Abstract
Seventy-five years ago, the geneticist Richard Goldschmidt hypothesized that single mutations affecting development could result in major phenotypic changes in a single generation to produce unique organisms within animal populations that he called “hopeful monsters”. Three decades ago, Sarah P. Gibbs proposed that photosynthetic unicellular micro-organisms like euglenoids and dinoflagellates are the products of a process now called “secondary endosymbiosis” (i.e., the evolution of a chloroplast surrounded by three or four membranes resulting from the incorporation of a eukaryotic alga by a eukaryotic heterotrophic host cell). In this article, we explore the evidence for Goldschmidt’s “hopeful monster” concept and expand the scope of this theory to include the macroevolutionary emergence of organisms like Euglena and Chlorarachnion from secondary endosymbiotic events. We argue that a Neo-Goldschmidtian perspective leads to the conclusion that cell chimeras such as euglenids and dinoflagellates, which are important groups of phytoplankton in freshwater and marine ecosystems, should be interpreted as “successful monsters”. In addition, we argue that Charles Darwin had euglenoids (infusoria) in mind when he speculated on the “primordial intermediate form”, although his Proto-Euglena-hypothesis for the origin of the last common ancestor of all forms of life is no longer acceptable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Russian botanist Constantin S. Merezhkowsky (1855–1921) and the German geneticist Richard Goldschmidt (1878–1958) were unusually gifted natural scientists whose ranges of knowledge in biology were of tremendous scope. Mereshkowsky is essentially remembered as the founder of the then heretical symbiogenesis-hypothesis, a concept that evolved into the serial endosymbiosis theory for the origin of eukaryotes. However, he also published numerous papers on algal systematics, anthropology and many other topics (Geus and Höxtermann 2007). Goldschmidt likewise had the reputation of being a heretic (Dietrich 2003) who deliberately published articles and books that were designed to undermine the classical gene concept as well as basic tenets of the Neo-Darwinian theory of biological evolution largely developed by August Weismann (1834–1914) and Alfred Russel Wallace (1823–1913) (see Kutschera and Niklas 2004). Merezhkowsky and Goldschmidt shared another important attribute. They both proposed novel modes and mechanisms for macroevolutionary events (i.e., phylogenetic changes occurring above the species level). In two key papers, Merezhkowsky (1905, 1910) deduced the symbiogenesis-theory with the explicit goal of explaining the evolutionary development of land plants from lower, algae-like forms of life. Goldschmidt (1933), on the other hand, introduced the “hopeful monster” concept to account for the abrupt origin of novel animal body plans although his ideas were equally applicable to plants. In addition, both men were anti-Darwinists—neither accepted natural selection as a major driving force in organismic evolution, at least with respect to phylogenetic processes that occur above the species level (macroevolution).
In this contribution, which is a sequel to our recent article on primary endosymbiosis and cell evolution (Kutschera and Niklas 2005), we first summarize the current status of Goldschmidt’s concept (with special reference to a 2006 publication of Theißen) and the secondary endosymbiotic theory. We then show that these two models for macroevolution can be combined to create a novel “symbiogenesis-monster theory” that may explain the evolutionary origin of certain photosynthetic eukaryotes (kingdom protoctista) via secondary endosymbiotic events by means of processes postulated thirty years ago by Gibbs (1978). Our account is based in part on observations on populations of freshwater flagellates of the genus Euglena (Figs. 1 and 2) maintained under laboratory conditions described previously (Scherp et al. 2001).
Euglenid species diversity illustrated for European freshwater ecosystems. Euglena gracilis has been used since ca. 1950 as a model organism for laboratory studies. The large species E. ehrenbergi is known as Ehrenberg’s eyeball organism (Adapted from Streble and Krauter 1973)
The place of hopeful monsters in evolutionary biology
In a review article published two years ago in this journal, Theißen (2006) made six claims that are relevant to the topics discussed here:
1. The modern theory of biological evolution is identical with the “Synthetic Theory (or Modern Synthesis)” that was developed during the 1930 and 1940s by Theodosius Dobzhansky (1900–1975), Ernst Mayr (1904–2005), Julian Huxley (1887–1975), George G. Simpson (1902–1984), Ledyard Stebbins (1906–2000) and Bernhard Rensch (1900–1990). 2. The (classical) Synthetic Theory of 1950 has serious shortcomings, because it is based on Darwin’s (1872) principle of gradualism that, according to Theißen (2006), cannot “fully explain the mechanism and mode of macroevolution” because the “empirical basis of gradualism is weak at best”. 3. The Modern Synthesis does not provide an explanation for the “origin of . . . eukaryotes, plants or animals from prokaryotes” on the basis of our knowledge of the 1950s (i.e., when the Synthesis was finished) because it cannot explain why “bacteria not just give rise to… better and better adapted bacteria forever, rather than giving rise to mushrooms, monkey flowers and man” [italics added]. 4. The architects of the “Synthetic Theory made over-extended claims, and hence left the realm of science and developed (evolutionary biology) into an ideology”. Moreover, “all scientific knowledge is hypothetical and preliminary” such that “in the natural sciences there is no such thing as a proven fact”. 5. The “hopeful monster”-concept of Goldschmidt (1933, 1940) may account for the evolutionary origin of certain body plans by means of “saltational evolution” wherein “hopeful monsters…acted as first steps”. 6. Finally, in several contexts, Theißen (2006) characterized modern evolutionary biology as “dogmatic” and argues that “the rejection of Intelligent Design and other versions of creationism is not based on the comprehensive explanatory power of any existing evolutionary theory” but rather must be grounded on a full recognition of the “limits of the scientific method”.
These six “statements of conviction” show how easily the classical synthetic theory (worked out between 1940 and 1950, see Reif et al. 2000; Junker 2004) can be confused with the modern theory of biological evolution of the late 1990s (Carroll 2000; Kutschera and Niklas 2004) whose “system of theories” incorporates concepts such as Merezhkowsky’s symbiogenesis model for the phylogenetic development of eukaryotic cells and the many insight provided by molecular evolutionary and developmental biology, which treats numerous phenomena such as gene duplication and divergence in function that were unknown to the architects of the Modern Synthesis. Within the framework of this “expanded synthesis”, the mechanisms of macroevolutionary transitions become apparent, notably the origin of eukaryotic organisms (animals, plants, and fungi) from prokaryotes via primary endosymbiotic events (Niklas 1997; Kutschera and Niklas 2004, 2005, 2006), which among many other “proven facts” refutes Theißen’s (2006) claim that evolutionary biology is a “dogmatic ideology” (see Kutschera and Niklas 2007 for the rejection of a dogmatic view in plant physiology). Finally, in the following section, we argue against the questionable postulate that Goldschmidt’s “hopeful monster theory” sensu stricto should be re-vitalized.
Richard Goldschmidt’s hopeful monsters
Goldschmidt first outlined his now famous “hopeful monster”-concept in a paper presented at the general meeting of the American Association for the Advancement of Science that was published 75 years ago. At that time, he argued that “rare but extremely consequential mutations affecting rates of decisive embryonic processes… might give rise to what one might term hopeful monsters, monsters which would start a new evolutionary line if fitting into some empty environmental niche” (Goldschmidt 1933, p. 539). However, this concept was much more fully explicated in his influential book entitled The Material Basis of Evolution (Goldschmidt 1940, p. 390) wherein he argued that, while small mutations are sufficient to account for phenomena such as intra-species variation (i.e., microevolution), they could also contribute to the origin of new species or taxa above the species level (i.e., macroevolution). However, in this book, Goldschmidt proposed two mechanisms rather than one to account for the sudden appearance of novel phenotypes—“systemic mutations” that result from large chromosomal re-arrangements and “developmental macromutations” that occur in developmentally important genes that result in large phenotypic effects. Successful systemic mutations (i.e., those that yield viable phenotypes) occur when well-integrated chromosomal arrangements are shifted to produce other stable modifications. This concept, which was an extension of Goldschmidt’s clear distaste of the classical definition of the gene, was grounded in his theories about physiological genetics wherein gene action was explained in terms of rates of reactions and thresholds among substances produced by genes (see Dietrich 2000, 2003).
The concept of the “hopeful monster” also drew heavily from Goldschmidt’s ideas about physiological genetics. However, as Dietrich (2003, p. 71) points out, it is clear that the “idea of a hopeful monster was not tied to [that] of systemic mutation” and that “Goldschmidt used hopeful monsters to argue, by analogy, for evolution by [means of] systemic mutations”. Nevertheless, Goldschmidt firmly believed that a single mutational step affecting the right developmental process at the right moment could accomplish everything (Goldschmidt 1940).
Unlike the concept of systemic mutations, the idea of “hopeful monsters” via the mutation of important developmental genes was not rejected by all of Goldschmidt’s contemporaries. Indeed, Sewall Wright (1889–1988) accepted the notion that macromutations of developmentally important genes are one possible mechanism for evolutionary change and integrated this idea into his shifting balance theory (see Wright 1982; Dietrich 2000). However, other architects of the Modern Synthesis were less sanguine about the evolutionary importance of “hopeful monsters” or, indeed, their very existence. For example, in a book-chapter entitled “Prologue: Some Thoughts on the History of the Evolutionary Synthesis”, Ernst Mayr wrote that “Goldschmidt belittled selection in a different way, when admitting that ‘selection may wipe out one type or isolate a new type’ but saying nothing about the possibility of selection leading to the construction of a new type… Goldschmidt thought of the origin of new types in terms of proximate causations …. In 1952 I asked Goldschmidt how the population in which a new hopeful monster occurred would react to it. He answered, after a considerable pause, ‘I have never thought of it that way’“ (Mayr and Provine 1980, p. 15). Mayr’s criticism was simple. By placing emphasis on the importance of genetic structure and developmental processes, Goldschmidt neglected the importance of population dynamics, which were appearant to the majority of the early architects of the Modern Synthesis (Gould 2002).
Clearly, the major criticism that has often been levelled against the evolutionary significance of “hopeful monsters” resulting from single-gene mutations is the low probability that any allele of this kind will become fixed in a population. Kimura (1962) showed that a new mutant arising as a single copy in a diploid population of size N has a probability of fixation P given by the formula P = (1 − e−2Nes/N)/(1 − e −4Nes), where Ne is the effective population size and s is the allele’s selective advantage. This formula reduces to P = (1 − e−2 s)/(1 − e−4 N s) when N = Ne, and becomes P = 1/2 N when s = 0.0 (i.e., a neutral mutation). Therefore, in the case of a hypothetical mutant with a selective advantage of s = 0.01 arising in a population of 1,000 individuals, P = (1 − e −0.002)/(1 − e −4) = 0.00199 (i.e., the fixation probability is roughly 0.2%), and, if the mutant is selectively neutral, P = 0.05%. This hypothetical case illustrates that “hopeful monsters” created even by advantageous or neutral allelic changes, which represent “best case scenarios”, have exceedingly low probabilities of becoming fixed in a population. Indeed, if our hypothetical mutant is even slightly deleterious (e.g., s = 0.001), we see that P = 0.004% when N = 1,000. Note that these calculations are based on relatively small natural populations of reproducing organisms (Niklas 1997).
Examples of multicellular hopeful monsters in the history of life
As noted above, Goldschmidt (1940) proposed that a “hopeful monster” occurs when one or more genes mutate to produce a significant phenotypic difference between the parent and its progeny. Strictly speaking, this phenomenon is predicted to occur in one generation and therefore excludes phenotypic changes resulting from non-heritable teratologies, hybridization or those caused by recessive allele mutations phenotypically expressed in the F1 when they occur in germ cell precursors followed by self-fertilization involving gametes bearing the mutated genome. Such a strict interpretation of the concept similarly excludes comparatively rapid phenotypic changes occurring over two or more generations in small populations experiencing parapatry, vicariance leading to allopatry, evolutionary scenarios involving neutral theory predictions, or even those resting on Sewall Wright’s shifting balance theory (Wright 1982). Also, “hopeful monsters” sensu stricto cannot be easily evoked to explain paleontological phenomena such as “punctuated equilibrium” or saltational evolution because the fossil record rarely provides sufficient evidence to justify the claim that phenotypic shifts occurring over even comparatively very short geological time periods (e.g., ~1 million years) took place in one or a few generations. This caveat is not an argument against the reality of punctuated equilibria or saltational evolution, which has been postulated to account for geologically rapid phenotypic changes in particular lineages (Eldredge 1989, 2008) (for paleobotanical examples, see Bateman and DiMichele 2002), but rather do emphasize what we believe Goldschmidt actually had in mind (in contrast to what some authors interpret to be his meaning).
Equally important, Goldschmidt’s “hopeful monsters” need not be viable proto-species. All that is required of them is that they are phenotypes that can survive and perhaps reproduce. Using this yardstick, the existence of “hopeful monsters” sensu stricto is irrefutable (case studies reviewed by Bachmann 1983; Garcia-Bellido 1983; Gottlieb 1984; Hilu 1983; Niklas 1997; Akam 1998; Gould 1977, 2002; Angelini and Kaufman 2005). The phyletic consequences of these macro-phenotypic changes are less clear. But an examination of the available literature, especially that devoted to mutations altering floral morphology, suggests that “hopeful monsters” have the potential to evolve rapidly into new kinds of organisms, because gene flow within populations of angiosperms are maintained in large part by the ability of flowers to attract specific pollinators and because a change in floral phenotype may result in shifts in pollination biology, which can genetically fragment and isolate reproductively viable populations. For example, Galen (1996) showed that populations of the alpine wildflower Polemonium viscosum can rapidly adapt to abrupt changes in pollinator assemblages. Her data indicate that the broadly flared flowers of the bumblebee pollinated P. viscosum could have evolved from narrower ones in a single generation because corolla flare increased by 12% from populations pollinated by a wide assemblage of insect pollinators to those pollinated only by bumblebees. Although this shift in floral phenotype is not the result of mutation, it demonstrates that mutations have the potential to result in rapid changes in gene flow within plant populations. By way of another example, species within the family Asteraceae are distinguished in part by whether their inflorescences contain radially symmetrical “disk” flowers, bilaterally symmetrical “ray” flowers, or both ray and disk flowers. Yet, by performing artificial crosses between two species of Haplopappus that have rayed and rayless florets (H. aureus and H. venetus subspecies venetus, respectively), Jackson and Dimas (1981) discovered that the presence or absence of ray flowers is controlled by a single gene, which can mutate to effect phenotypic differences reflected by the two species. Along similar lines, Singh and Jha (1978) examined X-ray induced mutants of soybean (Glycine max) that result in phenotypes bearing flowers with two or more carpels rather than one organ, which characterizes the family Fabaceae.
Perhaps the best known examples of single gene mutations with significant floral phenotypic effects are those altering homeotic genes (i.e., DNA-sequences that contain the genetic information required to direct development along a particular morphogenetic pathway, see Arthur 2002; Gilbert and Levit 2007), which have the ability to shift the developmental fate of cells, tissues, or entire organs. In the majority of cases, mutations of homeotic loci change the type (rather than the number) of organs produced, which suggests that the developmental patterns affected by these mutations involve genes that regulate organ identity and those that regulate organ number. Some of the best known floral homeotic mutations occur in the mouse-ear cress, Arabidopsis, and the snapdragon, Antirrhinum. Like many angiosperms with “perfect flowers”, these plants have four whorls of floral organs of which the outermost develop into sepals and the innermost develop into carpels. Mutations of AP3 and PI genes of Arabidopsis or the DEF gene of Antirrhinum cause petals to be replaced by sepals and stamens by carpels (Koorneef et al. 1983; Bowman et al. 1989), which results in “imperfect” flowers incapable of self-fertilization. Mutations in the AG gene of Arabidopsis and the PLENI gene of the snapdragon convert stamens into petals and carpels into sepals (Carpenter and Coen 1990), which are also incapable of self-fertilization. Because homeotic mutations such as these have the potential to establish reproductive barriers, they can serve as a genomic vehicle for character displacement, genetic divergence, and the eventual appearance of new species (Cubas et al. 1999; Albert et al. 2002; Fröhlich 2003, Rudall and Bateman 2003).
Neo-Goldschmidtian “hopeful monsters”: a less stringent definition
As noted, Goldschmidt’s “hopeful monster” concept focuses on systemic mutations that evoke large heritable phenotypic changes affecting embryological processes in a single generation. The stipulation that these changes occur in a single generation is arguably very stringent in light of numerous mechanisms that post-Goldschmidtian research has shown can result in rapid phenotypic departures from the parental condition but that require a few, rather than one, generation. For example, polyploidy resulting from mitotic or meiotic mis-division or interspecific hybridization is argued to be an important factor contributing to the rapid evolution and ecological success of the angiosperms, which appear to have genomes that are particularly tolerant of changes in chromosome number, insertions, deletion, or epigenome restructuring (for a recent review, see Leitch and Leitch 2007). Likewise, when we take a broad perspective of the factors responsible for some of the most significant changes in the course of organic evolution, it is impossible to ignore the importance of the primary endosymbiotic events that resulted in the lateral gene transfers that gave rise to the first eukaryotic life forms or those attending secondary and tertiary endosymbiotic events that engendered many new and important photoautotrophic clades such as the dinoflagellates and the euglenids (Niklas 1997; Cavalier-Smith 2000; Kutschera and Niklas 2004, 2005; Martin et al. 2001; Martin 2003; Embly and Martin 2006).
Indeed, in light of numerous evolutionarily important phenomena that result in significant genomic restructuring over the course of one or many generations, it is desirable to “relax” the temporal constraints that Goldschmidt imposed on the definition of a “hopeful monster” and to expand the repertoire of mechanisms that can (and have) resulted in dramatic phenotypic changes. Toward this end, we suggest that a “hopeful monster” should not be thought of as necessarily an individual within a population, but rather as any individual, type of organism or group of phyletically related organisms that demonstrates one or more significant heritable phenotypic departures from its ancestral condition over the course of one or a comparatively few generations. This Neo-Goldschmidtian perspective has three codicils: (1) it implies that the phenotypic changes resulting from single or multiple mutations are either adaptively neutral or potentially advantageous over the course of one or a few generations (i.e., the “hopeful monster” must survive but it need not be immediately reproductively competent), (2) it does not imply that the organism or its progeny be more ecologically or evolutionarily successful than its progenitors, although this may be (or become) the case, and (3) the effects of the mechanisms responsible for the initial genomic change(s) that result(s) in phenotypic modification(s) may set in motion a series of subsequent genomic restructurings occurring over the course of two or more generations.
Seen in this light, polyploid plants resulting from interspecific mating, or from mitotic or meiotic mis-divisions are “hopeful monsters” as are individuals or groups of organisms resulting from endosymbiotic events, which are associated with lateral gene transfer (Martin et al. 2001; Martin 2003; Embly and Martin 2006). To illustrate this point, we now pay particular attention to one group of organisms that resulted from secondary endosymbiosis, the euglenoids.
Euglenids: unicellular blue-light sensitive hopeful monsters
It has long been known that free-living green freshwater flagellates respond to a reduction in light intensity by characteristic changes in their motile behaviour. These unicellular organisms were first described by Antony von Leeuwenhoek (1632–1723), who wrote in 1675 that “the motion of most of these animalcules in the water was so swift, and so various, upwards, downwards, and round about, that was wonderful to see” (Wolken 1967). However, Christian G. Ehrenberg (1795–1876) was the first to systematically study and classify Leeuwenhoek’s “animalcules”. In 1830, he assigned these “infusoria” to the genus Euglena (“eyeball organism”). In his most important monograph he referred to the euglenids and characterized them as “green infusoria” that display plant- and animal-like characteristics (Ehrenberg 1838).
Up to the 1950s, biology students were taught that the ~1,000 described freshwater and marine morphospecies of euglenids (Figs. 1 and 2) collectively reflect the archaic condition at which point the plant and animal kingdom began to depart (Wolken 1967). This perspective was reasonable at the time because the taxonomic assignment of these organisms was difficult owing to the lumping of heterotrophic and photosynthetic species in the same genus (see Margulis and Schwartz 1998; Leander et al. 2007). Some euglenids, which live interstitially within sediments, are phagotrophic and ingest microbes such as bacteria or other eukaryotic cells. These species differ from free-living flagellated photosynthetic euglenids with respect to their gliding-mode of locomotion, which is achieved by means of two heterodynamic flagella that bear a row of hairs (cilia-like mastigonemata) and that are reinforced by a paraxial rod (Leander et al. 2007). The green, photosynthetic euglenids have chlorophyll a and b and were classified until 1968 by some workers as “green algae” (Chlorophyta). Other scientists placed all Euglena species in a separate division, the Euglenophyta. The erection of a new division was justified, as noted by Gibbs (1978), in part because the cell walls of Euglena consist of a proteinaceous pellicle (exoskeleton) as opposed to the carbohydrate-rich cell walls of green algae. Likewise, Euglena’s nuclear structure and mode of mitosis do not correspond to that of any other member of the Chlorophyta.
Recently, the confused taxonomy of the genus was resolved once it was appreciated that the photosynthetic euglenids evolved as a result of secondary endosymbiosis (Leander et al. 2007). Species such as E. gracilis (Figs. 1 and 2) are chimeric eukaryotic cells that occurred when a heterotrophic eukaryotic “host cell” acquired a green algal endosymbiont that subsequently assumed the functional role of a chloroplast (Fig. 3). This historical event was postulated by Gibbs (1978) on the basis of ultrastructural details among which the single most revealing line of evidence is the presence of a third membrane around the euglenoid chloroplast. This outer membrane represents the host cell’s plasmamembrane, which surrounded the endosymbiont and became invaginated.
Diagrammatic rendering of the symbiogenetic origin of the chloroplast in freshwater flagellates such as Euglena. “Ingestion” and survival of an algal cell (grey) via phagocytosis by a flagellated eukaryotic host cell (a) and subsequent reduction of the endosymbiont to a chloroplast surrounded by three membranes, which finalizes the secondary endosymbiotic “event” (b). c chloroplast, cy cytoplasm of the host cell, m mitochondrion, n nucleus (Adapted from Sitte 1989)
Gibbs’ hypothesis has been confirmed by numerous more recent investigations (Sitte 1989; Maier et al. 2000; Cavalier-Smith 2000; Moreira and Philippe 2001; Bhattacharya et al. 2003; Keeling 2004; Falkowski et al. 2004; Leander et al. 2007). Five lines of evidence are particularly revealing in terms of the endosymbiotic origin of photosynthetic euglenoids.
First, comparative morphology (cladistic analysis), combined with molecular phylogenetic data, reveal that the closest sister taxa to photosynthetic euglenids are eukaryophorous euglenozoa of the genus Paranema which has a cell ultrastructure consistent with the transition from a phagotrophic to a photosynthetic mode of nutrition (Leander et al. 2007). Also, it is now well established that the host cells of the “green Euglenophyta” are sister to members of the kinetoplastida (Trypanosoma, Leishmania) (Keeling 2004; Leander 2004; Takahashi et al. 2007) based in part on the general similarity between the cell morphologies of Euglena and Trypanosoma, which has long been recognized in the classical literature (Wolken 1967).
Second, in contrast to heterotrophic species, photosynthetic euglenids have an enlarged flagellar pocket or “reservoir” and a photo-sensing organelle near at the base of the flagellum. This “paraflagellar body” lies in the vicinity of the so-called “eyespot”, a carotinoid-rich shading structure involved in the step-up or step-down photophobic response of the “animal-like” micro-organism (Fig. 1). It is well established that the photoavoidance (or photoaccumulation) of green flagellates is mediated by blue light (Wolken 1967). Detailed studies have shown that the step-up photophobic response in Euglena gracilis is mediated by a blue-light receptor flavoprotein that is a photoactivated adenylyl cyclase. After light activation, an enhancement in cyclic AMP within the paraflagellar body promotes the activity of the flagellum as in sperm cells of metazoa (Iseki et al. 2002). Hence, with respect to the mode of the step-up photophobic response, E. gracilis displays a striking similarity to motile haploid cells of animals. In this context, it is worth noting that the mechanism of cytokinesis in Euglena is reminiscent to that of dividing heterotrophic “Protozoa” such as ciliates (Leander et al. 2007), but not analogous to the cell-plate associated mode of cell separation observed in many members of the green algae (Chlorophyta) (Niklas 2000, 2004; Scherp et al. 2001).
Third, studies of the feeding apparatus in heterotrophic euglenids reveal that those species that actively hunt and digest particulate food are characterized by an elaborate cytostome that is lacking in photosynthetic Euglena. The ultrastructure of this “siphon” has been studied in detail in several eukaryovorous (phagotrophic) euglenids. A vestigial feeding apparatus (cryptic cytostome) that serves no apparent function was discovered in Euglena more than two decades ago (Surek and Melkonian 1986). However, the phylogenetic significance of this relict feeding system was only fully appreciated within the context of more recent studies on the surface patterns and features of the cytoskeleton of various euglenids (Leander et al. 2007).
Fourth, the secondary plastids of Euglena are very similar to the corresponding organelles of land plants (embryophytes), but the nature of the “green cells” ingested and “enslaved” by the trypanosoma-like host organism remained unclear. A phylogenetic analysis based on a plastid-targeting, nuclear-encoded gene from a variety of green organisms revealed that the euglenophyte plastids may have originated relatively recently from a member of the basal lineage of the embryophyta (Takahashi et al. 2007). Hence, the trypanosoma-like ancestors of extant euglenids must have fed on large green prey cells for the subsequent chloroplast acquisition via secondary endosymbiosis.
Finally, a phylogenetic tree, based on rDNA sequences, reveals that the photosynthetic members of the genus Euglena comprise a monophyletic group (Triemer et al. 2006). This suggests that the secondary endosymbiotic event that gave rise to the “green” euglenids occurred only once during the evolutionary history of these chimeric unicellular “hopeful monsters”, which displayed little morphological change over the past 150 million years (Martin-Gonzalez et al. 2008).
Amoebic algae and other endosymbiotic “hopeful monsters”
The photosynthetic euglenids are not the only group of Goldschmidian-like “hopeful monsters”. Eighty years ago, “monster-like” marine green filamentous plasmodia were discovered by Geitler (1930) and named Chlorarachnion reptans (Fig. 4). The motile cells of Chlorarachnion capture prey organisms such as diatoms or bacteria via their filamentous pseudopodia whose structure is revealed with the aid of differential interference contrast microscopy (McFadden and Gilson 1995) (Fig. 5a). Like members of the genus Euglena (Figs. 1 and 2), the amoeboflagellate Chlorarachnion has green (chlorophyll b containing) plastids. However, in contrast to the euglenoid chloroplast, the chlorarachniophyte plastid has four membranes. In addition, it contains a degenerate nucleus, the “nucleomorph” (Figs. 5b and 6), a feature that also occurs in the Cryptophyta (McFadden and Gilson 1995; Maier et al. 2000; McFadden 2001; Keeling 2004; Gilson et al. 2006). The presence of a nucleomorph, which is all that remains of the nucleus of green algal endosymbionts (Fig. 6), has led to the general acceptance of the theory of secondary endosymbiosis (Cavalier-Smith 2000; Maier et al. 2000; McFadden 2001; Knoll 2003; Moreira and Philippe 2003; Dyall et al. 2004; Gibbs 2006; Archibald 2005, 2007; Leander et al. 2007; Braun and Phillips 2008).
The web-like creeping amoeba alga Chlorarachnion reptans. Light micrograph of a colony (a) and schematic drawing of three individuals (b) that have captured and engulfed diatoms (arrows) (Adapted from Geitler 1930)
Differential inference contrast light micrograph of the amoebic alga Chlorarachnion reptans (a) and diagram showing its chloroplast surrounded by four membranes within which the vestigial remains of the eukaryotic endosymbiont nucleus (the nucleomorph) is sandwiched between membranes two and three (b). c chloroplast, cy cytoplasm of the host cell, m mitochondrion, n nucleus, nm nucleomorph (Adapted from McFadden and Gilson 1995)
The symbiogenetic origin of the chloroplast in the amoeba alga Chlorarachnion reptans. A green algal cell is engulfed by a larger phagotroph to create a photosynthetic cell chimera. This process is associated with the lateral transfer of genes from the nucleus of the unicellular prey organism to that of the host (dashed lines) c chloroplast, cy cytoplasm of the host cell, m mitochondrion, n nucleus, nm nucleomorph (vestigial enslaved nucleus)
Although neither the green euglenids nor the green chlorarachniophytes play major ecological roles in marine or freshwater environments, “hopeful monsters” harbouring red (chlorophyll c containing) plastids as a consequence of secondary (as well as tertiary) endosymbiosis are ecologically dominant members of the world’s phytoplankton (Fig. 7). The differential ecological success of “green” versus “red” “hopeful monsters” in today’s oceans is curious. Certainly, when viewed in the context of the geologic time scale, the Paleozoic phytoplankton was dominated by cyanobacteria and green algae (Niklas 1997; Cowen 2000; Kutschera 2008; Kutschera and Niklas 2005), which stands in marked contrast to the dominant eukaryotic “red” phytoplankton of today (cryptophytes, haplophytes, diatoms, peridinium-containing dinophytes). Gryzebyk et al. (2003) have proposed that these phytoplankton clades rose to ecological prominence after the end-Permian mass extinction when the introduction of “red” secondary plastids into the cytoplasm of different heterotrophic host cells gave them an ecological advantage compared to their counterparts with “green” endosymbiotic plastids. This hypothesis is described in detail in a recent monograph edited by Falkowski and Knoll (2007).
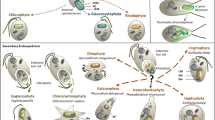
Diversity of extant unicellular eukaryotic organisms that document evolutionary secondary and tertiary endosymbiotic events. Chlorarachniophyta and Euglenophyta contain plastids (derived from green algae) that are surrounded by four or three membranes, respectively. Cryptophyta, Heterokontophyta and Haptophyta contain red plastids that are surrounded by four membranes (secondary endosymbiosis). The dinoflagellate Dinophysis (red lineage) contains plastids surrounded by more than four membranes (tertiary endosymbiosis). The chimeric cells of Chlorarachnion (green lineage) and Cryptomonas (red lineage) contain nucleomorphs
The dinoflagellates (division Pyrrhophyta, class Dinophyta) are one of the most important groups of phytoplankton in both marine and freshwater environments (Fig. 8). Nearly half of the described taxa are photoautotrophic; the remaining species are hetero- or mixotrophic flagellates. Some marine dinoflagellates are blue-light sensitive microbes that display intraspecific “cannibalistic” predation (Brunelle et al. 2007; Martel and Flynn 2008). Like the euglenid plastid, the red dinoflagellate plastid (which is characterized by the carotenoid peridinin) is surrounded by three membranes, which indicates these “hopeful monsters” where the result of secondary endosymbiosis (e.g., Lepidodinium). In addition, the plastids of several dinoflagellates have more than four membranes, indicating that these photosynthetic animals were the products of tertiary endosymbiosis (e.g., Dinophysis) (Figs. 7 and 8). These dinoflagellates are important primary oceanic producers (Graham and Wilcox 2000; Knoll 2003; Falkowski et al. 2004; Falkowski and Knoll 2007). They are the most common source of bioluminescent “red tides”, which has earned them the nickname of “fire plants” (Pyrrophyta); some dinoflagellates called “zooxanthellae” established a symbiosis with reef-forming coralls (Hackett et al. 2004; Falkowski et al. 2004).
Drawings of eleven species of dinoflagellates (dinoprotista), inclusive of members of the genera Peridinium and Dinophysis, reproduced from the classical monograph of Haeckel (1904)
Finally, it should be pointed out that successful primary and secondary endosymbiotic events (i.e., symbiogeneses sensu Merezhkowsky 1905, 1910 and Wallin 1927) that created novel unicellular cell chimera or “meta-algae” and subsequently gave rise to new classes of organisms are rare evolutionary phenomena. According to Cavalier-Smith (2000, 2006) only four to six endosymbiotic events had far-reaching consequences for the diversification of life on Earth: The origins of mitochondria and chloroplasts via primary endosymbiosis and at least two major secondary transfers of “green” and “red” chloroplasts from one photosynthetic eukaryote to a heterotrophic host cell. These key events in the history of life are depicted in Fig. 9.
Scheme depicting primary and secondary (palaeo)endosymbiotic events leading to novel multi- and unicellular body plans (macroevolution) in the phylogeny of the Monera (prokaryotic microbes), Fungi, Animalia, Plantae and Protoctista (eukaryotic micro-organisms). The placement of euglenids and dinoflagellata (Dinophyta) is indicated. These photosynthetic microbes are important members of the phytoplankton of freshwater and marine ecosystems. mya million years ago
Infusoria, Euglena, and Darwin’s primordial form
Although briefly discussed in the first (1859) edition of his seminal work, Darwin (1872) speculated at length on the common descent of all organisms on Earth in the 6th and definitive edition of The Origin of Species: “Analogy would lead me one step further, namely, to the belief that all animals and plants are descended from some one prototype… If we look even to the two main divisions—namely, to the animal and vegetable kingdoms—certain low forms are so far intermediate in character that naturalists have disputed to which kingdom they should be referred… Therefore, on the principle of natural selection with divergence of character, it does not seem incredible that, from some such low and intermediate form, both animals and plants may have been developed; and, if we admit this, we must likewise admit that all the organic beings which have ever lived on this earth may be descended from some one primordial form” (Darwin 1872, pp. 424–425). We do not know to which taxon Darwin referred when he talked about “low and intermediate” life forms. However, he mentioned “lower algae” in this context and knew the pertinent scientific literature of his time. It is therefore likely that he had “infusoria” in mind.
Certainly, Darwin was aware of Ehrenberg’s (1838) important monographic summary of what was then known about the “lower organisms” (i.e., bacteria, amoebae, unicellular algae etc.). These “Aufguss-Thierchen” or infusoria emerge in the liquid phase when grass or soil is covered by rain water and incubated for some time; they have a degree of internal organization similar to that of some “higher” animals. Unfortunately, Ehrenberg (1838) argued erroneously that the contractile vacuole and the nucleus of some “infusoria” correspond to the seminal vesicle and testis, respectively, and that the large granules in the peripheral cytoplasm were eggs. Hence, infusorians were regarded as complex unicellular organisms that were intermediate forms between “animals” (amoebae) and “plants” (lower algae). Nevertheless, because Ehrenberg (1838) described the genus Euglena for the first time (E. ehrenbergi was named in his honour; see Fig. 2), we suggest that these heterotrophic-photoautotrophic (“animal-plant”-like) protoctista served as the organic model for Darwin’s hypothetical “primordial, intermediate form”. Because Darwin (1872) was not aware of plant-microbe interactions and the associated principle of cell evolution via integration and cooperation (endosymbiosis) (Fig. 9), his “Proto-Euglena-hypothesis” for the origin of all forms of life is no longer acceptable. However, it should be stressed that Darwin’s general conclusion as to the common root of the tree of life has been confirmed by numerous cytological, biochemical and molecular studies (Kutschera and Niklas 2004).
Conclusions
In this article, we have juxtaposed Goldschmidt’s concept of “hopeful monsters” with the process of endosymbiosis. Goldschmidt (1933, 1940) proposed two mechanisms that can account for the sudden appearance of novel phenotypes—“systemic mutations” that result from large chromosomal re-arrangements and singe “macro”-mutations occurring in developmentally important genes that produce large phenotypic differences from the ancestral condition. Although the “hopeful monster”-concept was initially criticized severely (in large part because the low probability that mutations would be fixed in populations), numerous lines of evidence have since mounted in its favour (Theißen 2006).
Nonetheless, Goldschmidt’s original conceptualization stipulates that the phenotypic effects of single gene mutations or systemic genomic re-arrangements must occur in a single generation to constitute a legitimate “hopeful monster”. Unfortunately, this requirement immediately disqualifies many examples of dramatic phenotypic departures from the ancestral condition occurring over a few generations as examples of “hopeful monsters”. For this reason, we have advocated in this article a less stringent “Neo-Goldschmidian” perspective, one that permits novel phenotypes evoked by genomic changes to qualify as “hopeful monsters” even if the phenotypic effects of these changes occur over the course of two or more generations. This perspective immediately draws attention to the most profoundly important “hopeful monsters”, which evolved as a consequence of lateral gene transfer attending ancient and more recent endosymbiotic events.
Without doubt, the ecological and evolutionary importance of (endo)symbiosis, which was unknown to Darwin (1872) and ignored by the “architects” of the synthetic theory (see Reif et al. 2000), cannot be overstressed (Carroll 2001; Kutschera 2002, 2007). It gave rise to the first eukaryotic organisms during the Precambrian and it continued to give rise to numerous novel lineages of “unicellular photosynthetic animals” as a consequence of secondary and tertiary endosymbiotic events occurring throughout the Phanerozoic (Fig. 9). Unfortunately, the recognition that many “algae” were indeed not only “hopeful” but profoundly successful “monsters” came well after Goldschmidt’s time. Three decades ago, Gibbs (1978) suggested that flagellates of the genus Euglena were originally colourless unicellular eukaryotic animals that may have obtained their plastids by the ingestion and subsequent reduction of free-living green algae, based primarily on the fact that the Euglena chloroplasts have three rather than two membranes. However, the hypothesis that the third Euglena chloroplast membrane is derived from the cell membrane of the green algal endosymbiont (Fig. 3) was only accepted after the discovery of the nucleomorph in cryptomonads, such as Cryptomonas (Fig. 7). Today, primary, secondary (and tertiary) endosymbiosis is a well-established biological reality supported by a large body of empirical data dawn from a variety of eukaryotic microorganisms, including examples of early stages in the evolutionary development of ongoing endosymbiotic events (extant intermediate forms such as Paulinella, Cyanophora, Chlorarachnion etc., see McFadden 2001; Okamoto and Inouye 2005; Archibald 2005, 2006, 2007; Nowack et al. 2008).
We are not the first to suggest that a wider range of evolutionary mechanisms should be considered as agents capable of producing dramatic and sudden differences between ancestral and derived phenotypic states (see, for example, Bateman and DiMichele 2002). At issue is not the extent to which a broader range of mechanism so dilutes Goldschmidt’s “hopeful monster” concept as to make it meaningless. Rather, we believe the real issue is the time-scales over which these mechanisms operate. Single gene mutations can have immediate phenotypic effects as can gene duplication en masse through polyploidy. Therefore, these mechanisms are capable of producing “hopeful monsters” as Goldschmidt originally conceived them to be. In contrast, the phenotypic ancestral-to-derived changes attending natural selection operating over thousands or even hundreds of years cannot, in our opinion, be classified as “hopeful monsters” even if these changes appear to be “saltational” in the context of geological time-scales (Eldredge 1989, 2008). The importance of time-scales immediately draws attention to the phenotypic “immediacy” of endosymbiotic events—how long do they take do produce a “dramatic” phenotypic departure from the ancestral condition?
We would argue that any endosymbiont-host cell consortium that survives the initial “fusion” of two organisms represents an “immediate” phenotypic change even if the reproductive competency of one or both components is impaired. Goldschmidtian “hopeful monsters” need not be reproductive, nor need they possess an adaptive edge to qualify as such. They need only manifest a different phenotype within a single generation. Thus, we argue that any organism that survives an endosymbiotic event qualifies as a true “hopeful monster”, even if the process by which it eventually attains the status of a true species occurs over the course of geological time-scales. By the same token, any polyploid organism, such as a flowering plant, that survives en masse chromosome duplications is a “hopeful monster”, even if it is sterile and therefore an evolutionary “dead end”. This juxtaposition of Goldschmidt’s “hopeful monster” concept and the phenomenology of endosymbiotic events leads us to the centre of the arguably artificial divide between macro- and micromutation, that is, whether evolutionary transitions result from the modification of many genes with small effects (gradualism), or the modification of a few genes that result in profound phenotypic changes (saltationalism). The introduction and survival of a foreign organism in a host cell can be legitimately classified as a macromutation that nevertheless does not necessarily involve significant genomic modifications of either the endosymbiont or its host (i.e., a saltational event). In contrast, a complete functional integration of the two genomes within this “hopeful monster” may progress gradually over the course of thousands of generations and involve many genomic modifications. Hence, in the world of unicellular protoctista (“algae”), the continuity between macro- and microevolution postulated for multicellular organisms (Simons 2002) appears to exist, although more work is required to further corroborate this general conclusion (Cavalier-Smith 2006; Butterfield 2007).
References
Archibald JM (2005) Jumping genes and shrinking genomes–probing the evolution of eukaryotic photosynthesis with genomics. IUBMB Life 57:539–547
Archibald JM (2006) Algal genomics: exploring the imprint of endosymbiosis. Curr Biol 16:R1033–R1035
Archibald JM (2007) Nucleomorph genomes: structure, function, origin and evolution. Bioessays 29:302–402
Angelini DR, Kaufman TC (2005) Comparative developmental genetics and the evolution of arthropod body plans. Annu Rev Genet 39:95–199
Akam M (1998) Hox genes, homeosis and the evolution of segment identity: no need for hopeless monsters. Int J Dev Biol 42:445–451
Albert VA, Oppenheimer DG, Lindqvis C (2002) Pleiotropy, redundancy and the evolution of flowers. Trends Plant Sci 7:297–301
Arthur W (2002) The emerging conceptual framework of evolutionary developmental biology. Nature 415:757–764
Bachmann K (1983) Evolutionary genetics and the genetic control of morphogenesis in flowering plants. Evol Biol 16:157–208
Bateman RM, DiMichele WA (2002) Generation and filtering major phenotypic novelties: neoGoldschmidtian saltation revisited. In: Cronk QCB, Bateman RM, Hawkins JA (eds) Developmental genetics and plant evolution. Taylor & Francis, London, pp 109–159
Bhattacharya D, Yoon HS, Hackett JD (2003) Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26:50–60
Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1:37–52
Braun EL, Phillips N (2008) Phylogenomics and secondary plastids: a look back and a look ahead. J Phycol 44:2–6
Brunelle SA, Hazard ES, Sotka EE, Van Dolah FM (2007) Characterization of a Dinoflagellate crypochrome blue-light receptor with a possible role in circadian control of the cell cycle. J Phycol 43:509–518
Butterfield NJ (2007) Macroevolution and macroecology through deep time. Palaeontology 50:41–55
Carroll RL (2000) Towards a new evolutionary synthesis. Trends Ecol Evol 15:27–32
Carroll SB (2001) Chance and necessity: the evolution of morphological complexity and diversity. Nature 409:1102–1109
Cavalier-Smith T (2000) Membrane heredity and early chloroplast evolution. Trends Plant Sci 5:174–182
Cavalier-Smith T (2006) Cell evolution and Earth history: stasis and revolution. Phil Trans R Soc B 361:969–1006
Carpenter R, Coen ES (1990) Floral homeotic mutations proposed by transposon-mutagenesis in Antirrhinum majus. Genes Dev 4:1483–1493
Cowen R (2000) History of life, 3rd edn. Blackwell, Oxford
Cubas P, Vincent C, Coen E (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401:157–161
Darwin C (1872) The origin of species by means of natural selection, or the preservation of favoured races in the struggle for life, 6th edn. John Murray, London
Dietrich MR (2000) From hopeful monsters to homeotic effects: Richard Goldschmidt’s integration of development, evolution and genetics. Am Zool 40:738–747
Dietrich MR (2003) Richard Goldschmidt: hopeful monsters and other ‘heresies’. Nature Rev Genet 4:68–74
Dyall SD, Brown MT, Johnson PJ (2004) Ancient invasions: from endosymbionts to organelles. Science 304:253–257
Ehrenberg CG (1838) Die Infusionsthierchen als Vollkommene Organismen. Ein Blick in das tiefere organische Leben der Natur. Leopold Voss, Leipzig
Eldredge N (1989) Macroevolutionary dynamics. Species, niches, and adaptive peaks. McGraw-Hill, New York
Eldredge N (2008) The early evolution of punctuated equilibria. Evo Edu Outreach 1:107–113
Embly TM, Martin W (2006) Eukaryotic evolution: changes and challenges. Nature 440:623–630
Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR (2004) The evolution of modern eukaryotic phytoplankton. Science 305:354–360
Falkowski PG, Knoll AH (eds) (2007) Evolution of primary producers in the sea. Elsevier, Amsterdam
Fröhlich MW (2003) An evolutionary scenario for the origin of flowers. Nature Rev Genet 4:559–566
Galen C (1996) Rates of floral evolution: Adaptation to bumblebee pollination in an alpine wildflower Polemonium viscosum. Evolution 50:120–125
Garcia-Bellido A (1983) Comparative anatomy of cuticular patterns in the genus Drosophila. In: Godwin BC, Holder N, Wylie CC (eds) Development and evolution. Cambridge University Press, Cambridge, pp 227–255
Geitler L (1930) Ein grünes Filarplasmodium und andere neue Protisten. Arch Protistenkunde 69:615–637
Geus A, Höxtermann E (Hg) (2007) Evolution durch Kooperation und Integration. Zur Entstehung der Endosymbiosetheorie in der Zellbiologie. Basilisken-Presse, Marburg
Gibbs SP (1978) The chloroplast of Euglena may have evolved from symbiotic green algae. Can J Bot 56:2883–2889
Gibbs SP (2006) Looking at life: from binocular to the electron microscope. Annu Rev Plant Biol 57:1–17
Gilbert SF, Levit GS (2007) The national roots of evo-devo. Theory Biosci 126:115–116
Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI (2006) Complete nucleotide sequence of the chlorarachinophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci USA 103:9566–9671
Goldschmidt R (1933) Some aspects of evolution. Science 78:539–547
Goldschmidt R (1940) The material basis of evolution. Yale University Press, New Haven
Gottlieb LD (1984) Genetic and morphological evolution in plants. Am Nat 123:689–709
Gould SJ (1977) The return of hopeful monsters. Nat Hist 86:24–30
Gould SJ (2002) The structure of evolutionary theory. Harvard University Press, Cambridge
Graham LE, Wilcox LM (2000) Algae. Prentice-Hall, New Jersey
Grzebyk D, Schofield O, Vetriani C, Falkowski PG (2003) The mesozoic radiation of eukaryotic algae: the portable plastid hypothesis. J Phycol 39:259–267
Hackett DJ, Anderson DM, Erdner DL, Bhattacharya D (2004) Dinoflagellates: a remarkable evolutionary experiment. Am J Bot 91:1523–1534
Haeckel E (1904) Kunstformen der Natur. Bibliographisches Institut Leipzig, Wien
Hilu KW (1983) The role of single-gene mutations in the evolution of flowering plants. Evol Biol 16:97–128
Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M (2002) A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415:1047–1051
Jackson RC, Dimas CT (1981) Experimental evidence for systematic placement of the Haplopappus phyllocephalus complex. Syst Bot 6:8–14
Junker T (2004) Die zweite Darwinsche Revolution. Geschichte des Synthetischen Darwinismus in Deutschland 1924 bis 1950. Basilisken-Presse, Marburg
Keeling PJ (2004) Diversity and evolutionary history of plastids and their hosts. Am J Bot 91:1481–1493
Kimura M (1962) On the probability of fixation of mutant genes in populations. Genetics 47:713–719
Knoll AH (2003) Life on a young planet: The first billion years of evolution on earth. Princeton University Press, Princeton
Koorneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ (1983) Linkage map of Arabidopsis thaliana. J Hered 74:265–272
Kutschera U (2002) Bacterial colonization of sunflower cotyledons during seed germination. J Appl Bot 76:96–98
Kutschera U (2007) Plant-associated methylobacteria as co-evolved phytosymbionts: a hypothesis. Plant Signal Behav 2:74–78
Kutschera U (2008) Evolutionsbiologie. 3. Auflage. Verlag Eugen Ulmer, Stuttgart
Kutschera U, Niklas KJ (2004) The modern theory of biological evolution: an expanded synthesis. Naturwissenschaften 91:255–276
Kutschera U, Niklas KJ (2005) Endosymbiosis, cell evolution, and speciation. Theory Biosci 124:1–24
Kutschera U, Niklas KJ (2006) Photosynthesis research on yellowtops: macroevolution in progress. Theory Biosci 125:81–92
Kutschera U, Niklas KJ (2007) The epidermal-growth-control theory of stem elongation: an old and a new perspective. J Plant Physiol 164:1395–1409
Leander BS (2004) Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol 12: 251–258
Leander BS, Esson HJ, Breglia SA (2007) Macroevolution of complex cytoskeletal systems in euglenids. Bioessays 29:987–1000
Leitch AR, Leitch IJ (2007) Genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Maier U-G, Douglas SE, Cavalier-Smith T (2000) The nucleomorph genomes of cryptophytes and chlorarachinophytes. Protist 151:103–109
Margulis L, Schwartz KV (1998) Five kingdoms. An illustrated guide to the phyla of life on earth, 3rd edn. WH Freeman, New York
Martin W, Hoffmeister M, Rotte C, Henze K (2001) An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem 382:1521–1539
Martin W (2003) Gene transfer from organelles to the nucleus: Frequent and in big chunks. Proc Natl Acad Sci USA 100:8612–8614
Martin-Gonzalez A, Wierzchos JM, Gutierrez JC, Alonso J, Ascaso C (2008) Morphological stasis of protists in lower Cretaceous amber. Protist 159:251–257
Martel CM, Flynn KJ (2008) Morphological controls on cannibalism in a planktonic marine phagotroph. Protist 159:41–51
Mayr E, Provine,WB (eds) (1980) The evolutionary synthesis. Harvard University Press, Cambridge
McFadden G (2001) Primary and secondary endosymbiosis and the origin of plastids. J Phycol 37:951–959
McFadden G, Gilson P (1995) Something borrowed, something green: Lateral transfer of chloroplasts by secondary endosymbiosis. Trends Ecol Evol 10:12–17
Merezhkowsky C (1905) Über Nature und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralblatt 25: 593–604, 689–691
Merezhkowsky C (1910) Theorie der zwei Plasmaarten als Grundlage der Symbiogenese, einer neuen Lehre von der Entstehung der Organismen. Biol Centralblatt 30: 278–288, 289–303, 321–347, 353–367
Moreira D, Philippe H (2001) Sure facts and open questions about the origin and evolution of photosynthetic plastids. Res Microbiol 152:771–787
Niklas KJ (1997) The evolutionary biology of plants. The University of Chicago Press, Chicago
Niklas KJ (2000) The evolution of plant body plans–a biomechanical perspective. Ann Bot 85:411–438
Niklas KJ (2004) The walls that bind the tree of life. Bioscience 54:831–841
Nowack ECM, Melkonian M, Glockner G (2008) Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by Eukaryotes. Curr Biol 18:410–418
Okamoto N, Inouye I (2005) A secondary symbiosis in progress? Science 310:287
Reif W-E, Junker T, Hoßfeld U (2000) The synthetic theory of evolution: general problems and the German contribution to the synthesis. Theory Biosci 119:41–91
Rudall PJ, Bateman R (2003) Evolutionary change in flowers and inflorescences: evidence from naturally occurring terata. Trends Plant Sci 8:76–92
Scherp P, Grotha R, Kutschera U (2001) Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns and seed plants. Plant Cell Rep 20:143–149
Singh BB, Jha AN (1978) Abnormal differentiation of floral parts in a mutant strain of soybean. J Hered 69:143–144
Sitte P (1989) Phylogenetische Aspekte der Zellevolution. Biol Rundschau 28:1–18
Simons AM (2002) The continuity of microevolution and macroevolution. J Evol Biol 15:688–701
Streble H, Krauter D (1973) Das Leben im Wassertropfen. Mikroflora und Mikrofauna des Süßwassers. 4. Auflage. Frankh’sche Verlageshandlung, Stuttgart
Surek B, Melkonian M (1986) A cryptic cytostome is present in Euglena. Protoplasma 133:39–49
Takahashi F, Okabe Y, Nakada T, Sekimoto H, Ito M, Kataoka H, Nozaki H (2007) Origins of the secondary plastids of Euglenomorpha and Chlorarachniophyta as revealed by an analysis of the plastid-targeting, nuclear-encoded gene psbO. J Phycol 43:1302–1309
Theißen G (2006) The proper place of hopeful monsters in evolutionary biology. Theory Biosci 124:349–369
Triemer RE, Linton E, Shin W, Nudelman A, Monfils A, Bennett M, Brosnan S (2006) Phylogeny of the Euglenales based upon combined SSU and LSU rDNA sequence comparisons and description of Discoplastis Gen. nov. (Euglenophyta). J Phycol 42:731–740
Wallin IE (1927) Symbionticism and the origin of species. Bailliere, Tindall & Cox, London
Wolken JJ (1967) Euglena. An experimental organism for biochemical and biophysical studies, 2nd edn. Meredith Publishing, New York
Wright S (1982) The shifting balance theory and macroevolution. Annu Rev Genet 16:1–19
Acknowledgments
This article is dedicated to Prof. Sarah P. Gibbs, who discovered the principle of secondary endosymbiosis 30 years ago. The cooperation of the authors is supported by the Alexander von Humboldt-Stiftung (AvH), Bonn (Germany).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kutschera, U., Niklas, K.J. Macroevolution via secondary endosymbiosis: a Neo-Goldschmidtian view of unicellular hopeful monsters and Darwin’s primordial intermediate form. Theory Biosci. 127, 277–289 (2008). https://doi.org/10.1007/s12064-008-0046-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12064-008-0046-8