Abstract
Clonal selection algorithms are computational methods inspired by the behavior of the immune system which can be applied to solve optimization problems. However, like other nature inspired algorithms, they can require a large number of objective function evaluations in order to reach a satisfactory solution. When those evaluations involve a computationally expensive simulation model their cost becomes prohibitive. In this paper we analyze the use of surrogate models in order to enhance the performance of a clonal selection algorithm. Computational experiments are conducted to assess the performance of the presented techniques using a benchmark with 22 test-problems under a fixed budget of objective function evaluations. The comparisons show that for most cases the use of surrogate models improve significantly the performance of the baseline clonal selection algorithm.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Artificial Immune Systems (AISs) are computational methods inspired by the biological immune system and have found applications in many domains. One of them, optimization, is a key ingredient to design and operational problems in all types of engineering, as well as a tool for formulating and solving inverse problems such as system identification in scientific and engineering situations. From the optimization perspective, the AISs are stochastic population-based search methods which do not require a continuous, differentiable, or explicit objective function, and do not get easily trapped in local optima. However, AISs, as well as other nature-inspired search techniques, usually require a large number of objective function evaluations in order to reach a satisfactory solution. As new applications require the development of increasingly complex and computationally expensive simulation models, this becomes a serious drawback to the application of AISs in areas such as Structural Mechanics, Reservoir Simulation, Fluid Dynamics, Molecular Dynamics, and others. As a result, the user’s computational budget places a strong upper limit on the number of calls to the expensive simulation model adopted. It is then necessary to modify the search process in order to increase its convergence speed in the sense of attaining a given level of solution quality with less calls to the expensive simulation model. The strategy considered here is the use of a surrogate model (or metamodel), capable of providing a relatively inexpensive approximation of the objective function, thus replacing the computationally intensive original simulator evaluation.
The idea of reducing the number of computationally expensive function evaluations appeared early in the evolutionary computation literature [22] and efforts in this direction continue (see, for example [17, 18, 28, 34, 36, 38]). One may also mention additional reasons for using surrogate models in evolutionary algorithms: (a) to smooth the fitness landscape [47], (b) to alleviate user fatigue in interactive design systems [41], and (c) in noisy environments [24].
Many surrogate models are available in the literature, such as polynomial models [22], artificial neural networks [15], Kriging or Gaussian processes [14], Radial Basis Functions [19, 21], and support vector machines [25]. Alternatively, several surrogates may be derived from physical or numerical simplifications of the original model, in which case they are more strongly problem-dependent.
In this paper we extend the results of a previous conference paper [5] by proposing and comparing the performance of an artificial immune system (AIS) assisted by (1) a similarity-based surrogate model (SBSM) based on the nearest-neighbors idea, and (2) a locally defined linear approximation model.
The idea is to use a pre-defined number of expensive simulations and allow the AIS to perform additional (approximate) objective function evaluations (via surrogate model), in order to (hopefully) obtain a final solution which is better than the one the baseline algorithm would find using only that fixed number of expensive simulations.
This paper is organized as follows. AISs for optimization are discussed in Sect. 2. Sections 3 and 4 present the surrogate models and the surrogate-assisted AIS, respectively. A discussion of the computational results is given in Sect. 5, and the paper ends with some concluding remarks and suggestions for future work.
2 Artificial immune systems for optimization
When exposed to antigens organisms develop an efficient immune response where specific antibodies are produced to attack these antigens. The best performing immunological cells multiply (by cloning) and are improved (by hypermutation and replacement) while new cells, produced by the bone marrow, are generated. This scheme of adaptation is known as clonal selection and affinity maturation by hypermutation or, more simply, clonal selection [20].
Artificial immune systems for optimization problems [6] are inspired by natural immune systems concepts and mechanisms, such as clonal selection, immune network theory, vaccination, and others. In general, an AIS optimization algorithm will have a population of antibodies (candidate solutions) and another set composed by the antigens (objectives) that the antibodies attempt to reach or match (optimize). The main differences among the AIS techniques applied to optimization reside in which natural immune mechanism is considered to evolve the antibodies, i.e., how the candidate solutions evolve.
According to the clonal selection theory—the immune mechanism used by the algorithm considered here—there is a selection process which leads to the evolution of the immune system repertoire during the lifetime of the individual. Furthermore, on binding with a suitable antigen, activation of lymphocytes occurs and clones of the activated lymphocyte are produced expressing receptors identical to the original one that first encountered the antigen. The clonal selection culminates in the increase in the average affinity between the antibodies and antigens due to the somatic hypermutation and selection mechanisms of clonal expansion. That is responsible for the fact that upon a subsequent exposure to the antigen, a stronger immune response is produced [3]. The affinity maturation, as it is also known, is a mutation of the individuals occurring at a high rate, which is inversely proportional to the fitness of the antibody (antibody-antigen affinity), unlike the standard mutation of Evolutionary Algorithms (EAs). Thus, inferior individuals are more strongly modified than the better ones, which require a finer tuning. To avoid a random search, a selection method is necessary to keep the good solutions, eliminate the worst ones, and maintain diversity.
2.1 Clonal selection algorithm
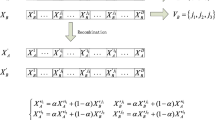
Inspired by the clonal selection theory, de Castro and von Zuben proposed an AIS algorithm (CLONALG [9]) that performs computational optimization and pattern recognition tasks. In this method, each antibody is cloned, hypermutated, and those with higher affinity are selected. The main features of this technique are (1) the mutation rate, usually inversely proportional to the affinity of the antibody with respect to the antigens, and (2) the absence of a recombination operator (such as crossover in GAs). Algorithm 1 shows CLONALG’s pseudocode which is inspired by the algorithm presented in [6].
In Algorithm 1, affinities contains the values of the objective function to be optimized (fitness) and antibodies contains the population of candidate solutions. Also, β is the number of clones that each antibody will be allowed to generate, ρ is a parameter used to compute the mutation rate, nAffinities (\(\bar f\)) contains the affinities of the antibodies normalized (by function “normalize”) in the interval \([0,1],\; pRandom\) is the percentage of new cells that are randomly generated (representing the mesenchymal stem cells generated by the bone marrow), clones contains the population of clones generated by the function “clone”, and cA contains the corresponding affinities. The function “initializePopulation” initializes, usually randomly, a population of cells, “evaluate” calculates the antibody-antigen affinity, “hypermutate” performs the somatic hypermutation, “select” selects the best candidate solution among an antibody and its clones to compose the antibodies population [11], and finally, “genNew” randomly generates, evaluates and includes new antibody cells in the candidate solutions population.
3 Surrogate models
Surrogate modeling, or metamodeling, can be viewed as the process of replacing the original evaluation function (a complex computer simulation) by a substantially less expensive approximation. The surrogate model should be simple, general, and keep the number of control parameters as small as possible [8].
In contrast to “eager” learning algorithms, such as neural networks, which generate a model and then discard the inputs, the Similarity-Based Surrogate Models (SBSMs) store their inputs and defer processing until a prediction of the affinity value of a new candidate solution is requested. Thus, SBSMs can be classified as “lazy” learners or memory-based learners [2] because they generate the output value by combining their stored data using a similarity measure. Any intermediate structure or result is then discarded.
In the proposed algorithm, the individuals evaluated by the original function (i.e., solutions evaluated exactly) are memory cells which are stored in a database. In the biological immune system, memory cells are kept (even without an antigen invasion) and play a relevant role in speeding up the response of the immune system against a similar invader. The population of memory cells is used to construct a surrogate, based on similarity, which is used along the optimization process to perform extra (surrogate) evaluations, resulting in a larger number of total (surrogate plus exact) evaluations. Those extra surrogate evaluations involve a simple procedure, with a negligible computational cost when compared to the cost of an exact evaluation using the simulation model.
The following section describes in detail the surrogate models used here.
3.1 Nearest neighbors
Two variants of the nearest neighbors technique are studied here as a relatively inexpensive surrogate of the original expensive objective function. The difference between these variants is the way of obtaining the nearest neighbors, that is, the querying algorithm used. Here we have:
-
1.
k-Nearest Neighbors (k-NN) [37], in which the k nearest candidate solutions are selected for use in the surrogate evaluation;
-
2.
r-Nearest Neighbors (r-NN) [23], where the set of neighbors is taken from a certain hyperbox in the search space.
The idea of using a Nearest Neighbors technique to assist an evolutionary algorithm was previously explored in [16, 32, 33] to reduce the number of exact function evaluations required during the search, and in [10, 17, 18] to extend the number of generations guiding the search towards improved solutions.
Given a candidate solution x h and the archive \({{\mathcal D}=\{(x_i, f(x_i)), i=1,\ldots,\eta\}}\) containing η exactly evaluated solutions x i , then the corresponding value \(\widehat{f}({x^h}) \approx f({x^h})\) to be assigned to x h is
where s(x h, x i) is a similarity measure between x h and x i and p is set to 2. When k-NN is used, \({{\mathcal I}^h_j}\) is the j-th element of the list that stores the individuals in the set \({{\mathcal D}}\) most similar to x h and k is the number of neighbors used to build the surrogate function.
When the r-NN technique is used then \({{\mathcal I}^h_j}\) is the j-th element of the list that stores the individuals from the set \({{\mathcal D}}\) which belong to the hyperbox centered in x h such that its i-th “side” has length \(2 r \left( x_i^u-x_i^l \right)\). Thus, for the r-NN case, 2r is the fraction of the search space range in each dimension used to build the hyperbox, and the number of elements found inside such hyperbox is automatically attributed to k.
The Nearest Neighbor technique does not require a predefined functional form nor a probability distribution. The variables can be either continuous or discrete and the database is updated whenever it is necessary to add a candidate solution. The computational cost for evaluating an individual is mainly due to the search for the nearest neighbors.
In the adopted real-coded CLONALG, the similarity measure is given by
where d E (x h, x i) is the Euclidean distance between x h and x i.
It is easy to see that, for both querying techniques considered here, only one parameter is needed to be set by the user: the number of nearest neighbors k or the length of the “size” of the hyperbox given by the fraction \(2r \in \left[0; 1\right]\) of each variable range. Also, it may occur that there are no candidate solutions inside the hyperbox. In this case, the two nearest neighbors are used by the surrogate function, and the 2-NN variant is recovered.
3.2 Simple and weighted local linear regression
Another kind of metamodel technique is local linear regression [46], considered here in both its simple and weighted variants.
A general form of a simple linear regression is given by
or, in matrix format,
where the superscript T denotes transposition and one must compute \(\varvec{\theta} \in R^{n+1}\). Given a matrix X with a set of candidate solutions in \({{\mathcal D}}\) and \(\user2{y}\) their corresponding objective function values, the least squares estimation leads to
were (X T X) must be a non-singular matrix.
It is assumed that given the set of a query point’s neighbors it is possible to obtain a linear approximation \(\widehat{f}({x^h}) \approx f({x^h})\). The number of neighbor points used by the model is the minimum number of the nearest neighbors such that (X T X) is a non-singular matrix.
Considering that the closest points/solutions should contribute more to the surrogate model, then we also used the weighted version of the linear regression. In this case, the model is weighted using a diagonal matrix W with elements given by
It is important to notice that for all surrogate models considered here, x h is searched in the memory cells before its fitness value is calculated by the surrogate model. Thus \(d_{E}\left( x^h, x^i \right) \neq 0\) and w ii can always be calculated.
The weighted least squares estimator reads now
and
Figure 1 shows an example of simple and weighted linear regression applied over three points in which the exact function is sin(x). When using the weighted linear regression, the matrix W is not the same for different points, leading to different models for each new point. Thus, for this case, we also plot a curve corresponding to the model generated at x = 1.
It is important to notice that higher order (say quadratic) polynomial regressors can also be considered in principle, but, as the number of unknown coefficients in \(\varvec{\theta}\) grows fast (quadratically for a quadratic polynomial regressor), here only linear ones were considered due to the high dimensionality of the problems (n ≫ 2).
4 Surrogate-assisted artificial immune system
The next crucial step is to define how the surrogate model will be incorporated into the search mechanism. Some possibilities discussed in the literature include incorporating the surrogate models as (1) local approximators [31], (2) surrogate-guided evolutionary operators [30], (3) surrogate-assisted local search [27, 44], and (4) a pre-selection technique [21, 29], as well as the use of multiple surrogates [1, 27, 35].
To the best of our knowledge, there are few surrogate-assisted immune algorithms in the literature. In [7], the authors introduce a surrogate model into the immune inspired algorithm cycle by means of a stochastic management procedure which, in each iteration, uses both surrogate and exact models in a cooperative way. For that first proposal, called Random Selection, a candidate solution is evaluated by the exact function according to a user specified probability.
According to [7] the use of a surrogate model allows for increasing the total number of iterations of the algorithm and, for almost all problems considered, leads to solutions which are better than those provided by the baseline clonal selection algorithm (CLONALG). However, the authors suggested that, for some objective functions, the surrogate model increases the exploitation of the baseline CLONALG, inducing a faster convergence to local optima.
A different model management, referred as Deterministic Selection (DS), was adopted in [5]. In that approach, all candidate solutions from the initial population are evaluated exactly and compose the initial archive which defines the surrogate model. When performing clonal expansion, all generated clones are evaluated by the surrogate model and only the best clone of each antibody is evaluated by the exact function. If the exact value is better than the affinity of the original antibody then the parent is replaced by this clone.
One should notice that, while the model management used in [7] leads to a population containing both exactly and approximately evaluated individuals, using DS the population contains only exactly evaluated solutions. Also, in [5] the surrogate model was only used to indicate ways of improving the candidate solutions. While in [7] the surrogate model should evaluate as accurately as possible a candidate solution to be compared with the exact objective function, in [5] what really matters is that the approximately evaluated affinity be accurate enough to correctly sort the hypermutated clones from each antibody. It is not necessary that the surrogate model accurately approximates the true function as long as it is able to correctly sort the generated hypermutated clones from each antibody.
When DS is considered, the use of the surrogate model can be controlled by the number β of clones defined by the user. As more clones are generated by each antibody, more approximate evaluations are performed. It is easy to see that CLONALG is recovered when the antibodies generate only one clone (β = 1). Also, if the number of exact objective function evaluations is used as the stop criterion, the number of final generations is equal for all β values as only the better hypermutated clone from each antibody will be evaluated exactly.
In both cases, using either Random or Deterministic Selection, every individual evaluated by the exact function is immediately stored in the archive used by the surrogate model. In the immunological paradigm, this database of antibodies corresponds to the memory cells: a set of representative cells stored with the objective of improving the immune system response to subsequent attacks.
The surrogate model used by references [5, 7] was the k-NN. Although the use of this simple method improved the performance of the baseline search technique in most cases, in a few problems the results obtained by the algorithm using this surrogate model were much worse than those achieved by the baseline technique alone.
Here we study alternative surrogate models trying to maintain simplicity, namely the r-NN variant, where the neighbors are those within a certain hyperbox, and linear regression, considering both simple and weighted versions.
It should be noticed that, as the computational experiments considered a maximum number of exact simulations as the stop criterium for the clonal selection algorithm, the use of surrogate models actually increases the total computational time. The greater the number of solutions in the database, the more expensive is the surrogate model. However, as it is assumed that the objective function is computationally expensive, this delay should be insignificant. Figure 2 shows the delay as a function of the number of exact simulations for the cases SLR β = 16 (WLR β = 16 is similar), {r = 0.001, β = 2}, and {k = 16, β = 16}. It can be seen that the delay is, at most, of a few seconds. As in expensive real-world situations only a limited number of exact objective function evaluations are allowed, the computational cost of the approximations is really insignificant.
5 Computational experiments
We will consider here the standard optimization problem which consists in finding \(x\in R^n\) that minimizes the objective function f(x), subject to lower and upper bounds x l ≤ x ≤ x u. We are particularly interested in the situation where obtaining the value of f for a given x requires a computationally expensive simulation.
5.1 Definition of the test-problems
In order to assess the performance of the surrogate-assisted clonal selection algorithms proposed here for expensive optimization problems, a set F of test-functions \(f_j, j\in \{1,\ldots,n_f\}\) was collected from [40, 42, 43, 45, 48]. The functions are specified in column “Function” from Table 1 while their search space bounds (the same one for all dimensions) and number of exact function evaluations used by the algorithms are presented in columns “Bounds” and “nfe”, respectively. Complementary information for functions \(f_{14},\; f_{15}\), and f 16 can be found in Table 2.
Functions f 01 to f 13 and f 17 to f 22 correspond to high-dimensional problems (n = 30). Functions f 01 to f 05 are unimodal while f 08 to f 13 and f 18 to f 22 are multimodal ones. Function f 06 is the step function, which has one minimum. Function f 07 is a noisy quartic function. They appear to be the most difficult class of problems for many optimization algorithms [48]. Functions \(f_{14},\; f_{15}\), and f 16 are low-dimensional functions (\(n=4,\; n=6\), and n = 4, respectively) which have only a few local minima. Finally, Function f 17 is a Fast Fractal “DoubleDip” Function.
It is clear that these problems do not require expensive simulations. The idea is to assess the relative performance of the proposed technique, when a fixed number of objective function evaluations is allowed, in a larger number of test-problems than it would have been possible in the case of using expensive real-world problems.
5.2 User defined parameters
The baseline CLONALG, defined by Algorithm 1 from Sect. 2.1, has four user defined parameters, namely, the number of clones (β), the decay of the mutation potential (ρ), the number of new candidate solutions randomly generated in each iteration (pRandom), and the population size (populationSize). All CLONALG variants (see Sect. 5.2.2) use the same values for these parameters which are: \(\rho=5,\; pRandom=0\), and popula-tionSize = 50. The exception is the parameter β which has its sensibility tested in the experiments assuming the values 2, 4, 8, or 16 (the value 1 is also considered by the baseline CLONALG)
5.2.1 Hypermutation
Hypermutation is the operator responsible for modifying the candidate solutions in the search for improvements. The hypermutation operator used here is similar to the one presented by [12] where the mutation potential must be calculated first. We used the one proposed by [9], where the mutation potential is given by \(\alpha = e^{-\rho \bar f}\) and \(\bar f\) is the normalized affinity of the original antibody. The number of mutations is given by αn. Once that is calculated, a randomly chosen variable i from the clone x C is modified as
where ν is a random number uniformly distributed in (0,1) and j is another randomly chosen design variable. Although that seems to be a good operator for most objective functions considered here, since they have optimal solutions with variables assuming the same value, that is not usually true. Thus, here we modify a clone in the following way
where y is an antibody randomly chosen from the population.
It can be noticed that the hypermutation is fully random, although weighted by the quality of the candidate solution. We also generate another clone which is symmetrically placed with respect to the parent solution (also known as two-way mutation [26]): while one clone is hypermutated by the procedure above, the other one is modified as
Except for the baseline CLONALG, which uses β = 1, the hypermutation process, including the symmetric clone, is the same for all CLONALG variants in our experiments.
5.2.2 Variants
A set of CLONALG variants corresponding to different values of β and other parameters for each surrogate model was considered.
-
Baseline—since the only parameter is β they are referred to as β = 2i, where \(i \in \{0, \ldots, 4 \}\);
-
k-NN—as in this case the parameters are β and the number of nearest neighbors to be used by the surrogate model, these variants are defined as {k = 2i, β = 2j }, where \(i,j \in \{1, \ldots, 4 \}\);
-
r-NN—the parameters are β and the range for the neighbors of the surrogate model. Thus, they are labeled as {r = 10−i, β = 2j }, where \(i \in \{2, 3, 4\}\) and \(j \in \{1, \ldots, 4 \}\);
-
Linear Regression—it is referred to as SLR β = 2i, when simple linear regression is used, and WLR β = 2i, when we have the weighted one instead. For both cases \(i \in \{1, \ldots, 4 \}\).
The sensibility analysis of the surrogate-assisted parameters is important because as this technique improves exploitation with respect to the baseline CLONALG, it also makes it easier to get trapped in local minima. Then a good balance between exploration and exploitation should be achieved. Also a sensibility analysis with respect to the baseline’s β parameter is made to verify the performance of the symmetric mutation.
5.3 Performance profiles
The most common way of assessing the relative performance of a set V of variants \(v_i, i\in \{1,\ldots,n_v\}\) is to define a set F of representative test-functions \(f_j, j\in \{1,\ldots,n_f\}\) and then test all variants against all problems measuring the performance t f,v of variant \(v\in V\) when applied to function \(f\in F\). The performance indicator to be maximized here is the inverse of the minimum objective function value found by variant v in test-function f averaged over 30 runs.
Now a performance ratio can be defined as
It is interesting to be able to assess the performance of the variants in V on a potentially large set of test-functions F in a compact graphical form. This can be attained following Dolan & Moré [13] and defining
where \(\left| . \right|\) denotes the cardinality of a set. Then ρ v (τ) is the probability that the performance ratio r f,v of variant \(v\in S\) is within a factor τ ≥ 1 of the best possible ratio. If the set F is large and representative of problems yet to be tackled then variants with larger ρ s (τ) are to be preferred. The performance profiles thus defined have a number of useful properties [4, 13]:
-
1.
ρ v (1) is the probability that variant v will provide the best performance in F among all variants in V. If ρV1(1) > ρV2(1) then variant V1 was the winner in a larger number of problems in F than variant V2,
-
2.
the area under the ρ v curve (\(AUC_v = \int \rho_v(t) dt \)) is an overall performance indicator/measure for variant v in the problem set F: the larger the AUC the higher the variant efficiency, and
-
3.
a measure of the reliability of variant v is its performance ratio in the problem where it performed worst: \(R_v = \sup \{ \tau: \rho_v(\tau) < 1 \}\). As a result, the most reliable variant is the one that minimizes R v ; that is, it presents the best worst performance in the set F:
$$ v^* =\arg\min_{v\in V} R_v = \arg\min_{v\in V} \sup \{ \tau: \rho_v(\tau) < 1 \} $$
5.4 Results
The CLONALG variants from Sect. 5.2.2 are evaluated against the functions presented in Sect. 5.1 The best variants with respect to each surrogate model used by the search mechanism and the baseline are selected to simplify the final comparisons. We used performance profiles (see Sect. 5.3) to find those best variants
The performance profiles which compare the baseline CLONALG variants are presented in Fig. 3. It is easy to see that the variant β = 2 performs better than the other ones. Also notice that the β = 2 variant uses the two-way mutation (see Sect. 5.2.1) unlike the one with β = 1, indicating the efficiency of this mutation operator which keeps the idea, usually adopted in immune algorithms found in the literature, that few clones should be generated.
Figure 4 presents the comparison between surrogate-assisted CLONALG variants with k-NN. In this case a more complex discussion must be made because two variants have shown superior performance, namely: {k = 16, β = 8} and {k = 16, β = 16}. The {k = 16, β = 8} variant is the most reliable since it is able to find solutions for all problems within about fourteen times worst than the best performer in any problem. The {k = 16, β = 16} variant is the most efficient, finding the best results for about 50% of the objective functions. We have chosen the {k = 16, β = 16} variant as the “winner” because it presents the largest area under the curve (see the item (2) in Sect. 5.3).
As in the k-NN case, the comparison between surrogate-assisted CLONALG variants which use r-NN leads to a choice of the best one. The performance profiles for the r-NN variants can be shown in Fig. 5 in which the variants {r = 0.01, β = 2} and {r = 0.001, β = 2} present the best performances with respect to efficiency (finding the best results in about 55% of the objective functions) and reliability (average results are at maximum about six times worse than the best performer), respectively. However, considering the area under the curve, it is easy to verify the superiority of the {r = 0.001, β = 2} variant.
Figure 6 shows the performance profiles comparing surrogate-assisted CLONALG variants with simple linear regression. The best number of clones for this scenario is β = 16, while the baseline CLONALG performs better with β = 2. It is important to notice here that the performance is proportional to the use of the surrogate model. It means that the more calls to the approximated objective function evaluations are made, the better the results are, in general.
As with the simple linear surrogate-assisted clonal selection variants, Fig. 7 shows that the best number of clones β for WLR is sixteen. Also in this case, the performance is proportional to the use of the surrogate model. That is important because it shows the stability of the algorithm with respect to its user defined parameters.
Considering the individual analyses with respect to each surrogate model we have chosen the following ones to be used in the comparisons between the different approaches: β = 2, SLR β = 16, WLR \(\beta=16,\; \{k=16, \beta=16\}\), and {r = 0.001, β = 2}. The performance profiles comparing the results found by these algorithms are presented in Fig. 8. Some important conclusions can be made from this plot: (1) all surrogate-assisted CLONALG variants perform better than the baseline considering all metrics derived from the performance profiles (efficiency, reliability, or area under the curve); (2) WLR β = 16 is the most reliable; (3) {k = 16, β = 16} is the most efficient; and (4) WLR β = 16’s curve presents the greatest area under the curve.
Also, we compared the performance of the baseline CLONALG directly with WLR β = 16 and {k = 16, β = 16} variants. The performance profiles are presented in Fig. 9 where we can see that while the WLR β = 16 variant found results about 1.2 times worst than the best ones, the solutions of the {k = 16, β = 16} were about 3 times the best ones. In addition, Fig. 9 shows that WLR β = 16 found results better than the baseline CLONALG for more than 80% of the objective functions while the {k = 16, β = 16} variant achieved about 55%. Summarizing, the WLR β = 16 variant performs better than the baseline CLONALG that {k = 16, β = 16}, showing a superior efficiency and reliability with respect to the improvement of the {k = 16, β = 16} variant over the same baseline.
We also analyzed each test-problem individually and the results found by the best variants and the baseline CLONALG are presented by boxplots in Figs. 10–14.
The boxplots standing for the results found in functions f 01 to f 06 are presented by Fig. 10. Here we can see that the CLONALG variants assisted by simple and weighted linear regression outperform the baseline one for these objective functions. The k-NN variant found worst results for the function f 05 while for the r-NN one, that happened for functions f 04 and f 05. That shows the superiority of the CLONALG variants assisted by linear regression, WLR and SLR, with respect to the baseline algorithm when solving high dimensional unimodal functions (functions f 01 to f 05), and also the improvement of all surrogate-assisted clonal selection algorithms used in the comparisons when the high dimensional step function is considered. According to the statistical analysis presented in Table 3, all those differences are statistically significant (based in the non-parametric Wilcox test). The exception is the performance’s decrease caused by the use of the r-NN in the function f 05.
The same improvement due to the use of linear regressions in CLONALG can be seen in the Fig. 11 which presents the boxplots corresponding to the results found in functions f 07 to f 12. The CLONALG variants with r-NN only did not present good results for function f 08 while the use of k-NN decrease the performance of the baseline CLONALG in functions \(f_{07},\; f_{08}\), and f 09, that is in half of the functions shown in this figure. Except for the variant with k-NN, the baseline CLONALG is outperformed by the surrogate-assisted one in the high dimensional noisy quartic function and this superiority is significant (see Table 3).
Boxplots with the results found in functions f 13 to f 18 are shown in Fig. 12. Here we can see that although CLONALG variants assisted by linear regressions presented better results than the baseline one for most of these objective functions, the variants with WLR and SLR performed worse in functions f 14 and f 18, respectively. For function f 17, only the variant with k-NN presents a decrease with respect to baseline CLONALG. All surrogate-assisted variants are outperformed by the baseline CLONALG in function f 14. It is important to highlight that this is the only function in the benchmark where the baseline CLONALG was not statistically outperformed by the use of the WRL metamodel. Also, it should be noted that the superiority of the baseline CLONALG in function f 14, corresponds to an absolute difference of the means in the order of 10−4.
The boxplots corresponding to the solutions of the remaining functions (f 19 to f 22) are presented in Figs. 13 and 14 where we can see that the baseline CLONALG performs better than other variants in function f 19 despite the fact that the median of the results found by the variant with k-NN is superior. Also, this decrease by the use of a surrogate model is only significant in the variant with r-NN, as we can see in Table 3. Considering the variant with SLR and the remaining functions, the results are inferior to those achieved by the baseline CLONALG in function f 20 while the same happens for the WLR variant in functions f 20 and f 22. However, none of these observed decreases with respect to the use of linear regressors are statistically significant. A decrease in the performance can be verified with respect to the variant with the r-NN in the function f 21 (its median is also worst in function f 20). Finally, the baseline CLONALG found results better than the variant with the k-NN for all remaining functions.
The results found by the baseline CLONALG and the surrogate-assisted variants are summarized in Table 3. Also the statistical significance of the better results and of the results found by the variants with respect to the baseline CLONALG are highlighted. The best results are presented in boldface while * and + indicate that the difference observed is not statistically significant with respect to the best result and the baseline algorithm, respectively. Differences in the results are statistically significant when the p-value from the non-parametric Wilcox’s test is less than 0.05. It is important to notice that although the \(\{k=16, {\boldsymbol\beta}=16\}\) variant is the most efficient (best results in eight functions), their results are statistically worse than those found by the baseline CLONALG in seven functions, namely \(f_{05},\; f_{08},\; f_{09},\; f_{17},\; f_{20},\; f_{21}\), and f 22. Another crucial information is that in addition to the reliability of the WRL β = 16 variant, f 14 is the only function where that variant is statistically significantly worse than the baseline CLONALG. Also the difference between the means has an order of magnitude of 10−4 and the function f 14 is the one with the lowest dimensionality in our benchmark (n = 4).
6 Concluding remarks and future works
In this paper a real-coded clonal selection algorithm is enhanced by means of (1) the use of the two-way mutation, and (2) the use of similarity-based surrogate models, namely the k- and r-nearest neighbors techniques, and simple and weighted linear regressors. The objective is to improve the baseline CLONALG’s performance when solving optimization problems involving computationally expensive objective functions.
Computational experiments were conducted to assess the performance of the proposed procedure using a benchmark with 22 test-problems from the literature where a maximum number of objective function evaluations is prescribed. The results show that the use of surrogate models improve the results obtained by the baseline CLONALG for most test-problems. The WRL β = 16 variant showed to be the best one performing statistically significantly worse than the baseline CLONALG only for one (low-dimensional) objective function.
As a future work, the extension to constrained optimization problems is perhaps the most relevant one. Constrained optimization problems appear in engineering design situations, where the constraints are often complex implicit functions of the design variables.
Another idea that could be pursued in order to improve efficiency in the scenario of expensive affinity computation is the immune network theory, aiming at removing similar antibodies by means of a suppression technique. The resulting search algorithm would combine the memory and self-adaptation features of the immune system.
Finally, an adaptive choice of the better surrogate model for each different situation is an important research avenue.
References
Acar E, Rais-Rohani M (2009) Ensemble of metamodels with optimized weight factors. Struct Multidisc Optim 37(3):279–294
Aha DW (1997) Editorial. Artif Intell Rev 11(1–5):1–6 Special issue on lazy learning
AISWeb (2008): the online home of artificial immune systems. http://www.artificial-immune-systems.org., accessed 11/09/2008
Barbosa HJC, Bernardino HS, Barreto AMS (2010) Using performance profiles to analyze the results of the 2006 CEC constrained optimization competition. In: IEEE world congress on computational intelligence. Barcelona, Spain
Bernardino HS, Barbosa HJ, Fonseca LG (2010) A faster clonal selection algorithm for expensive optimization problems. In: Hart E, McEwan C, Timmis J, Hone A (eds) Artificial immune systems, lecture notes in computer science, vol 6209. Springer, Berlin / Heidelberg, pp 130–143
Bernardino HS, Barbosa HJC (2009) Artificial immune systems for optimization. In: Chiong R (ed) Nature-inspired algorithms for optimisation.. Springer, Berlin, pp 389–411
Bernardino HS, Fonseca LG, Barbosa HJC (2009) Surrogate-assisted artificial immune systems for expensive optimization problems. In: dos Santos WP (ed) Evolutionary computation. IntechWeb, pp 179–198
Blanning RW (1974) The source and uses of sensivity information. Interfaces 4(4):32–38
de Castro LN, von Zuben FJ (2002) Learning and optimization using the clonal selection principle. IEEE Trans Evol Comput 6(3):239–251
Custodio FL, Barbosa HJC, Dardenne LE (2010) Full-atom ab initio protein structure prediction with a genetic algorithm using a similarity-based surrogate model. In: IEEE world congress on computational intelligence. Barcelona, Spain
Cutello V, Narzisi G, Nicosia G, Pavone M (2005) Clonal selection algorithms: A comparative case study using effective mutation potentials. In: Proc. of the intl. conf. on artificial immune systems—ICARIS 2005, LNCS, vol 3627. Springer, Banff, Canada, pp 13–28
Cutello V, Nicosia G, Pavone M (2006) Real coded clonal selection algorithm for unconstrained global optimization using a hybrid inversely proportional hypermutation operator. In: Proc. of the ACM symposium on applied computing—SAC ’06. ACM Press, New York, pp 950–954
Dolan E, Moré JJ (2002) Benchmarcking optimization software with performance profiles. Math Program 91(2):201–213
Emmerich M, Giannakoglou K, Naujoks B (2006) Single- and multiobjective evolutionary optimization assisted by gaussian random field metamodels. Evol Comput 10(4):421–439
Ferrari S, Stengel RF (2005) Smooth function approximation using neural networks. IEEE Trans Neural Netw 16(1):24–38
Fonseca LG, Barbosa HJC, Lemonge ACC (2007) Metamodel assisted genetic algorithm for truss weight minimization. In: ICMOSPS’07. Durban, South Africa. CD-ROM
Fonseca LG, Barbosa HJC, Lemonge ACC (2009) A similarity-based surrogate model for enhanced performance in genetic algorithms. Opsearch 46:89–107
Fonseca LG, Barbosa HJC, Lemonge ACC (2010) On similarity-based surrogate models for expensive single- and multi-objective evolutionary optimization. In: Tenne Y, Goh CK (eds) Computational intelligence in expensive optimization problems. Springer, New York, pp 219–248
Forrester AI, Keane AJ (2009) Recent advances in surrogate-based optimization. Prog Aerosp Sci 45:50–79
Garrett SM (2004) Parameter-free, adaptive clonal selection. IEEE Congr Evol Comput 1:1052–1058
Giannakoglou KC (2002) Design of optimal aerodynamic shapes using stochastic optimization methods and computational intelligence. Prog Aerosp Sci 38(1):43–76
Grefenstette J, Fitzpatrick J (2009) Genetic search with approximate fitness evaluations. In: Proc. of the intl. conf. on genetic algorithms and their applications, pp 112–120
Hu H, Lee DL (2006) Range nearest-neighbor query. IEEE Trans Knowl Data Eng 18(1):78–91
Jin Y, Branke J (2005) Evolutionary optimization in uncertain environments-a survey. IEEE Trans Evol Comput 9(3):303–317
Kecman V (2001) Learning and soft computing: support vector machines, neural networks, and fuzzy logic models. Complex adaptive systems. MIT Press, Cambridge
Liang Y, Leung KS (2002) Two-way mutation evolution strategies, pp 789 –794
Lim D, Jin Y, Ong YS, Sendhoff B (2010) Generalizing surrogate-assisted evolutionary computation. IEEE Trans Evol Comput 14(3):329–355
Ong Y, Nair P, Keane A (2003) Evolutionary optimization of computationally expensive problems via surrogate modeling. AIAA J 41(4):687–696
Praveen C, Duvigneau R (2009) Low cost PSO using metamodels and inexact pre-evaluation: application to aerodynamic shape design. Comput Methods Appl Mech Eng 198(9–12):1087–1096
Rasheed K, Vattam S, Ni X (2002) Comparison of methods for using reduced models to speed up design optimization. In: Proc. of genetic and evolutionary computation conference. Morgan Kaufmann, New York, pp 1180–1187
Regis RG, Shoemaker CA (2004) Local function approximation in evolutionary algorithms for the optimization of costly functions. IEEE Trans Evol Comput 8(5):490–505
Runarsson T (2006) Approximate evolution strategy using stochastic ranking. In: Yen GG et al (eds) IEEE world congress on computational intelligence. Vancouver, Canada, pp 745–752
Runarsson TP (2004) Constrained evolutionary optimization by approximate ranking and surrogate models. In: Yao X et al (eds) Proc. of 8th parallel problem solving from nature. Springer, Heidelberg, pp 401–410
Salami M, Hendtlass T (2003) A fast evaluation strategy for evolutionary algorithms. Appl Soft Comput 2:156–173
Sanchez E, Pintos S, Queipo N (2007) Toward an optimal ensemble of kernel-based approximations with engineering applications. Structural and multidisciplinary optimization, pp 1–15
Sastry K, Lima CF, Goldberg DE (2006) Evaluation relaxation using substructural information and linear estimation. In: Proc. of the 8th annual conference on genetic and evolutionary computation. ACM Press, New York, pp 419–426
Shepard D (1968) A two-dimensional interpolation function for irregularly-spaced data. In: Proc. of the 1968 23rd ACM national conference. ACM Press, New York, pp 517–524
Smith RE, Dike BA, Stegmann, SA (1995) Fitness inheritance in genetic algorithms. In: Proc. of the ACM symposium on applied computing, pp 345–350
Suganthan PN (2010) Benchmarks for evaluation of evolutionary algorithms. http://www3.ntu.edu.sg/home/epnsugan/index_files/cec-benchmarking.htm, accessed in 2010
Suganthan PN, Hansen N, Liang JJ, Deb K, Chen YP, Auger A, Tiwari S (2005) Problem definitions and evaluation criteria for the cec 2005 special session on real-parameter optimization. Tech. Rep. 2005005, Nanyang Technological University
Sun XY, Gong D, Li S (2009) Classification and regression-based surrogate model-assisted interactive genetic algorithm with individual’s fuzzy fitness. In: Proc. of the 11th annual conference on genetic and evolutionary computation. ACM Press, New York, pp 907–914
Tang K, Li X, Suganthan PN, Yang Z, Weise T (2009) Benchmark functions for the cec’2010 special session and competition on large scale global optimization. Tech. rep., Nature Inspired Computation and Applications Laboratory
Tang K, Yao X, Suganthan PN, MacNish C, Chen YP, Chen CM, Yang Z (2007) Benchmark functions for the cec’2008 special session and competition on large scale global optimization. Tech. rep., Nature Inspired Computation and Applications Laboratory
Wanner EF, Guimaraes FG, Takahashi RHC, Lowther DA, Ramirez JA (2008) Multiobjective memetic algorithms with quadratic approximation-based local search for expensive optimization in electromagnetics. IEEE Trans Magn 44(6):1126–1129
Whitley D (2010) Test functions. http://www.cs.colostate.edu/~genitor/functions.html, accessed in 2010
Yan X, Su XG (2009) Linear regression analysis: theory and computing. World Scientific Publishing Company, Singapore
Yang D, Flockton SJ (1995) Evolutionary algorithms with a coarse-to-fine function smoothing. IEEE Intl Conf Evol Comput 2:657–662
Yao X, Liu Y, Lin G (1999) Evolutionary programming made faster. IEEE Trans Evol Comput 3:82–102
Acknowledgments
The authors acknowledge the support received from CNPq (308317/2009-2) and FAPERJ (grants E-26/102.825/2008 and E-26/100 .308/2010).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bernardino, H.S., Barbosa, H.J.C. & Fonseca, L.G. Surrogate-assisted clonal selection algorithms for expensive optimization problems. Evol. Intel. 4, 81–97 (2011). https://doi.org/10.1007/s12065-011-0056-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12065-011-0056-1