble-based classifier achieved the highest accuracy of 94.35% on the BreakHis dataset. However, Zou et al. [44] have proposed a deep learning based model, while Karthik et al. [21] have proposed a deep learning based ensemble for classification, and have achieved better accuracy than the SELF. On the other hand, the SELF has outperformed than other deep learning based ensemble classifiers as shown in Fig. 11. Figure 12 exhibits the comparison of the SELF with the various existing ensemble-based classifiers on the WBCD dataset. From Fig. 12, it is observed that the performance of SELF is better than the existing machine learning based ensemble classifiers on the WBCD dataset.
6 Conclusion
In this work, we proposed SELF, a stacked-based ensemble learning framework, using the five best-performing machine learning algorithms, i.e., Extra tree, Random Forest, AdaBoost, Gradient Boosting, and KNN9, to classify the breast cancer on BreakHis and WBCD datasets. The proposed classifier has shown great potential to increase the classification accuracy by improving the accuracy, precision, sensitivity, specificity, and F1-Score with values of 94.35%, 92.45%, 95.96%, 82.87%, and 94.17% respectively on the BreakHis dataset. After analysing the overall performance, we found that the SELF is performing better than several existing classifiers on the BreakHis dataset. Similarly, we have also evaluated the SELF on the WBCD dataset and analysed that it also performs well with improved accuracy of 99% approximately. Further, this work can be extended by including different optimization techniques in the proposed framework to enhance classification performance.
References
Abbas S, Jalil Z, Javed AR, Batool I, Khan MZ, Noorwali A, Gadekallu TR, Akbar A (2021) BCD-WERT: a novel approach for breast cancer detection using whale optimization based efficient features and extremely randomized tree algorithm. PeerJ Comput Sci 7:e390
Agarwal P, Yadav A, Mathur P (2022) Breast cancer prediction on BreakHis dataset using deep CNN and transfer learning model. In: Data engineering for smart systems. Springer, Berlin, pp 77–88
Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A (2020) Dataset of breast ultrasound images. Data Brief 28:104863
Al Rahhal MM (2018) Breast cancer classification in histopathological images using convolutional neural network. Int J Adv Comput Sci Appl 9(3):64
Alhayali RAI, Ahmed MA, Mohialden YM, Ali AH (2020) Efficient method for breast cancer classification based on ensemble Hoffeding tree and Naïve Bayes. Indones J Electr Eng Comput Sci 18(2):1074–1080
Alqudah A, Alqudah AM (2022) Sliding window based support vector machine system for classification of breast cancer using histopathological microscopic images. IETE J Res 68(1):59–67
Assegie TA, Tulasi RL, Kumar NK (2021) Breast cancer prediction model with decision tree and adaptive boosting. IAES Int J Artif Intell 10(1):184
Boyle P, Levin B et al (2008) World cancer report 2008. IARC Press, International Agency for Research on Cancer
Burçak KC, Baykan ÖK, Uğuz H (2021) A new deep convolutional neural network model for classifying breast cancer histopathological images and the hyperparameter optimisation of the proposed model. J Supercomput 77(1):973–989
Chudhey AS, Goel M, Singh M (2022) Breast cancer classification with random forest classifier with feature decomposition using principal component analysis. In: Advances in data and information sciences. Springer, Berlin, pp 111–120
Culjak I, Abram D, Pribanic T, Dzapo H, Cifrek M (2012) A brief introduction to OpenCV. In: Proceedings of the 35th international convention MIPRO. IEEE, pp 1725–1730
de Matos J, de Souza Britto A, de Oliveira LE, Koerich AL (2019) Texture CNN for histopathological image classification. In: IEEE 32nd international symposium on computer-based medical systems (CBMS). IEEE, pp 580–583
Deif M, Hammam R, Solyman A (2021) Gradient boosting machine based on PSO for prediction of leukemia after a breast cancer diagnosis. Int J Adv Sci Eng Inf Technol 11(2):508–515
Deniz E, Şengür A, Kadiroğlu Z, Guo Y, Bajaj V, Budak Ü (2018) Transfer learning based histopathologic image classification for breast cancer detection. Health Inf Sci Syst 6(1):1–7
George YM, Zayed HH, Roushdy MI, Elbagoury BM (2013) Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst J 8(3):949–964
Ghiasi MM, Zendehboudi S (2021) Application of decision tree-based ensemble learning in the classification of breast cancer. Comput Biol Med 128:104089
Golatkar A, Anand D, Sethi A (2018) Classification of breast cancer histology using deep learning. In: International conference image analysis and recognition. Springer, Berlin, pp 837–844
Hekal AA, Moustafa HE-D, Elnakib A (2022) Ensemble deep learning system for early breast cancer detection. Evolut Intell. https://doi.org/10.1007/s12065-022-00719-w
Jabbar MA (2021) Breast cancer data classification using ensemble machine learning. Eng Appl Sci Res 48(1):65–72
Jackins V, Vimal S, Kaliappan M, Lee MY (2021) AI-based smart prediction of clinical disease using random forest classifier and Naive Bayes. J Supercomput 77(5):5198–5219
Karthik R, Menaka R, Siddharth M (2022) Classification of breast cancer from histopathology images using an ensemble of deep multiscale networks. Biocybern Biomed Eng 42:963–976
Kaymak S, Helwan A, Uzun D (2017) Breast cancer image classification using artificial neural networks. Procedia Comput Sci 120:126–131
Khorshid SF, Abdulazeez AM (2021) Breast cancer diagnosis based on k-nearest neighbors: a review. PalArch’s J Archaeol Egypt Egyptol 18(4):1927–1951
Khourdifi Y, Bahaj M (2018) Applying best machine learning algorithms for breast cancer prediction and classification. In: International conference on electronics, control, optimization and computer science (ICECOCS). IEEE, pp 1–5
Lee S-J, Tseng C-H, Yang H-Y, Jin X, Jiang Q, Pu B, Hu W-H, Liu D-R, Huang Y, Zhao N (2022) Random RotBoost: an ensemble classification method based on rotation forest and AdaBoost in random subsets and its application to clinical decision support. Entropy 24(5):617
Liu M, Hu L, Tang Y, Wang C, He Y, Zeng C, Lin K, He Z, Huo W (2022) A deep learning method for breast cancer classification in the pathology images. IEEE J Biomed Health Inform 26:5025–5032
Liu Y, Ren L, Cao X, Tong Y (2020) Breast tumors recognition based on edge feature extraction using support vector machine. Biomed Signal Process Control 58:101825
Mahesh T, Vinoth Kumar V, Vivek V, Karthick Raghunath K, Sindhu Madhuri G (2022) Early predictive model for breast cancer classification using blended ensemble learning. Int J Syst Assur Eng Manag. https://doi.org/10.1007/s13198-022-01696-0
Mohapatra P, Panda B, Swain S (2019) Enhancing histopathological breast cancer image classification using deep learning. Int J Innov Technol Explor Eng 8:2024–2032
Nakach F.-Z, Zerouaoui H, Idri A (2022) Deep hybrid AdaBoost ensembles for histopathological breast cancer classification. In: World conference on information systems and technologies. Springer, pp 446–455
Nanglia S, Ahmad M, Khan FA, Jhanjhi N (2022) An enhanced predictive heterogeneous ensemble model for breast cancer prediction. Biomed Signal Process Control 72:103279
Naseem U, Rashid J, Ali L, Kim J, Haq QEU, Awan MJ, Imran M (2022) An automatic detection of breast cancer diagnosis and prognosis based on machine learning using ensemble of classifiers. IEEE Access 10:78242–78252
Park SH, Han K (2018) Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 286(3):800–809
Rezazadeh A, Jafarian Y, Kord A (2022) Explainable ensemble machine learning for breast cancer diagnosis based on ultrasound image texture features. Forecasting 4(1):262–274
Seo H, Brand L, Barco LS, Wang H (2022) Scaling multi-instance support vector machine to breast cancer detection on the BreakHis dataset. Bioinformatics 38(Supplement-1):i92–i100
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2015) A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng 63(7):1455–1462
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2015) A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng 63(7):1455–1462
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2016) Breast cancer histopathological image classification using convolutional neural networks. In: International joint conference on neural networks (IJCNN). IEEE, pp 2560–2567
Srinivas T, Karigiri Madhusudhan AK, Arockia Dhanraj J, Chandra Sekaran R, Mostafaeipour N, Mostafaeipour N, Mostafaeipour A (2022) Novel based ensemble machine learning classifiers for detecting breast cancer. Math Probl Eng. https://doi.org/10.1155/2022/9619102
Talatian Azad S, Ahmadi G, Rezaeipanah A (2021) An intelligent ensemble classification method based on multi-layer perceptron neural network and evolutionary algorithms for breast cancer diagnosis. J Exp Theor Artif Intell 1–21
Wild CP, Stewart BW, Wild C (2014) World cancer report 2014. World Health Organization, Geneva
William H (2018) Breast cancer wisconsin (original) data set
Zerouaoui H, Idri A (2022) Deep hybrid architectures for binary classification of medical breast cancer images. Biomed Signal Process Control 71:103226
Zou Y, Zhang J, Huang S, Liu B (2022) Breast cancer histopathological image classification using attention high-order deep network. Int J Imaging Syst Technol 32(1):266–279
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jakhar, A.K., Gupta, A. & Singh, M. SELF: a stacked-based ensemble learning framework for breast cancer classification. Evol. Intel. 17, 1341–1356 (2024). https://doi.org/10.1007/s12065-023-00824-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12065-023-00824-4