Abstract
This paper proposes an ensemble deep learning system for the early detection of breast cancer. Unlike traditional ensemble learning that processes the whole image, the proposed system processes only the Suspected Nodule Regions (SNRs), extracted using an optimal dynamic thresholding method, where the threshold varies corresponding to the details of each input image. The SNRs affords better performance and offer the ability to detect small size nodules. The ensemble is composed of four transfer learning Convolutional Neural Networks (CNNs) (i.e., AlexNet, ResNet-50, ResNet-101, and DenseNet-201). A binary Support Vector Machine (SVM) follows each CNN model to provide either a malignant or a benign score. To obtain the final system decision, the first-order momentum is derived over the four binary outputs of the ensemble CNNs and the final decision is guided by the four CNNs’ training accuracies. The proposed ensemble fusion scheme, tested on Region of Interest (ROI) images, using the public digital medical images Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM), achieves an accuracy of 94% to distinguish between Malignant (M) and Benign (B) classes and of 95% to distinguish between Malignant Mass (MM) and Benign Mass (BM) nodules. Comparison with related methods on the same data confirms the accuracy and advantages of the proposed ensemble system.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer is an epidemic health care problem all over the world, accounting for 627,000 women cancer deaths, according to the World Health Organization (WHO) [10, 28]. The standard clinical imaging protocol is mammography, to detect and treat cancer or pre-cancer patients [20]. However, a mammogram requires high skills in assessing the resulting images, especially in early cases. Computer Aided Detection (CAD) systems have been developed to reduce the workload and improve the detection accuracy of doctors and experts [22, 25, 32].
Machine and deep learning and artificial intelligence show a great success in solving medical problems [1, 14]. Recently, Convolutional neural networks (CNNs) show impressive performance in the field of pattern recognition, classification [32], object detection [29], and disease diagnosis [2, 7, 33], and more specifically in the field of breast cancer detection [3, 6, 8, 18, 24, 27, 30, 31], and [12]. For example, Lévy et al. [18] applied GoogLeNet to classify benign and malignant breast masses, using cropped Digital Database for Screening Mammography (DDSM) dataset, to achieve an accuracy of 92.9%. Yi et al. [31] used GoogleNet to classify cropped DDSM mammograms as benign or malignant with an accuracy of 85%. Chen et al. [8] used a fine-tuned ResNet model to classify ROI CBIS-DDSM images, into benign or malignant, achieving an accuracy of 93.15%. Xi et al. [30] used VGGNet to classify ROI CBIS-DDSM data into mass or calcification, achieving an accuracy of 92%. Castro et al. [6] used a CNN model to classify full mammogram mass nodules into benign or malignant, achieving a sensitivity of 80% on CBIS-DDSM database. Tsochatzidis et al. [27] fine-tuned the ResNet-101 model, achieving an accuracy of 75.3% on CBIS-DDSM database, to classify mass nodules into benign and malignant. Ragab et al. [24] achieved an accuracy of 87.2%, using Alexnet model and ROI CBIS-DDSM database, to classify mass nodules into benign and malignant. Ansar et al. [3] achieved an accuracy of 74.5% using MobileNet model and ROI CBIS-DDSM data, to classify mass nodules into benign and malignant. Hekal et al. [12] used AlexNet and ROI CBIS DDSM data to classify tumor like regions into benign and malignant, achieving an accuracy of 95%.
Although efficient methods for breast cancer detection were presented, more advances should be investigated to improve the accuracy of breast cancer detection. This paper develops a system for the early detection of breast cancer using the ROI CBIS-DDSM database [17]. Table 1 shows typical examples of the ROI CBIS-DDSM database, containing two categories, i.e., malignant and benign, and two types of nodules, i.e., mass and calcification. The main features/contributions of this work are as follows:
-
The proposed ensemble learning processes SNRs instead of the ROI images, achieving four-fold advantages: (i) ability to detect smaller size nodules within the SNRs, (ii) ability to help the model to focus only on the part to be classified (tumors), (iii) eliminate the overhead of processing the whole ROI image, and (vi) improve the detection accuracy.
-
The proposed ensemble learning applies transfer learning with a shallow classifier (SVM), achieving three-fold advantages: (i) eliminate the need for big data for training, (ii) transports the weighs of the convolutional layers without training, and (iii) eliminate the need to design and build new CNN models.
-
The proposed system applies a simple first-order momentum, guided by the achieved models’ training accuracies, to fuse the binary outputs of the ensemble, which further improves the accuracy of the system
-
The proposed system achieves superior performance over the state-of-the-art methods on the challenging standard ROI CBIS-DDSM dataset [17]
The rest of this paper is as follows: Sect. 2 shows the materials and methods; Sect. 3 explores the findings and relevant discussions; Finally, Sect. 4 concludes the paper.
2 Research methods
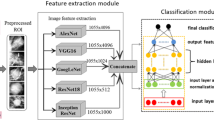
The proposed ensemble system (Fig. 1) is based on four processing steps: extracting suspected regions (SNR image), ensemble learning, shallow classification, decision fusion and final diagnosis. This section illustrates each of these steps.
2.1 Extracting suspected nodule regions (SNRs)
The proposed system extracts the SNRs from the ROI images based on an automated Otsu thresholding [21], where the threshold is dynamic and corresponds to each input ROI image. The proposed Otsu thresholding method processes a smoothed version of the input ROI image with a Gaussian kernel of a zero mean and a variance, which is selected to equal four to suppress the high frequency noise. Optimal Otsu threshold is estimated by minimizing intra-class intensity variance on the image histogram [9, 26]. Figure 2 illustrates the steps of extracting the SNR image. The SNR estimation algorithm can be summarized as in Algorithm I:

The main advantages of SNR extraction are the ability to detect smaller size nodules within the SNRs, less algorithmic overhead, and the improvement in the detection accuracy.
2.2 Ensemble learning
The ensemble is composed of four pretrained CNN networks: AlexNet, ResNet-50, ResNet-101, and DenseNet-201. We select these networks since they are more popular for data classification, especially for breast tumor classification (e.g., [24] and [12] used AlexNet [8] and [27] used ResNet, and [19] used DenseNet).
The input to the ensemble is the standard resized SNR image (i.e., 227 \(\times\) 227 for AlexNet, and 224 \(\times\) 224 for ResNet-50, ResNet-101, and DenseNet-201 (see Table 2). AlexNet [15] contains five convolution layers (conv), three max pooling layers, and three fully connected layers (Fc6, Fc7 ,and Fc8) (see Fig. 3). Each convolutional layer consists of convolutional filters and a nonlinear activation function ReLU. ResNet [27] is an abbreviation for Residual Network. The basic idea of a ResNet model is to skip blocks of convolutional layers by using shortcut connections. ResNet-50 contains 49 convolution layers, one max pooling layer, one average pooling layer and one fully connected layer (see Fig. 4). ResNet-101 contains 100 convolution layers, one max pooling layer, one average pooling layer, and one fully connected layer (see Fig. 5). DenseNet [13] is based on residual learning like ResNet. DenseNet201 contains 200 convolution layers, four max pooling layer, one average pooling layer, and one fully connected layer (see Fig. 6).
In the proposed system, transfer learning is adopted to decrease the training overhead. Therefore, the weights of the convolutional layers of the pretrained models are transferred without training, and only the fully connected layers are trained using the ROI CBIS-DDSM data. To apply transfer learning of the CNN models, the last fully connected layer of each pretrained model (FC8 layer in AlexNet or FC1000 layer in ResNet50, ResNet-101, and DenseNet-201) is replaced by a shallow classifier (namely, a (Supported Vector Machine) SVM classifier). The vectors of activities of the FC7 layer in AlexNet or the flatten layer (just before FC1000) in ResNet50, ResNet-101, and DenseNet-201, represent the feature descriptor of the input ROI CBIS-DDSM image. Features are further normalized between 0 and 1 before being fed to the input of the SVM classifier.
2.3 Shallow classifier
To classify the ROI images, a SVM classifier, with a binary kernel, is used, to account for the variability of the classes. The supported vector machine has the advantage of less risk of over-fitting [4, 16]. In addition, it has been repeatedly used in the literature to solve this problem (breast cancer classification) [12, 24]. The idea of SVM is to formulate an effective way of learning by separating hyper planes in a high dimensional feature space [11, 23]. The input of the classifier is the vectors of activities (FC7 in AlexNet or flatten layer in ResNet-50, Resnet-101, and DenseNet-201) and the output is the binary classification of the input image (e.g., Benign or Malignant).
2.4 Fusion
A binary Support Vector Machine (SVM) follows each CNN model to provide either a binary one, e.g., malignant class, or a binary zero, e.g., benign class. To obtain the final system decision, the first-order momentum is derived over the four outputs of the ensemble, taking into account the network training accuracies, following Algorithm II (see Fig. 7 for typical examples).

Four typical examples illustrating the proposed fusion of the four networks’ SVM binary outputs (left), multiplied by the accuracy of each network (right): If first-order momentum (mean) \(>0.5\), then decide class 1. If mean < 0.5, then decide class 0. If mean \(=\) 0.5, a tie is reached and the system choose the class of the larger sum of accuracies
2.5 Performance evaluation
To test the proposed system, we used six standard metrics to evaluate the system performance, i.e., Accuracy (ACC), Sensitivity (SEN), Specificity (SPE), Positive Predictive Value (PPV), Negative Predictive Value (NPV), and F1 score (FSC), defined as follows [34]:
3 Results and discussions
This section explains, in details, the collected database, the experimental setup, and the results and related discussions.
3.1 Collected database (CBIS-DDSM)
To test the proposed CAD system, the ROI CBIS-DDSM [17] database is used, a standardized version of the Digital Database for Screening Mammography (DDSM) [5]. It contains 3549 ROI mammogram images, with 1852 calcification and 1697 mass images (typical examples are shown in Table 1).
3.2 Experimental setting
CBIS-DDSM ROI dataset is used to test and evaluate the proposed system. The data has been divided randomly into training set (70%) and testing set (30%), in order to train and test the proposed system. The Bayesian optimizer is used to minimize the binary cross entropy function through the training process of the deep transfer learning model, with a learning rate of \(10^{-4}\). During training, the data is shuffled using a mini-patch size of 128. The maximum number of epochs is set to 20.
3.3 Comparison results
To evaluate the potential of the individual investigated learning system, performance metrics are derived for the AlexNet, ResNet-50, ResNet-101, and DenseNet-201, and compared to the proposed system, applying the same type of SNR prepossessing (see Algorithm I and Fig. 2 for the proposed SNR extraction method).
As illustrated in Fig. 8 and Table 3, all CNN models achieve acceptable accuracy (even if each CNN is used alone), since they apply the proposed SNR extraction method. To quantify the advantage and the need of applying SNR to the system, Fig. 9 presents the improvement in the accuracy for all individual transfer learning systems. It is clear from the figure the role of applying SNR, which significantly improves the performance for all investigated systems, e.g., the accuracy increases by around 50% for the proposed system.
Quantitative comparison between five systems (i.e., AlexNet, ResNet-50, ResNet-101, DenseNet-201, and the proposed system); using the SNRs extracted from ROI CBIS-DDSM dataset and a binary SVM classifier into, i.e., benign (B) or malignant (M), mass (MA) or calcification (CA), benign mass (BM) or malignant mass (MM), or benign calcification (BC), or malignant calcification (MC)
To further highlight the advantages of the proposed system, visual and quantitative assessments of the ensemble learning and our fusion algorithm (Algorithm II) have been carried out. Samples for the classification of four mass images are shown in Fig. 10. The Ground Truth (GT) diagnosis for the first two columns are benign “B” and for the last two columns are malignant “M”). Figure 10 demonstrates the advantage of the proposed ensemble fusion algorithm (Algorithm II) to produce better classification results: Even if there is an error on one or two CNN outputs, the proposed system shows the potential to achieve the correct output. The visual results are verified quantitatively in Table 3. It is remarkable that the proposed system achieves the best performance for all the three investigated metrics overall competing for individual CNN systems. These results highlight the advantages of the proposed system. Furthermore, the comparison results in Table 4 show the advantage of the proposed system over other related competing methods. This is due to the inclusion of the SNR extraction step, which limits the search area to the tumor-like regions and enables the system to find the small nodules correctly.
3.4 Computational complexity
All results on this paper are obtained using an ordinary laptop, Intel core I5-6200U @2.30 GHz, 6GRAM. The summary of time performance is detailed below in Table 5.
The computational complexity of each model lies in the number of convolutional layers (CL) of each model. Table 2 summarizes the number of convolutional layers (CL) for each model. As shown in the table, the AlexNet consists of the least number of layers, so it takes the least time (i.e., the mean time between 1.8 and 8.2 s, based on the problem, as shown in Table 5). On the other hand, DensNet-201 contains the largest number of layers so it takes more time (i.e., the mean time is between 1.9 and 48 s, based on the problem). The fusion step depends on a simple first order momentum, so it takes a small amount of time (1.7 s, as shown in Table 5). The overall mean time of the proposed system is around 15–60 s per each test image, based on the problem solved. The current processing time (0.25–1 min) is still sufficient in this medical application. However, since the proposed system speed is not near real time, in the future, we will replace using Matlab, in the current work, with Python and depend on the GPU parallel processing algorithms in order to minimize the processing time of the test image.
4 Conclusion
In this work, we proposed a CAD system to early detect breast cancer based on deep learning. Unlike related work, the utilized CNN models extract features from the SNRs based on automated adaptive Otsu thresholding, in order to improve the training capabilities of the deep learning model. An ensemble is used for feature extraction followed by SVM classifiers. The final decision is taken by fusing the binary outputs of the SVM classifiers, taking into account the training accuracy of each classifier. Experiments results on the ROI CBIS-DDSM data confirm the superior of the proposed method over the related work. In the future, other public databases for breast cancer detection will be investigated to test the robustness of the proposed system. In addition, other features/models will be tested in order to improve the performance.
Availability of data and materials
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Alizadehsani R, Roshanzamir M, Hussain S, Khosravi A, Koohestani A, Zangooei MH, Abdar M, Beykikhoshk A, Shoeibi A, Zare A, et al. (2021) Handling of uncertainty in medical data using machine learning and probability theory techniques: a review of 30 years (1991–2020). Ann Oper Res 1–42. https://doi.org/10.1007/s10479-021-04006-2
Altaf F, Islam SM, Akhtar N, Janjua NK (2019) Going deep in medical image analysis: Concepts, methods, challenges, and future directions. IEEE Access 7:99540–99572
Ansar W, Shahid AR, Raza B, Dar AH (2020) Breast cancer detection and localization using mobilenet based transfer learning for mammograms. In: International symposium on intelligent computing systems. Springer, pp 11–21
Bennett K, Demiriz A (1998) Semi-supervised support vector machines. Adv Neural Inf Process Syst 11:368–374
Bowyer K, Kopans D, Kegelmeyer W, Moore R, Sallam M, Chang K, Woods K (1996) The digital database for screening mammography. In: 3rd international workshop on digital mammography, vol 58, p 27
Castro E, Cardoso JS, Pereira JC (2018) Elastic deformations for data augmentation in breast cancer mass detection. In: 2018 IEEE EMBS international conference on biomedical and health informatics (BHI). IEEE, pp 230–234
Chan HP, Samala RK, Hadjiiski LM, Zhou C (2020) Deep learning in medical image analysis. In: Deep learning in medical image analysis. Springer, pp 3–21
Chen Y, Zhang Q, Wu Y, Liu B, Wang M, Lin Y (2018) Fine-tuning resnet for breast cancer classification from mammography. In: The international conference on healthcare science and engineering. Springer, pp 83–96
Deepa S, Subbiah BV (2013) Efficient ROI segmentation of digital mammogram images using Otsu’s N thresholding method. Natl J Adv Comput Manag 4(1)
DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL (2019) Breast cancer statistics, 2019. CA Cancer J Clin 69(6):438–451
Gunn SR (1998) Support vector machines for classification and regression. ISIS Techn Rep 14(1):5–16
Hekal AA, Elnakib A, Moustafa HED (2021) Automated early breast cancer detection and classification system. Signal, Image Video Process 15:1–9
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Huang XL, Ma X, Hu F (2018) Machine learning and intelligent communications. Mob Netw Appl 23(1):68–70
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Lameski P, Zdravevski E, Mingov R, Kulakov A (2015) SVM parameter tuning with grid search and its impact on reduction of model over-fitting. In: Rough sets, fuzzy sets, data mining, and granular computing. Springer, pp 464–474
Lee RS, Gimenez F, Hoogi A, Rubin D (2016) Curated breast imaging subset of DDSM. Cancer Imaging Arch 8:2016
Lévy D, Jain A (2016) Breast mass classification from mammograms using deep convolutional neural networks. arXiv preprint arXiv:1612.00542
Li H, Zhuang S, Da Li, Zhao J, Ma Y (2019) Benign and malignant classification of mammogram images based on deep learning. Biomed Signal Process Control 51:347–354
Matheus BRN, Schiabel H (2011) Online mammographic images database for development and comparison of cad schemes. J Digit Imaging 24(3):500–506
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Patil RS, Biradar N (2020) Automated mammogram breast cancer detection using the optimized combination of convolutional and recurrent neural network. Evolut Intell 14:1–16
Pisner DA, Schnyer DM (2020) Support vector machine. In: Machine learning. Elsevier, pp 101–121
Ragab DA, Sharkas M, Marshall S, Ren J (2019) Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ 7:e6201
Suzuki S, Zhang X, Homma N, Ichiji K, Sugita N, Kawasumi Y, Ishibashi T, Yoshizawa M (2016) Mass detection using deep convolutional neural network for mammographic computer-aided diagnosis. In: 2016 55th annual conference of the society of instrument and control engineers of Japan (SICE). IEEE, pp 1382–1386
Swetha T, Bindu CH (2015) Detection of breast cancer with hybrid image segmentation and Otsu’s thresholding. In: 2015 international conference on computing and network communications (CoCoNet). IEEE, pp 565–570
Tsochatzidis L, Costaridou L, Pratikakis I (2019) Deep learning for breast cancer diagnosis from mammograms-a comparative study. J Imaging 5(3):37
WHO (2020) Breast cancer. https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/. Accessed 7 Nov 2020
Wu P, Li H, Zeng N, Li F (2022) FMD-Yolo: an efficient face mask detection method for COVID-19 prevention and control in public. Image Vis Comput 117:104341
Xi P, Shu C, Goubran R (2018) Abnormality detection in mammography using deep convolutional neural networks. In: 2018 IEEE international symposium on medical measurements and applications (MeMeA). IEEE, pp 1–6
Yi D, Sawyer RL, Cohn III D, Dunnmon J, Lam C, Xiao X, Rubin D (2017) Optimizing and visualizing deep learning for benign/malignant classification in breast tumors. arXiv preprint arXiv:1705.06362
Zeng N, Wang Z, Zineddin B, Li Y, Du M, Xiao L, Liu X, Young T (2014) Image-based quantitative analysis of gold immunochromatographic strip via cellular neural network approach. IEEE Trans Med Imaging 33(5):1129–1136
Zeng N, Li H, Peng Y (2021) A new deep belief network-based multi-task learning for diagnosis of Alzheimer’s disease. Neural Comput Appl 1–12. https://doi.org/10.1007/s00521-021-06149-6
Zhu W, Zeng N, Wang N (2010) Sensitivity, specificity, accuracy, associated confidence interval and roc analysis with practical SAS implementations. NESUG Proc: Health Care Life Sci, Baltimore, Maryland 19:67
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hekal, A.A., Moustafa, H.ED. & Elnakib, A. Ensemble deep learning system for early breast cancer detection. Evol. Intel. 16, 1045–1054 (2023). https://doi.org/10.1007/s12065-022-00719-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12065-022-00719-w