Abstract
PprA, a pleiotropic protein involved in radioresistance of Deinococcus radiodurans was detected in multiprotein DNA processing complex identified from this bacterium. pprA mutant expressing GFP-PprA could restore its wild type resistance of γ radiation. Under normal conditions, GFP-PprA expressing cells showed PprA localization on both septum trapped nucleoids (STN) and nucleoids located elsewhere (MCN). Cell exposed to 4 kGy γ radiation showed nearly 2 h growth lag and during this growth arrest phase, the majority of the cells had GFP-PprA located on MCN. While in late phase (∼120 min) PIR cells, when cells are nearly out of growth arrest, PprA was maximally found with STN. These cells when treated with nalidixic acid showed diffused localization of PprA across the septum. gyrA disruption mutant of D. radiodurans showed growth inhibition, which increased further in gyrA pprA mutant. Interestingly, gyrA mutant showed ∼20-fold less resistance to γ radiation as compared to wild type, which did increase further in gyrA pprA mutant. These results suggested that PprA localization undergoes a dynamic change during PIR, and its localization on nucleoid near septum and functional interaction with gyrase A might suggest a mechanism that could explain PprA role in genome segregation possibly through topoisomerase II.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Deinococcus radiodurans, a member of family Deinococcaceae, is extremely resistant to several abiotic stresses including radiations and desiccation (Slade and Radman 2011; Misra et al. 2013). An efficient DNA double strand break (DSB) repair by extended synthesis dependent strand annealing (ESDSA) mechanism (Zahradka et al. 2006) and the strong oxidative stress tolerance are amongst the mechanisms that are implicated in the extreme phenotypes of this bacterium (Culotta and Daly 2013). Another unique feature of this bacterium is that it has a multipartite genome, which is organized in the form of a highly condensed toroidal structure (Levin-Zaidman et al. 2003; Zimmerman and Battista 2005). Proteins associated with nucleoid of D. radiodurans are characterized and a number of SMC (structural maintenance of chromosomes) like proteins including TopoII have been detected (Makarova et al. 2001; Nguyen et al. 2009). DNA topoisomerases are ubiquitous enzymes that help to maintain the correct DNA topology very specific to an organism. Roles of these enzymes in resolution and segregation of newly replicated intertwined circular chromosome have been demonstrated in E. coli (Champoux 2001).
D. radiodurans genome encodes topoisomerase II (TopoII) and topoisomerase IB (TopoIB) enzymes. TopoIB has been structurally characterized (Krogh and Shuman 2002) and TopoII was found to be associated with nucleoid and its colocalization with the nucleoid has been shown in D. radiodurans (Nguyen et al. 2009). PprA is found in multiprotein complex characterized from D. radiodurans and could stimulate the DNA end-joining activity of deinococcal ATP type DNA repair ligase, another component of the complex (Kota and Misra 2008; Kota et al. 2010) as well as T4 DNA ligase (Narumi et al. 2004). Devigne et al. (2013) have reported the involvement of PprA in DNA segregation and cell division in D. radiodurans. But the mechanism(s) underlying this role of PprA and its implication in the radioresistance of this bacterium were not explained. In this paper, we demonstrate that PprA works with DNA gyrase and contributes to radioresistance mostly through genome maintenance. Further, we demonstrated that trans expression of GFP-PprA complemented pprA loss in wild type and its cellular localization changes during γ radiation stressed growth in D. radiodurans. Mislocalization of PprA in the presence of nalidixic acid (Nal) and lack of any additive effect of pprA disruption on the γ radiation resistance loss in gyrA mutant might suggest the involvement of both PprA and GyrA in radioresistance through common pathway(s), most likely through faithful segregation of duplicated genome elements in daughter cells.
Materials and methods
Bacterial strains, plasmids and media
D. radiodurans R1 (ATCC13939) and pprA:cat (hereafter referred as pprA mutant) mutant were gifts from Professor J. Ortner, DLR, Institute Aerospace Medicine, Köln, Germany (Schaefer et al. 2000) and Prof. I. Narumi, JAERI, Takasaki, Japan, respectively. These strains were maintained in TGY (0.5% Bacto tryptone, 0.3% Bacto yeast extract and 0.1% glucose) medium. E. coli strain HB101 was used for maintaining the cloned gene on plasmids while E. coli BL21 (DE3) pLysS was used for the expression of recombinant protein. Recombinant E. coli harbouring expression vectors and their derivatives were grown in the presence of required antibiotics as described in respective experiments. Shuttle expression vector pVHS559 (Charaka and Misra 2012) and its derivatives were maintained in E. coli strain HB101 in presence of spectinomycin (40 μg/mL), while in D. radiodurans these were maintained in presence of spectinomycin (75 μg/mL). All recombinant techniques were as described earlier (Sambrook and Russell 2001).
Construction of PprA-GFP expression plasmid
Genomic DNA of D. radiodurans was prepared using protocol as described earlier (Battista et al. 2001). In brief, the coding sequence of PprA was PCR amplified from the chromosomal DNA of D. radiodurans and cloned at pDSW209 vector (Weiss et al. 1999) at BamHI and HindIII sites, downstream to GFP protein to yield pDSpprA. The pDSpprA plasmid was transformed to E. coli DH5 α strain and the recombinant clones were induced with 100 μM IPTG and the expression of translation fusion was checked using antibodies against GFP and PprA separately. The gfp–pprA region was PCR amplified from pDSpprA and cloned at ScaI and XhoI sites in pVHS559 and pVHpprA was obtained. Both pVHS559 and pVHpprA were transformed into pprA mutant and transformants were screened on TYG agar plates supplemented with spectinomycin (75 μg/mL). For localization of PprA, the pVHpprA recombinant plasmid was transformed into D. radiodurans. Recombinant GFP-PprA was expressed by inducing the culture with 10 mM IPTG in case of D. radiodurans.
Generation of gyrase A disruption mutant
The coding sequence of gyrase A (DR_1913) subunit of D. radiodurans was PCR amplified using gyrAF (5\(^{\prime }\)CGCGG ATCCATGACCGGAATTCAACCTGT3\(^{\prime })\) and gyrAR (5\(^{\prime }\)CC CAAGCTTTTACAGCTCGTCTTCCAAGCGA3\(^{\prime })\) primers and cloned at BamHI and HindIII sites in pDSW209 to get pDSgyrA plasmid. The pDSgyrA was digested with AfeI and the ApaI-XbaI fragment containing complete expressing cassette of nptII (KanR) from pNOKOUT (Khairnar et al. 2008) was excised out and blunt end ligated at AfeI site in pDSgyrA. The resultant plasmid pGAnptII was linearized and transferred into both wild type and pprAmutant cells. The transformants were grown for several generations and dose of kanamycin was increased from 5 μg/mL to 25 μg/mL in successive subculture. The cells growing in the presence of chloramphenicol (5 μg/mL) and kanamycin (25 μg/mL) were checked for allelic replacement of gyrA with gyrA:nptII by PCR amplification. The cells conferring gyrA:nptII disruption in pprA mutant and wild type were named as gyrA pprA double mutant and gyrA single mutants, respectively.
Gamma radiation treatment
Bacterial cells were treated with required doses of γ radiation as described earlier (Misra et al. 2006). In brief, the overnight grown culture of pprA mutant expressing wild type GFP-PprA fusion was subcultured in fresh TYG medium containing spectinomycin (75 μg/ml). To it, 10 mM IPTG was added and these cells were allowed to grow for 14 h. For checking the functional complementation, the pprA mutant expressing wild-type GFP-PprA in trans pprA mutant containing pVHS559 vector and wild-type D. radiodurans containing pVHS559 plasmid were subjected to different doses of γ radiation (2, 4 and 6 kGy). Appropriate dilutions of these cultures were spread on TGY agar plate and the colony-forming units were recorded after 36 h incubation at 32∘C. For microscopic studies, pprA mutant expressing PprA-GFP were irradiated with 4 kGy γ radiation at a dose rate of 4.16 kGy/h in Gamma chamber (GC 5000, 60Co, Board of Radiation and Isotopes Technology, Mumbai, India). The irradiated cells were allowed to recover in TGY medium and samples were collected at different PIR points and used for further studies as required.
Microscopic examination
Fluorescence microscopic studies were carried out as described earlier (Charaka and Misra 2012). In brief, the Deinococcus cells containing pVHpprA (GFP-PprA) were induced with 10 mM IPTG for 18 h. Cells were stained with Nile red (4 μg/mL) and 4, 6 diamdino2-phenylodine dihydroide chloride (DAPI) (0.2 μg/mL) separately. For Nal effect, the cells expressing GFP-PprA were treated with γ radiation and incubated with and without Nal (20 μg/mL) for 2 h and in the presence of 10 mM IPTG. These cells were mounted on agarose-coated slides (1.2%) and observed under Axio Imager CM5 microscope as described in Charaka and Misra (2012). Nile red, DAPI and GFP fluorescence was recorded at 516 nm, 350 nm and 488 nm, respectively. Images were analysed using Axiovision 4.8 software, modified using Image J and Adobe photoshop CS3 software. Cell and nucleoid sizes were measured by Axiovision software and statistically analysed using graphpad Prism software.
Results and discussion
GFP-PprA expression complemented γ radiation resistance loss in pprA mutant
The effect of γ radiation on survival of pprA mutant was compared with wild type. The PprA involvement in γ radiation resistance has been known for nearly 10 years (Narumi et al. 2004) and its role in UV radiation and mitomycin C resistance in D. radiodurans have been demonstrated recently (Selvam et al. 2013). The mechanism(s) underlying PprA involvement in different types of DNA damage tolerance is not well-understood. To understand the real-time function(s) of PprA during post DNA damage recovery, GFP-PprA fusion was expressed in D. radioduransand its activity was ensured. We demonstrated that pprA mutant, which showed nearly 5-fold drop in γ radiation resistance as compared to wild type at 6 kGy, could restore the γ radiation resistance of mutant to almost wild type levels when GFP-PprA was expressed in trans (figure 1). This confirmed that PprA in the form of GFP-PprA was functionally active and could be used for cellular localization of PprA in D. radiodurans.
Functional complementation of GFP-PprA in pprA:cat mutant of Deinococcus radiodurans. Exponentially growing cells of D. radiodurans (wild type), its pprA:cat mutant (pprA) and mutant expressing GFP-PprA in trans (pprA+PprA) were treated with different doses of γ radiation and colony forming units were monitored on TGY agar plate.
Cellular localization of PprA shows dynamics during postirradiation recovery
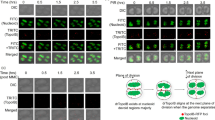
pprA mutant expressing GFP-PprA was examined microscopically and the localization of PprA during different stages of PIR was monitored. We found that GFP-PprA localizes as green fluorescent spot on septum-trapped nucleoid (STN) as well as on nucleoids located in the mid-cell position (MCN) (figure 2A). Although PprA localization on D. radiodurans genome was reported earlier (Devigne et al. 2013), here we noticed that the preference of PprA localization between STN and MCN was found to be cell-cycle stage dependent. In unirradiated heterogeneous population, the 47.1 ± 4.93% cells had GFP-PprA localized with MCN while 13.4 ± 2.52% cells were having GFP-PprA associated with STN. During early PIR when growth is completely arrested, larger fraction of cell showed PprA localization on MCN. However, after 2 h PIR, when cells resume growth, the majority of these cells showed the green fluorescent dot on STN (figure 2B). Interestingly, it was observed that the cells exposed to 4 kGy γ radiation showed delayed growth with ∼2 h lag period (figure 2C) and resumed its growth after 2 h PIR. Such types of dynamic shift in GFP-PprA localization from MCN to STN position in 2 h PIR (figure 2B), the time when these cells are nearly out of cell-cycle arrest phase (figure 2C), provided evidence though indirect to suggest that PprA has a role in molecular processes associated with cytokinesis. Devigne et al. (2013) have also shown the similar role of this protein albeit using immunostaining approach (Devigne et al. 2013), and earlier PprA was shown to be a nucleoid-binding protein in mammalian cells (Satoh et al. 2006). These results indicated that PprA binds to STN in cells ready for division and therefore, its role in faithful transmission of duplicated genome elements in daughter cells could be suggested.
Effect of γ induced DNA damage on genome dynamics in mutant expressing GFP-PprA. The pprA:cat mutant expressing PprA-GFP was treated with 4 kGy γ radiation and cells were collected at different PIR periods (60, 90, 120 and 180 min) and imaged for GFP fluorescence as described in materials and methods. Based on the localization of nucleoids, these were designated either at septum trapped position (STN) or located elsewhere in the cell (MCN). The area marked in red circles is shown as clips for STN and MCN (A). Nearly 500 of such cells were counted for distribution of GFP-PprA foci on nucleoid position segregated to either MCN or STN category and plotted as a function of recovery period (B). D. radiodurans R1 treated with 4 kGy gamma radiation and allowed to recover in fresh growth medium. Change in optical density at 600 nm was monitored online (C).
PprA works through DNA gyrase
Since, majority of the cells that showed PprA localization on STN during PIR, were out of post lag phase, the possibility of PprA playing an important role in the segregation of duplicated intertwined circular chromosomes in D. radiodurans similar to that is reported in E. coli (Champoux 2001) was hypothesized. There are three known mechanisms that contribute to segregation/resolution of duplicated bacterial genome elements prior to the cell division. These are (i) the classical genome partitioning system involving ‘Par’ proteins, (ii) through the involvement of DNA topoisomerase II (TopoII) and IV as in case of E. coli (Champoux 2001), and (iii) FtsK/XerCD recombinase in E. coli (Lesterlin et al. 2004) and SpoIIIE in Bacillus subtilis (Kaimer et al. 2009, 2011. We did not find any change in the localization pattern of PprA in cells lacking centromere-binding protein(s) i.e. ΔparB1, ΔparB2 and ΔparB3 mutants (data not shown), which may rule out the possibility of PprA’s working through classical ‘Par’ system in D. radiodurans. On the other hand, we observed that PprA localization pattern on STN was entirely different in cells treated with Nal an inhibitor of DNA topoisomerase II, as compared to both untreated (figure 3A) and 2 h PIR controls. For example, unlike controls, the Nal treated cells showed diffused GFP-PprA localization across the septum. Since, PprA is also characterized as a DNA-binding protein with its preference to dsDNA ends, its diffused binding could also be attributed to its interaction on the damaged site in the genome. These results suggested that the compact localization of PprA on STN requires an active DNA gyrase.
Functional interaction of DNA gyrase A with PprA. The pprA::cat cells expressing GFP-PprA were grown under normal conditions (control) and in the presence of Nal (50 μg/ml) for 2 h in the presence of 20 mM IPTG and cells were imaged for GFP fluorescence (A). gyrA gene was disrupted with nptII in wild type (gyrA) and pprA mutant (gyrApprA) backgrounds and the effect of gyrA disruption on growth characteristics of D. radiodurans was compared with wild type (WT) and pprA mutant (pprA) cells (B). Similarly, an equal OD 600 of wild type (WT) and gyrA (gyrA), pprA (pprA), and gyrApprA (gyrApprA) mutant cells were treated with 6.5 kGy γ radiation and the percent survival of these cells in response to γ radiation exposure was compared with respective untreated controls (C).
To ascertain if PprA and GyrA interactions contribute to the radioresistance and in the normal growth of this bacterium, gyrA gene was disrupted in both wild type and pprA mutant cells. These mutants were compared for normal growth as well as γ radiation response. gyrA mutant showed normal growth inhibition of D. radiodurans, which increased further in gyrA pprA double mutant (figure 3B, compare gyrA and gyrA pprA). However, the level of γ radiation resistance in pprA gyrA double mutant was nearly similar to gyrA single mutant at 6.5 kGy dose, which was ∼20 fold less as compared to wild type (figure 3C). This indicated that gyrA mutation is dominant over pprA mutation and also that GyrA and PprA proteins seem to function through common pathway(s) at least in radioresistance of this bacterium. These results suggested that PprA and GyrA are functionally interacting in this bacterium.
D. radiodurans is better known for its extreme radioresistance but its cytogenetic features are equally fascinating (Misra et al. 2013). Remarkably, it assembles its shattered genome quite efficiently by employing ESDSA mechanism (Zahradka et al. 2006) and protects its biomolecules, at least proteins, from oxidative damage (Daly et al. 2010). A pleiotropic mutation making D. radiodurans sensitive to various types of DNA damages was mapped in the genome of this bacterium and named as pprA (Kitayama et al. 1983; Narumi et al. 2004). The mechanisms underlying PprA role in radioresistance so far were suggested through the protection of damaged DNA from nucleolytic degradation and stimulation of DNA ends joining activity of DNA ligases (Narumi et al. 2004; Kota et al. 2010). Since, PprA was also found in the multiprotein DNA processing complex comprising of a few other important DNA-repair proteins (Kota and Misra 2008), the possibility of PprA working through protein–protein interaction could be believed. Recent observation though with limited mechanistic details, that PprA is also important in cell division and genome segregation (Devigne et al. 2013) indicated the possibility of a role for protein in these processes that are well-associated with radioresistance. Here, we report some interesting results suggesting that (i) the cellular localization of PprA on nucleoids either trapped between the septum (STN) or located elsewhere (MCN), during PIR depends upon the recovery stage of these cells from γ radiation damage and also on their preparedness for cell division, and (ii) PprA works through DNA gyrase because Nal treatment resulted in diffused localization of GFP-PprA in the cell, and gyrA inactivation in pprA background did not show additive effect in γ radiation response of gyrA single mutant. D. radiodurans genome is multipartite with ploidy and forms toroidal structure, which once upon a time was thought to be the basis for its extreme radioresistance (Levin-Zaidman et al. 2003). Although, its genome is annotated with the genes encoding TopoIB and both the subunits of TopoII (White et al. 1999), the roles of these enzymes in genome maintenance and radiation resistance are yet to be studied in D. radiodurans. Except that TopoIB has been structurally characterized (Krogh and Shuman 200) and TopoII was detected in the pool of proteins identified from the nucleoid of this bacterium (Nguyen et al. 2009) and now it is shown that the levels of TopoIB as well as both the subunits of TopoII were found to be significantly more in nucleoid of γ irradiated cells than unirradiated (de la Tour et al. 2013). This raises the possibility of DNA topoisomerase’s roles in radiation resistance and genome maintenance of this bacterium. Although, the molecular basis that would explain the involvement of PprA and DNA gyrase in genome segregation and cell division remains to be seen, PprA supports to TopoII in the decatenation of intertwined duplicated circular chromosomes may be speculated. Earlier, the roles of DNA topoisomerases in decatenation of duplicated intertwined circular chromosome of E. coli and bacterial genome segregation have been reported (Forterre et al. 2007; Champoux 2001). Regardless of the mechanism(s), it is important to emphasize that PprA is found to have distinct roles in nondividing (i.e. DSB repair) and dividing cells, where it seems to regulate the faithful distribution of duplicated genome, most probably through the involvement of DNA gyrase in D. radiodurans.
References
Battista J. R., Park M. J. and McLemore A. E. 2001 Inactivation of two homologous proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology 43, 133–139.
Champoux J. J. 2001 DNA topoisomerase: structure, function and mechanism. Annu. Rev. Biochem. 70, 369–413.
Charaka V. K. and Misra H. S. 2012 Functional characterization of chromosome I partitioning system in Deinococcus radiodurans for its role in genome segregation. J. Bacteriol. 194, 5739– 5748.
Culotta V. C. and Daly M. J. 2013 Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid. Redox Signal. 19, 933–944.
Daly M. J., Gaidamakova E. K., Matrosova V. Y., Kiang J. G., Fukumoto R., Lee D. Y. et al. 2010 Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5, e12570.
de la Tour C. B., Passot F. M., Toueille M., Mirabella B., Guérin P., Blanchard L. et al. 2013 Comparative proeomics reveals key proteins recruited at the nucleoid of Deinococcus after irradiation-induced DNA damage. Proteomics. 31, 933–944.
Devigne A., Mersaoui S., Bouthier-de-la-Tour C., Sommer S. and Servant P. 2013 The PprA protein is required for accurate cell division of γ-irradiated Deinococcus radiodurans bacteria. DNA Repair 12, 265–272.
Forterre P., Gribaldo S., Gadelle D. and Serre M. C. 2007 Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446.
Kaimer C., González-Pastor J. E. and Graumann P. L. 2009 SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis. Mol. Microbiol. 74, 810–825.
Kaimer C., Schenk K. and Graumann P. L. 2011 Two DNA translocases synergistically affect chromosome dimer resolution in Bacillus subtilis. J. Bacteriol. 193, 1334–1340.
Khairnar N. P., Kamble V. A. and Misra H. S. 2010 RecBC enzyme overproduction affects UV and γ radiation survival of Deinococcus radiodurans. DNA Repair 7, 40–47.
Kitayama S., Asaka S. and Totsuka K. 1983 DNA double stand breakage and removal of cross-links in Deinococcus radiodurans. J. Bacteriol. 155, 1200–1207.
Kota S. and Misra H. S. 2008 Identification of a DNA processing complex from Deinococcus radiodurans. Biochem. Cell. Biol. 86, 448–458.
Kota S., Kamble V. A., Rajpurohit Y. S. and Misra H. S. 2010 ATP-type DNA ligase requires other proteins for its activity in vitro and its operon components for radiation resistance in Deinococcus radiodurans in vivo. Biochem. Cell. Biol. 88, 783–790.
Krogh B. O. and Shuman S. 2000 Catalytic mechanism of DNA topoisomerase IB. Mol. Cell 5, 1035–1041.
Lesterlin C., Barre F. X. and Cornet F. 2004 Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 54, 1151–1160.
Levin-Zaidman S., Englander J., Shimoni E., Sharma A. K., Minton K. W. and Minsky A. 2003 Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance Science 299, 254–256.
Makarova K. S., Aravind L., Wolf Y. I., Tatusov R. L., Minton K. W., Koonin E. V. et al. 2001 Genome of extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspectives of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–49.
Misra H. S., Rajpurohit Y. S. and Kota S. 2013 Physiological and molecular basis of extreme radioresistance in Deinococcus radiodurans. Curr. Sci. 104, 194–205.
Misra H. S., Khairnar N. P., Kota S., Shrivastava S., Joshi V. P. and Apte S. K. 2006 An Exonuclease I sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol. Microbiol. 59, 1308–1316.
Narumi I., Satoh K., Cui S., Funayama T., Kitayama S. and Watanabe H. 2004 PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol. Microbiol. 54, 278–285.
Nguyen H. H., Bouthier de la Tour C., Toueille M., Vannier F., Sommer S. and Servant P. 2009 The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol. Microbiol. 73, 240–252.
Sambrook J. and Russell D. W. 2001 Molecular cloning: a laboratory manual, 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA.
Satoh K., Wada S., Kikuchi M., Funayama T., Narumi I. and Kobayashi Y. 2006 Method for detecting DNA strand breaks in mammalian cells using the Deinococcus radiodurans PprA protein. Mut. Res. 596, 36–42.
Schaefer M., Schmitz C., Facius R., Horneck G., Milow B., Funken K.-H. and Ortner J. 2000 Systematic study of parameters influencing the action of Rose Bengal with visible light on bacterial cells: comparison between biological effect and singlet-oxygen production. Photochem. Photobiol. 71, 514–523.
Selvam K., Duncan J. R., Tanaka M. and Battista J. R. 2013 DdrA, DdrD, and PprA: Components of UV and Mitomycin C Resistance in Deinococcus radiodurans R1. PLoS One 8, e69007.
Slade D. and Radman M. 2011 Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 75, 133–141.
Weiss D. S., Chen J. C., Ghigo J. M., Boyd D. and Beckwith J. 1999 Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181, 508–520.
White O., Eisen J. A., Heidelberg J. F., Hickey E. K., Peterson J. D., Dodson R. J. et al. 1999 Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286, 1571–1577.
Zahradka K., Slade D., Bailone A., Sommer S., Averbeck D., Petranovic M. et al. 2006 Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443, 569–573.
Zimmerman J. M. and Battista J. R. 2005 A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 31, 15–17.
Acknowledgements
Authors are grateful to Dr S. K. Apte, for his constructive criticism and encouragement while pursuing this work. We thank Prof. Narumi for pprA:cat mutant, Prof. Suzanne Sommer for p11559, and Dr (Mrs) Bhavani Shankar and Dr Jaya Kumar, RB and HSD, for their support in fluorescence microscopic studies. CVK thanks Department of Atomic Energy, Government of India, for research fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Kota S., Charaka V. K. and Misra H. S. 2014 PprA, a pleiotropic protein for radioresistance works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans. J. Genet. 93, xx–xx]
Rights and permissions
About this article
Cite this article
KOTA, S., CHARAKA, V.K. & MISRA, H.S. PprA, a pleiotropic protein for radioresistance, works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans . J Genet 93, 349–354 (2014). https://doi.org/10.1007/s12041-014-0382-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-014-0382-z