Abstract
Topoisomerases are crucial enzymes in genome maintenance that modulate the topological changes during DNA metabolism. Deinococcus radiodurans, a Gram-positive bacterium is characterized by its resistance to many abiotic stresses including gamma radiation. Its multipartite genome encodes both type I and type II topoisomerases. Time-lapse studies using fluorescently tagged topoisomerase IB (drTopoIB-RFP) and DNA gyrase (GyrA-RFP) were performed to check the dynamics and localization with respect to DNA repair and cell division under normal and post-irradiation growth conditions. Results suggested that TopoIB and DNA gyrase are mostly found on nucleoid, highly dynamic, and show growth phase-dependent subcellular localization. The drTopoIB-RFP was also present at peripheral and septum regions but does not co-localize with the cell division protein, drFtsZ. On the other hand, DNA gyrase co-localizes with PprA a pleiotropic protein involved in radioresistance, on the nucleoid during the post-irradiation recovery (PIR). The topoIB mutant was found to be sensitive to hydroxyurea treatment, and showed more accumulation of single-stranded DNA during the PIR, compared to the wild type suggesting its role in DNA replication stress. Together, these results suggest differential localization of drTopoIB-RFP and GyrA-RFP in D. radiodurans and their interaction with PprA protein, emphasizing the functional significance and role in radioresistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Topoisomerases are ubiquitous enzymes responsible for resolving the DNA topological problems that arise during DNA packaging, replication, repair, recombination, DNA–protein interaction, and other cellular processes. Topoisomerases are categorized into either type I or type II depending on the number of strands the enzyme cleaves during the course of action. Type I enzymes nick only one DNA strand and pass either one or both the strands of DNA through the break before the enzyme reseals it. Type II enzymes cleave both strands of the DNA making a double-strand break and passing both the strands through the break and then religating it. Unlike type II enzymes, type I topoisomerases are not dependent on ATP for their catalytic activity. Type I enzymes are further divided into type IA and type IB based on their structure and mode of action. The bacterial and archaeal topoisomerase 1 (TopI), topoisomerase III, and reverse gyrase are included in type IA family enzymes and they form a covalent bond with the 5'-end of the cut DNA and catalytic tyrosine (Champoux 2001, Chen et al. 2013). The catalytic eukaryotic nuclear topoisomerase I and poxvirus topoisomerases belong to the type IB family. Type IB family enzymes form covalent bonds with the 3'-end of the cut DNA and the catalytic tyrosine, which relaxes both positive and negative supercoiled DNA. A few bacteria like Deinococcus, Mycobacterium, Agrobacterium, Bordetella, and Sphingomonas also contain type IB-like topoisomerases which are more similar to those of poxvirus topoisomerases (Krogh and Shuman 2002). In general, cellular organisms contain more than one type of topoisomerases to solve a variety of topological problems. For example, in humans, six topoisomerases are present, belonging to both type I and type II, acting in different cellular processes for stable genome maintenance (Pommier et al. 2022).
Deinococcus radiodurans is an extremophile, known for its resistance to many DNA-damaging agents including ionizing radiation. Efficient DNA repair pathways, strong anti-oxidant mechanisms along with genomic features like condensed nucleoid, multipartite genome, high GC content, and polyploidy contribute to the extreme phenotype of this bacterium (Cox and Battista 2005; Slade and Radman 2011; Misra et al. 2013). The D. radiodurans genome encodes two type I topoisomerases and one type II topoisomerase and lacks topoisomerase IV enzyme, generally found in E. coli (Zechiedrich et al.2000) for maintaining the correct topology of the genome (White et al. 1999). The type I topoisomerases belong to both type IA (topA, encoded by E5E91_RS06960) and type IB (topoisomerase IB, encoded by E5E91_RS03550) classes whereas DNA gyrase is the only type II enzyme that performs supercoiling and decatenation activities in this bacterium. DNA gyrase was shown as one of the main proteins involved in nucleoid architectural organization. DNA gyrase and topA were also found significantly in higher amounts on nucleoids upon irradiation (de la Tour. et al. 2013). Topoisomerase IB of D. radiodurans (drTopoIB) resembles more to poxvirus TopIB than the eukaryotic counterpart in structure, size, and overall domain organization. The crystal structure of drTopoIB with DNA has revealed that the enzyme has two DNA binding sites and the secondary DNA binding site assists in DNA synopsis and plectonomic supercoiling (Patel et al. 2010). Although, the functional characterization of drTopoIB's role in genome maintenance through its interaction with genome segregation proteins has been shown earlier (Krogh and Shuman 2002; Patel et al. 2010, Kota and Misra 2015; Kota et al. 2021); the real-time in vivo dynamics of DNA topoisomerases and their interacting partners in D. radiodurans grown under normal and gamma stressed conditions have not been studied yet and would be worth studying.
In this paper, the time-lapse studies on the in vivo localization of fluorescently tagged DNA gyrase and drTopoIB under normal and post-irradiation conditions suggested the dynamic movement of both the topoisomerases with respect to DNA duplication and genome segregation processes in D. radiodurans. Further, the kinetic studies of DNA gyrase and its interacting partner, PprA, a pleiotropic protein involved in DNA damage repair, suggested the colocalization of these proteins on the nucleoid during the post-irradiation recovery period (PIR). The nalidixic acid treatment had perturbed the dynamics of these proteins indicating the functional interaction between DNA Gyrase and PprA in vivo. Further, the topoIB deletion mutant (ΔtopoIB) was more sensitive to hydroxyurea (HU) treatment, and showed delayed recovery and more single-stranded DNA accumulation during the PIR period as compared to the wild type. These results suggest the differential roles of DNA gyrase and drTopoIB in D. radiodurans in different molecular events associated with DSB repair and in combating different stresses in this bacterium.
Materials and methods
Bacterial strains, plasmids, and materials
Deinococcus radiodurans R1 (ATCC13939) strain was obtained as a kind gift from Professor J Ortner, Germany (Schäfer et al. 2000). Wild type and its derivatives were grown in TGY medium (0.3% Bacto yeast extract, 0.5% Bacto tryptone, 0.1% glucose) aerobically with a shaking speed of around 150–160 rpm, at 32 °C in the presence of antibiotics at the required concentrations. The Escherichia coli strains DH5α and Novablue were used to maintain the recombinant plasmids. To maintain plasmids and recombinant constructs, the antibiotic concentrations used were; E. coli—kanamycin (25 µg/ml), spectinomycin (100 µg/ml), ampicillin (100 µg/ml), D. radiodurans—kanamycin (10 µg/ml), spectinomycin (70 µg/ml), chloramphenicol (5 µg/ml).
Construction of expression plasmids
The drGyrA-RFP fusion protein was generated by cloning the coding sequence of drGyrA subunit with histidine tag from the plasmid pETgyrA using the primers FP5' CCCCCGGATCCGGCAGCTATGACCGGAATTCAACC3' and RP5' CCCCCGGTACCGCGGCCGCCAGCTCGTCTTCCTCCTTGCG3' in pDsRed at the upstream of red fluorescent protein coding region to generate pDsRedgyrA (Kota et al.2016). Then PCR amplification of his-gyrA-rfp region was performed taking pDsRedgyrA as a template and cloned in the plasmid pRadgro and transformed in D. radiodurans cells. Other fluorescently tagged proteins used in this study- TopoIB-RFP, GFP-PprA, FtsZ-GFP, and D. radiodurans topoIB mutant strain were constructed/generated earlier (Kota et al. 2021; Kota et al. 2014a; Modi et al. 2014).
Cell survival studies
The wild-type Deinococcus radiodurans, topoIB mutant, and cells expressing fluorescently tagged proteins were exposed to different genotoxic agents as described below (Misra et al. 2006). For gamma radiation exposure, late log phase cells were pelleted, suspended in fresh medium, and subjected to 6 kGy γ-radiation at a dose rate of 1.5 kGy/h (Gamma Cell 5000, 60Co, Board of Radiation and Isotopes Technology, DAE, India). An equal number of unirradiated and irradiated cells were spotted on the TYG medium containing Petri dishes and incubated at 32 °C or the cells were grown in TYG medium with appropriate antibiotics in 96 well microtiter plates and growth was monitored by measuring the optical density at 600 nm at 32 °C for 17 h in a Synergy H1 Hybrid multi-mode microplate reader. For microscopic studies, samples at regular intervals were collected and processed. For mitomycin C (MMC), nalidixic acid (NDA), and rifampicin treatments, late log phase cells were pelleted suspended in fresh medium and treated with MMC (10 µg/ml), NDA (20 µg/ml) or rifampicin (30 µg/ml) for 30 min. After the treatment, the cells were pelleted, resuspended in a fresh medium, and allowed for recovery. At regular time points samples were collected and processed for further experiments. Similarly, the cells were treated with hydroxyurea as described earlier (Maurya et al. 2021). In brief, the late log phase culture cells were pelleted, suspended in fresh medium, and divided into two sets. One set was treated with 500 mM HU for 60 min and the other set was kept as a control. Both treated and untreated samples were centrifuged and suspended in a fresh medium. The samples were aliquoted and one set was exposed to gamma radiation as described above and the other was kept as control. These samples were either spotted on the TYG plates or growth was monitored in a multimode plate reader as described above. The generation time of wildtype and mutants in all conditions was calculated as mentioned (Maier et al. 2009). All the data obtained were analyzed and plotted using GraphPad Prizm6 software.
Microscopic studies
The dynamics and localization of TopoIB and DNA gyrase with respect to cell growth were observed using time-lapse imaging as described earlier (Mishra et al. 2022). The D. radiodurans cells expressing either TopoIB-RFP or GyrA-RFP were stained for genome with 150 nM of SYTO 9 dye, washed twice with PBS, and placed on an agarose pad prepared in 2X TYG. Before placing the cells, air holes were constructed on the pads for the oxygenation of cells. Studies were performed using confocal microscopy on IX3SVR with an Olympus IX83 inverted microscope, having a focal plane of a 100 × 1.40 NA oil-immersion apochromatic objective lens (Olympus) as described earlier (Maurya et al. 2019). Using DM-488/561 dichroic mirror and corresponding single-band emission filters the fluorescence emission was recorded at regular time intervals for a period of 4 h at very low laser power (561 nm and 488 nm). To monitor the dynamics after gamma radiation exposure and MMC treatment, the 6 kGy irradiated cells or MMC treated cells were recovered for 1 h in a fresh medium with constant shaking and then cells were stained, placed on agarose pads, and visualized as described above. The in-built Cellsens software was used to process the images. For kinetic studies, the sample cells collected at regular intervals were washed and suspended in PBS, and stained with DAPI (4’, 6-diamidino-2-phenylindole dihydrochloride) (0.2 μg / ml) for nucleoid visualization. The excess stain was removed by washing thrice with PBS and then mounted on agarose beds on the glass slides. The cells were observed under TRITC (tetramethylrhodamine isothiocyanate), DAPI, and FITC channels depending on the requirements. The images were processed with in-built Cellsens software and images obtained under different channels were shown individually or as merged. We used Adobe Photoshop 7.0 for the adjustment of image brightness and contrast. Nearly 250–300 cells for each experiment were analyzed and data were plotted using GraphPad Prizm 5 software. All the co-localization studies were done using built-in CellSens Software and the represented co-localization patterns showed Pearson’s correlation coefficient (R(r)) > 0.7–1.0 and overlap coefficient (R) of 0.9–1.0. Co-localization was represented as white foci in the images. All the microscopic experiments were performed a minimum of three times and representative pictures were shown.
Immunofluorescent imaging to detect the single-strand DNA
To monitor the rate of conversion of single-stranded DNA to double-strand DNA we performed immunofluorescence experiments using the technique of 5-Bromo-2′-deoxyuridine (BrdU) incorporation into the DNA (Slade et al. 2009). For this, D. radiodurans wild type (R1), topoIB mutant cells (Δtopo) treated and untreated with HU in different combinations were subjected to 6 kGy radiation and were recovered with fresh medium for 1 h. Then the cells were pulse-labeled with BrdU for 15 min and transferred to a fresh medium and after 2 h of growth the amount of newly synthesized single-strand DNA in the cells was detected by anti-BrdU antibodies as described earlier (Devigne et al. 2013). In brief, the cells were fixed with 37% paraformaldehyde and subsequently washed with PBS. Later, the cells were permeabilized by treatment with lysozyme (3 mg/ml) for 1 h at 37 ◦C, then with 0.1% Triton X-100 in PBS at 37 ◦C for 5 min. The cells were washed with PBS, resuspended, and around 5 µl was spread onto a poly-L-lysine pretreated slide and allowed to air dry. Fixation was done by treating the slides with 4% PFA at 37 ◦C for 30 min. Blocking was done with 2% BSA in PBS-T (0.05% Tween 20 in PBS) at 37 ◦C for 2 h and then incubated with anti-BrdU antibody (Sigma) overnight in a blocking solution. Cells were washed with PBS-T for 20 min twice and then incubated with a secondary antibody, anti-mouse IgG conjugated with Alexa flour 594 (Sigma) for 2 h in a blocking solution. Cells were again washed with PBS-T for 20 min twice and slides were mounted using a mounting medium containing antifade DAPI. Microscopic studies were carried out as described in the earlier section. The average intensity of ssDNA accumulations in the cells was calculated using the ImageJ software and the arbitrary units obtained were plotted using the GraphPad Prizm6 software.
Bioinformatic analysis
Prediction of putative DNA binding motifs of D. radiodurans DNA Gyrase A was done. For this, sequence alignment of D. radiodurans DNA Gyrase A C- terminal domain (CTD) with M. tuberculosis DNA Gyrase A CTD was performed using the PROMALS3D online server.
Statistical analysis
All the error bars in the graphs denote standard deviations. The statistical analysis of the immunofluorescence data was done using- Student’s t-test. The significance value (P value) obtained at 95% confidence intervals is depicted as *** for a P value < 0.001.
Results
Deinococcal Topo IB and DNA gyrase showed dynamic sub-cellular localization in vivo
Previously, the expression and localization of drTopoIB-RFP under constitutive groESL promoter were reported (Kota et al. 2021). Here, the GyrA-RFP was expressed under groESL promoter on a low copy number plasmid pRADGro, and the distribution of DNA gyrase and drTopo IB in the cell was monitored under normal and post-DNA damaging treatments-gamma radiation and mitomycin (Fig. 1, 2). Both GyrA-RFP and drTopoIB-RFP expressing strains showed similar phenotypes and ionizing radiation resistance as that of the wild type indicating episomal expression of proteins has no/minimal effect on the growth of this bacterium (Fig S1). However, a clear difference in the dynamics of these two topoisomerases was observed during time-lapse studies under normal growth conditions. For example, the drTopoIB was seen localized on the nucleoid, though a significant proportion of this protein was found in the periplasmic space or the septal regions. As shown in Fig. 1a at time- 0 h, the drTopoIB-RFP fluorescence was mostly seen on nucleoid-devoid regions at the old septum. At 0.5 h, drTopoIB was seen dispersed in the cell both on the nucleoid and other parts of the cell. At 1.5 h, this protein was noticed aligning along with the separating site in the genome, which became more apparent at the 2.5 h period. At 3.5 h, the protein was seen along the space where the segregation of the duplicated genome would have almost completed. Interestingly, this stage coincides with the cell wall constriction as observed in the DIC picture. The dynamics of drTopoIB were also observed in the cells exposed to gamma radiation and mitomycin albeit with slightly different patterns (Fig. 1b, c). At 0 h PIR, the drTopoIB-RFP was observed at the genome-constricted position as well as in other regions in the cell (Fig. 1b). At the later stage of PIR, the protein had aligned at the genome separation site. The drTopoIB-RFP dynamics were also monitored in a population of the cells under time-lapsed growth and were found to be almost the same as observed in isolated cells (Fig S2). Thus, the dynamicity and distributive pattern of the drTopoIB-RFP protein observed during time-lapse studies suggested its role in maintaining DNA topology and genome stability during normal growth and post-irradiation recovery periods probably through its interaction with other cellular proteins in D. radiodurans (Kota et al. 2021).
Cellular localization dynamics of deinococcal TopoIB-RFP in normal untreated (UT) and DNA damaging conditions. Time-lapse confocal microscopic images show the cells expressing RFP fusion of TopoIB (red foci) stained for genomes with SYTO 9 green dye (green) during the untreated condition (a), post-irradiation recovery (PIR) (b) and post-mitomycin C (MMC) treatment (c). All the cells were allowed to divide on microscopic slides and observed at different time points (t = hrs). Images were taken in differential inference contrast (DIC), FITC (nucleoid- green), and TRITC (TopoIB-RFP- red) channels and are presented in planar view. The scale bar for (a) panels is 1 μm and for (b) and (c) panels- 500 nm. A representative set of data from a reproducible experiment is shown. TopoIB-RFP shows association on the nucleoid as well as on the membrane with maximum foci in the nucleoid-devoid regions. A general pattern of the localization dynamics of TopoIB-RFP foci during cell growth and genome segregation as observed in the majority of cells is schematically represented for clear understanding
Cellular localization dynamics of deinococcal DNAgyrA-RFP in normal untreated (UT) and DNA damaging conditions. Time-lapse confocal microscopic images show cells expressing RFP fusion of DNAgyrA (red foci) stained for genomes with SYTO 9 green dye (green) during the untreated condition (a), post-irradiation recovery (PIR) (b) and post-mitomycin C (MMC) treatment (c). All the cells were allowed to divide on microscopic slides and observed at different time points (t = hrs). Images were taken in differential inference contrast (DIC), FITC (nucleoid- green), and TRITC (DNAgyrA-RFP- red) channels and are presented in planar view. The scale bar for all the panels is 500 nm. The images shown here are representative pictures of the experiments conducted at least three times. DNAgyrA forms foci majorly on the nucleoid and aligns at the genome-separating site. A general pattern of the localization dynamics of DNAgyrA-RFP foci during cell growth and genome segregation as observed in the majority of cells is schematically represented for clear understanding
In the case of DNA Gyrase, GyrA-RFP foci were mostly found on the nucleoid regions under normal growth conditions (Fig. 2a). After 2 h incubation time, a significant number of foci were noticed on the genome separation site. After 4 h, the protein was noticed on both nucleoid and near the constriction sites of the genome that mark the segregation of duplicated genome. Unlike in normal growth conditions, the dynamics of GyrA-RFP foci in PIR cells were found to be different. For instance, during PIR, the majority of GyrA-RFP foci were observed on the nucleoid till 4 h (monitored in this study). A few GyrA-RFP fluorescent accumulations showing dynamics were also noticed during PIR. In general, the cell division in cells exposed to gamma radiation gets arrested for approximately 4 h. During this period, extensive DSB repair and reassembly of the genome takes place before the cell division resumes. The localization pattern of GyrA-RFP until 4 h of PIR in irradiated cells (Fig. 2b) and in the unirradiated cells (Fig. 2a) suggests a role of DNA gyrase in genome segregation and cell division in this bacterium. Since, DNA gyrase is a DNA binding protein with decatenation function, and is also known to interact with many proteins including DNA damage repair protein PprA in D. radiodurans (Kota et al. 2016; Devigne et al. 2016), a possibility of GyrA-RFP foci sitting on the duplicated catenated circular DNA cannot be ruled out. The localization pattern of GyrA-RFP in the cell exposed to mitomycin was found to be similar to gamma-irradiated cells (Fig. 2c). Further, a similar result was noticed in time-lapse microscopic studies with the population of the cells (Fig S3). Here, we noticed that the majority of GyrA-RFP foci were present at probable sites of genome separation. The protein was seen in parallel to the old septum and perpendicular to the new cell wall constriction, which is a prerequisite for cytokinesis. As mentioned earlier, DNA gyrase is the single type II topoisomerase in D. radiodurans. It can be speculated that its localization on the nucleoid might be important for maintaining the steady-state superhelicity of the genome while its localization at the site of genome separation might be necessary for performing decatenation of duplicated circular chromosomes. Thus, based on time-lapse studies under normal growth conditions and PIR, we may conclude that the dynamicity of both the topoisomerases coincides with the genome maintenance, duplication, and segregation processes in D. radiodurans.
drTopoIB-RFP and drFtsZ-GFP sparingly co-localize in D. radiodurans
Time-lapse microscopic studies showed that drTopoIB-RFP also localizes at nucleoid-devoid regions, the potential cellular space for FtsZ ring formation. Hence, a possible interaction of drTopoIB with drFtsZ and colocalization of both proteins on the septum was hypothesized. The cells co-expressing drTopoIB-RFP and drFtsZ-GFP were examined in time-lapse studies. (Fig. 3a). Results showed that only small fractions of drTopoIB-RFP and drFtsZ-GFP foci co-localize in the cells while the majority of foci were seen separate (Fig. 3a, 0.5 h & 1 h). This indicated either a direct interaction of drFtsZ-GFP with drTopoIB-RFP as a result of spatial proximity or through common partners. Recently, it has been reported that both drTopoIB and drFtsZ interact with drFtsK, which is a known multifunctional DNA translocase having roles in both genome segregation and cell division (Mishra et al. 2022). Further, it was observed that a large population of the cells have drTopoIB-RFP localized in parallel to the new division septum, besides on nucleoid (Fig. 3b). A similar observation was reported earlier when drTopoIB-RFP expressing cells were stained with vancomycin (Kota et al. 2021). Results suggest that drTopoIB interacts with drFtsZ either temporarily or through their common partners which seems to be worth pursuing separately.
Co-expression studies of deinococcal TopoIB-RFP and FtsZ-GFP. A Time-lapse microscope was carried out to check the co-localization of TopoIB-RFP (red) and deinococcal FtsZ-GFP (green) in untreated (UT) conditions (a). Cells were allowed to grow on the slides and microscopic images were taken at regular time points (t = hrs). Images taken in different channels DIC-differential inference contrast, FITC (green), and TRITC (red) were shown separately and also merged images (scale bar- 1 μm). A microscopic image of a representative population of cells expressing both TopoIB-RFP and FtsZ-GFP growing under normal conditions was depicted (b). All the microscopic experiments were conducted a minimum of three times and representative pictures were shown here. Deinococcal TopoIB-RFP and FtsZ-GFP do not co-localize in the cells and exist in different planes. A typical pattern of cellular localization of TopoIB-RFP and FtsZ-GFP as analyzed from the microscopic pictures was schematically shown
Deinococcal PprA protein co-exists with DNA gyrase during the PIR period
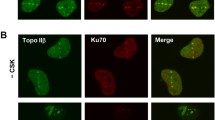
Previously, it was reported that DNA gyrase and topoisomerase IB of D. radiodurans interact with PprA (Kota et al. 2016; Devigne et al. 2016). Besides intense GyrA-RFP foci were observed during PIR in time-lapse microscopy in this study which indicates the formation of multiprotein complexes. Hence, the possible co-localization of PprA with topoisomerases in D. radiodurans cells was checked by co-expressing PprA-GFP with GyrA-RFP or drTopoIB-RFP, and the localization and dynamics of fluorescent proteins were monitored during PIR period. Under normal conditions, GyrA-RFP and PprA-GFP formed intense foci on the nucleoid whereas drTopoIB-RFP produced faded and fewer foci on the nucleoid and localization on the nucleoid-devoid regions (Fig. 4a). During PIR, PprA-GFP foci number increased around 1 h after treatment, and the co-localization of PprA and GyrA on nucleoid (depicted as white foci in the images) was observed up to 3 h in a majority of the cells (Fig. 4a). The period of colocalization of PprA with GyrA in the PIR sample coincides with the time that PIR cells take for the reassembly of the shattered genome in D. radiodurans. Interestingly, when the cells were treated with nalidixic acid, the number of intense foci increased (Fig. 4b). In these cells, PprA-GFP also co-localized with DNA gyrase, but the dynamicity of both proteins had disappeared (Fig. 4b). In cells co-expressing drTopoIB-RFP and PprA-GFP, the majority of them showed dispersed localization of TopoIB-RFP during PIR, and very little co-localization of Topo IB and PprA was observed (Fig. 4c). Interestingly, a similar trend was observed in untreated cells (at the time- 1 h). Thus, the results so far indicate that PprA and DNA gyrase proteins may act together on the nucleoid to repair the genome during the PIR period and the interaction between PprA and drTopoIB may be transient.
Localization studies of DNA topoisomerases with PprA in D. radiodurans post-irradiation and nalidixic acid (NDA) treatment. Cells expressing both PprA-GFP and DNAgyrA-RFP were grown in broth post-irradiation and observed under the confocal microscope at different time intervals (t = hrs). DNAgyrA and PprA both showed intense foci formation post-irradiation. The percentage of cells showing different numbers of intense localized DNAgyrA-RFP foci per cell was counted and a quantitative data set was presented. Also, the total number of DNAgyrA-RFP foci, PprA-GFP foci, and GyrA-PprA co-occurring foci in a population of ~ 200 cells at specific time points were calculated and plotted (a). The panel shows the cells expressing DNAgyrA-RFP and PprA-GFP under untreated conditions (UT) and post-NDA treatment at specific t. The total number of DNAgyrA-RFP foci, PprA-GFP foci, and GyrA-PprA co-occurring foci post-NDA treatment in a population of ~ 200 cells was calculated and plotted (b). A similar experiment was conducted with cells expressing TopoIB-RFP and PprA-GFP. TopoIB-RFP showed dispersed localization during the post-irradiation recovery period at the time points monitored. At t = 1 h, some cells showed co-occurrence of TopoIB-RFP and PprA-GFP foci as shown in zoomed view (c). The images shown here are representative pictures of the experiments conducted at least three times and the scale bar for all the images is 0.2 μm. Error bars depict standard errors
Topo IB is involved in DNA replication stress in D. radiodurans
Recently, it was reported that treatment of D. radiodurans with hydroxyurea (HU) before ionizing radiation has improved PIR and PprA protein plays a role in the regulation of replication initiation in this bacterium (Maurya et al. 2021). HU inhibits replication by depleting deoxyribonucleotide and imposes replication arrest stress in the cells. It was hypothesized that TopoIB can counter replication arrest imposed by HU. The wild-type D. radiodurans (R1) and topoIB (Δtopo) mutant cells were treated with HU and ionizing radiation and cell survival was monitored. Under normal growth conditions, both R1 and Δtopo responded nearly similarly to ionizing radiation (Fig. 5a, Fig S4). Even after HU treatment, there was no significant difference in the growth response of R1 and Δtopo (Fig. 5b, Fig S4). However, exposure to both HU and ionizing radiation together produced different responses in Δtopo and wild-type cells. For instance, Δtopo showed less survival when HU treatment was given before exposure to ionizing radiation as indicated by a drastic increase in the generation time of these cells whereas wild-type cells survived better under this condition (Fig. 5b).
Assessing the role of TopoIB in replication stress. Cell survival studies of D. radiodurans (R1) and topo:nptII mutant (Δtopo) under unirradiated (UIrr), post-irradiated (Irr) conditions. The growth curve was plotted and the generation time (doubling time) was calculated as described in the methodology (a). A similar growth study was done post-hydroxyurea (HU) treatment (HU-UIrr) and post-HU treatment followed by irradiation (HU-Irr) (b) Immunofluorescence assay was carried out to detect single-stranded DNA in WT and Δtopo using anti-BrdU antibodies and Alexa flour 594 secondary antibodies giving red fluorescence (c). Δtopo showed affected survival post-HU and irradiation treatment as well as more BrdU incorporation indicating slower recovery. Cells treated with HU showed localization of TopoIB-RFP as intense foci which were not seen after rifamycin treatment (d). Cell population kinetics studies were done post-HU treatment and irradiation. The number of TopoIB-RFP foci, PprA-GFP foci, and TopoIB-PprA co-occurring foci post-HU treatment and irradiation in a population of ~ 200 cells was calculated and plotted (e). Error bars denote standard errors. All these experiments indicate the role of TopoIB during DNA replication
Extended Synthesis Dependent Strand Annealing (ESDSA) is shown to be a crucial process in DNA double-strand break repair in D. radiodurans (Zahradka et al. 2006). It has been shown that early PIR cells have a high density of ssDDNA, that gets converted into dsDNA, which eventually results in a decrease in the intensity of BrdU incorporated into DNA. This concept was used to detect single-strand DNA as a measure of DSB repair and the newly synthesized BrdU-ssDNA was estimated by immunofluorescence using anti-BrdU antibodies. The results showed that after 3 h PIR, the HU-treated topoIB mutant cells showed more single-stranded DNA than the wild-type population indicating a slow recovery of the shattered genome in the absence of drTopoIB in the cells (Fig. 5c).
Further, the HU treatment of drTopoIB-RFP expressing cells produced very intense RFP foci at 0 h which at a later stage got diffused (Fig. 5d, e). However, when drTopoIB-RFP expressing cells were treated with rifampicin, the number of intense drTopoIB-RFP foci observed at 0 h was less than that observed in HU-treated cells (Fig. 5d). The HU-treated cells expressing both PprA-GFP and drTopoIB-RFP, showed a higher density of foci, and colocalization of these proteins on the nucleoid at 0 h. However, after gamma radiation in cells, drTopoIB-RFP produced more number of localized drTopoIB-RFP foci at ~ 3 h PIR compared to untreated cells, and an increased co-localization with PprA protein at around 3 h (Fig. 5e). It was observed that unlike in the case of TopoIB, HU treatment did not induce intense foci formation of GyrA-RFP (Fig S5), which is intriguing. Observation of more drTopoIB-RFP foci and co-localization of TopoIB and PprA at later PIR time suggests the functional significance of these interactions in genome maintenance when post-DSB repaired cells would be ready for division. But the exact mechanisms underlying drTopoIB role in the maintenance of reassembled genome and the significance of its interaction with PprA are not clear yet. In the ΔtopoIB mutant, a significant decrease in the copy number of all the four genome elements, and a higher frequency of anucleated cells were noticed compared to wild type under normal growing conditions previously (Kota et al. 2021). It may suggest the possibility of drTopoIB association with the replisome complex and assisting the replication fork progression in this bacterium and if so, then the inability of topoIB mutant to recover from the replication stress caused by HU could be explained, and will be pursued independently. Topoisomerase IB is a swivelase, known for relaxing both positive and negative supercoils to facilitate DNA replication and transcription in the cells (Soren et al. 2020). Altogether these results suggest that the action of topoisomerases in response to different genotoxic stresses seems to be different depending upon types of DNA damage and PprA protein differentially interacts with topoIB and DNA gyrase in this bacterium.
Discussion
In general, the nucleoid-associated proteins, structural maintenance of chromosome complexes (SMC), and topoisomerases maintain the architecture of the genome in the cell (Ptacin and Shapiro 2013). The collective action of these proteins along with external (epigenetic) factors decide the fate of the genome availability to cellular processes. The genome of D. radiodurans is multipartite, tightly coiled, and exists in various shapes like ring, toroidal, or crescent based on the growth stage of the cell (Floc’h et al. 2019). The role of the highly condensed nucleoid in radioresistance in this bacterium was also proposed in a way that could facilitate the accurate repair of broken DNA ends (Levin-Zaidman et al. 2003). D. radiodurans genome encodes three topoisomerases and numerous nucleoid-associated proteins like HU to maintain its nucleoid architecture (White et al. 1999). In E. coli, the DNA topology of its genome is maintained by two type I topoisomerases and two type II topoisomerases (Zechiedrich et al. 2000). D. radiodurans has many peculiarities associated with genome maintenance such as (i) ESDSA, a unique mechanism for DNA double-strand break repair, involving several steps of topology prone DNA metabolism (Zahradka et al. 2006), (ii) single type II DNA topoisomerase (Zechiedrich et al. 2000), (iii) packaging of multipartite polyploid genome into doughnut-shaped toroidal nucleoid (Levin-Zaidman et al. 2003) and (iv) true rapid kinetics of genome assembly during PIR (Misra et al. 2006). Therefore, the role of DNA Gyrase and topoisomerase IB would be very crucial in the maintenance of host-specific DNA topology in the genome. Studies on DNA topoisomerases that can specifically address these concerns are not adequately done at in vivo levels. Furthermore, since these proteins interact with other proteins in this bacterium, how the real-time function dynamics of these proteins get influenced by their interacting partners had not been shown earlier. This study has brought forth some findings to suggest that both GyrA and drTopoIB play important roles in genome maintenance and regulation of replication stress induced with HU under normal conditions and /or gamma radiation exposure.
The time-lapse microscopic studies under normal and PIR conditions showed that drTopoIB-RFP and GyrA-RFP were mostly localized on the nucleoid indicating their role in the maintenance of DNA supercoiling. At the same time, there exists a clear difference in the subcellular localization between the two topoisomerases studied depending on the growth stage of the cell. For example, drTopoIB-RFP was noticed both on nucleoid and outside nucleoid regions in significant proportions in time-lapse studies and population studies while GyrA-RFP was seen majorly on the nucleoid. When the co-localization of drTopoIB-RFP and FtsZ-GFP on the septum and peripheral membrane was checked, they localized in different planes in the cells suggesting the role of drTopoIB in the molecular events occurring perpendicular to the plane of the FtsZ ring. The drTopoIB is a monomeric enzyme, with a bipartite domain structure having amino-terminal (N) consisting of beta-sheets and carboxyl-terminal (C) mostly made of alpha helixes. The crystal structure has revealed that the N and C terminal domains are separated in an open confirmation exposing the active site in the catalytic region for the DNA binding (Patel et al. 2006). Further, this enzyme has two DNA binding sites, which are functionally independent as the mutations in the secondary DNA binding site do not affect the supercoil relaxation activity but affect the plectonemic supercoil formation (Patel et al. 2010). On the other hand, the human topoisomerase IB is divided into four domains- N-terminal, core, linker, and C-terminal domain. The core domain contains all the catalytic residues except the catalytic active tyrosine which is present in the C-terminal domain. The linker region is the most flexible region in the protein and the N-terminal region was shown to interact with various cellular proteins like WRN protein which stimulates its relaxation activity (Takahashi et al. 2022, Soren et al. 2020). The drTopoIB enzyme was also shown to interact with other DNA metabolic proteins and activities of the drTopoIB seem to be modulated by PprA, and others in D. radiodurans (Kota et al. 2014b, Kota and Misra 2015; Kota et al. 2021). Studies also revealed that deinococcal proteins like PprA, FtsK, etc. are dynamic inside the cell and present both on the nucleoid and the septum. (Mishra et al. 2022, Kota et al. 2014b). Thus, the cellular localization pattern observed during time-lapse microscopic experiments indicates the functional role of the enzyme in stable genome maintenance in association with other DNA metabolic proteins in this bacterium. Though, TopoIB may not be essential in simple eukaryotes like yeast studies have shown that this protein is required in multicellular organisms even at developmental stages, and tissue and cell-specific localization and functionalities have been reported (Wuqiang et al. 2021).
When the in vivo localization and dynamics of DNA gyrase were monitored with respect to DNA damage and repair in D. radiodurans, most of the drDNAgyrA-RFP was observed on the nucleoid in time-lapse studies, which concurs with its function of producing DNA-negative supercoils, relaxation of positive supercoils and decatenase activity i.e. maintaining steady-state supercoiling. A few organisms like D. radiodurans and Mycobacterium tuberculosis, contain a single type II enzyme i.e., DNA gyrase, and lack topoisomerase IV. In these bacteria, DNA gyrase itself performs both the supercoiling and decatenase functions (Aubry et al. 2006). DNA gyrase enzyme supercoils DNA by cleavage and strand passage mechanism at the three molecular interfaces called “gates” (N-gate, DNA-gate, and C-gate) formed within the holoenzyme (heterotetramer; A2B2) (Kampranis et al. 1999). The GyrA-box (QKRGGKG) present at the C-terminal domain of the GyrA subunit wraps the DNA and performs the supercoiling reaction (Kramlinger et al. 2006). Previously in Mycobacterium tuberculosis, it was shown that the DNA gyrase contains a second potential DNA binding motif (GyrA-box-I, QGRGGKG) at the C terminal domain and this motif may help the enzyme to function as a better decatenase than its E. coli counterpart (Bouige et al. 2013). When searched for the second putative DNA binding motif in D. radiodurans DNA gyrase, the sequence analysis revealed that it contains the GyrA-box-I (KGRGGLG) apart from the canonical GyrA-box (QKRGGRG (Fig S6). Studies using single-molecule imaging techniques in E. coli it was calculated that ∼12 gyrase enzymes would be present around the replication fork to tackle the topological challenges during DNA replication while the majority of the enzyme molecules (∼ 300) are present on the chromosome to maintain DNA topology (Mathew et al. 2019). Microscopic observations in the present study showed that the number of GyrA-RFP foci has increased during post-irradiation recovery. Earlier transcriptome analysis of post-irradiated D. radiodurans cells revealed that both gyrA and gyrB were upregulated during the recovery period (Liu et al. 2003). Hence, the role of DNA gyrase during the ESDSA process to maintain DNA topology could be envisaged. In E. coli topoisomerase IV acts behind the replication fork and after completion of replication during the process of sister chromosomes separation to unlink the molecules (Zawadzki et al. 2015). Further in E. coli, topoisomerase IV interaction with MukBEF (a class of SMC) is essential for efficient decatenation and segregation of ori sequences that are newly replicated, whereas FtsK dependant XerCD tyrosine recombinases acting at dif sites are required for dimer resolution at ter region (Galli et al. 2017). In the absence of topoisomerase IV in Deinococcus radiodurans, DNA gyrase has to perform decatenation activity during replication and genome segregation processes. Few of the GyrA-RFP foci observed on the separating genome during time-lapse studies, could be the ones engaged in the process of unlinking the intertwined circular genetic materials (Fig. 2a, b). Also, the possibility of other mechanisms involved in such processes cannot be ruled out. Similar observations were reported in Mycobacterium tuberculosis, where the chromatin immunoprecipitation studies revealed that DNA gyrase occupied at/near the ter region of the chromosome indicating its functional significance as a decatenase (Ahmed et al. 2017). Recently, we have shown that deinococcal FtsK is active and the genome of this bacterium encodes putative tyrosine recombinases (Mishra et al. 2022). It would be worth understanding how the topoisomerases and FtsK-dependent site-directed recombination systems could work in the maintenance of complex genome architecture in this bacterium.
Earlier reports revealed that the PprA protein interacts directly with DNA gyrase and regulates its catalytic activity (Kota et al. 2016; Devigne et al. 2016). The co-expression studies during PIR showed the co-existence of these two essential proteins on the nucleoid and their cellular dynamics at the time of DNA damage repair in D. radiodurans suggesting the interaction of these two proteins in vivo. Further during the PIR, it was noticed that GyrA-RFP formed fluorescent accumulations in time-lapse studies and intense foci in kinetic studies. These fluorescent foci may represent the DNA gyrase molecules that are loaded on the nucleoid that has accumulated supercoils. A similar observation was recorded in Bacillus subtilis (Tadesse et al. 2006), where the authors showed that DNA gyrase, topoisomerase I, and IV have distinct sub-cellular localization patterns. They demonstrated that DNA gyrase is highly dynamic and mostly co-localized with the replication fork while Topo I formed discrete centers near to SMC complex. The possibility of DNA gyrase and PprA forming repair complexes in cells along with other proteins to increase the catalytic efficiency and functional diversity during the DNA repair cannot be ruled out. Previously, a DNA processing complex characterized in this bacterium was found to have PprA, DNA topoisomerase, and other proteins together (Kota and Misra 2008).
In summary, the time-lapse microscopic studies reported here facilitated the understanding of the in vivo dynamics and roles of DNA gyrase and TopoIB in a radioresistant bacterium Deinococcus radiodurans. The two topoisomerases showed distinct cellular localization and dynamics respective to genome segregation and cell division processes and showed differential responses to DNA damaging agents in this bacterium (Fig. 6). Further, the PprA protein co-localizes with DNA gyrase on the nucleoid and also seems to transiently associate with drTopoIB during the genome maintenance processes.
Schematic model of differential localization dynamics of topoisomerases- TopoIB and GyrA during cell division in D. radiodurans. DNA gyrase, a single type II enzyme in this bacterium is found to be localized on the nucleoid and at the genome separation sites whereas TopoIB localizes on the nucleoid and septal regions but does not co-localize with the cell division protein FtsZ. Cellular localization and dynamics of both the topoisomerases depend on the cell growth stage
References
Ahmed W, Sala C, Hegde SR, Jha RK, Cole ST, Nagaraja V (2017) Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet 13(5):e1006754. https://doi.org/10.1371/journal.pgen.1006754. (PMID:28463980;PMCID:PMC5433769)
Aubry A, Fisher LM, Jarlier V, Cambau E (2006) First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem Biophys Res Commun 348(1):158–165. https://doi.org/10.1016/j.bbrc.2006.07.017. (Epub 2006 Jul 13 PMID: 16876125)
Bouige A, Darmon A, Piton J, Roue M, Petrella S, Capton E, Forterre P, Aubry A, Mayer C (2013) Mycobacterium tuberculosis DNA gyrase possesses two functional GyrA-boxes. Biochem J 455(3):285–294. https://doi.org/10.1042/BJ20130430. (PMID: 23869946)
Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413
Chen SH, Chan NL, Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170
Cox MM, Battista JR (2005) Deinococcus radiodurans—the consummate survivor. Nat Rev Microbiol 3(11):882–892
de la Tour CB, Passot FM, Toueille M, Mirabella B, Guérin P, Blanchard L, Servant P, de Groot A, Sommer S, Armengaud J (2013) Comparative proteomics reveals key proteins recruited at the nucleoid of Deinococcus after irradiation-induced DNA damage. Proteomics 23–24:3457–3469. https://doi.org/10.1002/pmic.201300249. (Epub 2013 Dec 4 PMID: 24307635)
Devigne A, Mersaoui S, Bouthier-de-la-Tour C, Sommer S, Servant P (2013) The PprA protein is required for accurate cell division of γ-irradiated Deinococcus radiodurans bacteria. DNA Repair (Amst). 12(4):265–72. https://doi.org/10.1016/j.dnarep.2013.01.004
Devigne A, Guérin P, Lisboa J, Quevillon-Cheruel S, Armengaud J, Sommer S, Bouthier de la Tour C, Servant P (2016) PprA protein is involved in chromosome segregation via its physical and functional interaction with DNA gyrase in irradiated deinococcus radiodurans bacteria. mSphere 1(1):e00036-15. https://doi.org/10.1128/mSphere.00036-15. (PMID: 27303692; PMCID: PMC4863600)
Floc’h K, Lacroix F, Servant P, Wong YS, Kleman JP, Bourgeois D, Timmins J (2019) Cell morphology and nucleoid dynamics in dividing deinococcus radiodurans. Nat Commun 10(1):3815. https://doi.org/10.1038/s41467-019-11725-5.PMID:31444361;PMCID:PMC6707255
Galli E, Midonet C, Paly E, Barre FX (2017) Fast growth conditions uncouple the final stages of chromosome segregation and cell division in escherichia coli. PLoS Genet 13(3):e1006702. https://doi.org/10.1371/journal.pgen.1006702.PMID:28358835;PMCID:PMC5391129
Kampranis SC, Bates AD, Maxwell A (1999) A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci U S A 96(15):8414–8419. https://doi.org/10.1073/pnas.96.15.8414.PMID:10411889;PMCID:PMC17530
Kota S, Misra HS (2008) Identification of a DNA processing complex from Deinococcus radiodurans. Biochem Cell Biol. 86(5):448–58. https://doi.org/10.1139/o08-122
Kota S, Misra HS (2015) Topoisomerase IB of Deinococcus radiodurans resolves guanine quadruplex DNA structures in vitro. J Biosci. 40:833–843. https://doi.org/10.1007/s12038-015-9571-z
Kota S, Charaka VK, Misra HS (2014a) PprA, a pleiotropic protein for radioresistance, works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans. J Genet 93(2):349–354. https://doi.org/10.1007/s12041-014-0382-z. (PMID: 25189229)
Kota S, Charaka VK, Misra HS (2014) PprA, a pleiotropic protein for radioresistance, works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans. J Genet. 93(2):349–54
Kota S, Rajpurohit YS, Charaka V, Satoh K, Narumi I, Misra HS (2016) DNA gyrase of deinococcus radiodurans is characterized as type II bacterial topoisomerase and its activity is differentially regulated by PprA in vitro. Extremophile 20:195–205
Kota S, Chaudhary R, Mishra S, Misra HS (2021) Topoisomerase IB interacts with genome segregation proteins and is involved in multipartite genome maintenance in Deinococcus radiodurans. Microbiol Res 242:126609
Kramlinger VM, Hiasa H (2006) The, “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J Biol Chem 281(6):3738–3742. https://doi.org/10.1074/jbc.M511160200. (Epub 2005 Dec 5 PMID: 16332690)
Krogh BO, Shuman S (2002) A poxvirus-like type IB topoisomerase family in bacteria. Proc Natl Acad Sci USA. 99(4):1853–8. https://doi.org/10.1073/pnas.032613199. (Epub 2002)
Levin-Zaidman S, Englander J, Shimoni E, Sharma AK, Minton KW (2003) A: Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299(5604):254–256. https://doi.org/10.1126/science.1077865
Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, Gaidamakova EK, Zhai M, Makarova KS, Koonin EV, Daly MJ (2003) Transcriptome dynamics of deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci USA. 100(7):4191–6. https://doi.org/10.1073/pnas.0630387100
Maier RM (2009) Bacterial growth. In: Maier RM, Pepper IL, Gerb CP (eds) Environmental toxicology, part I: review of basic microbiological concepts, 2nd edn. Elsevier Academic Press, San Diego, CA, pp 37–54
Maurya GK, Kota S, Misra HS (2019) Characterisation of ParB encoded on multipartite genome in Deinococcus radiodurans and their roles in radioresistance. Microbiol Res 223:22–32
Maurya GK, Chaudhary R, Pandey N, Misra HS (2021) Molecular insights into replication initiation in a multipartite genome harboring bacterium Deinococcus radiodurans. J Biol Chem 296:100451. https://doi.org/10.1016/j.jbc.2021.100451. (Epub 2021 Feb 21)
Mishra S, Misra HS, Kota S (2022) FtsK, a DNA Motor protein, coordinates the genome segregation and early cell division processes in deinococcus radiodurans. mBio. 13(6):e0174222. https://doi.org/10.1128/mbio.01742-22
Misra HS, Khairnar NP, Kota S, Shrivastava S, Joshi VP, Apte SK (2006) An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol Microbiol 59(4):1308–1316
Misra HS, Rajpurohit YS, Kota S (2013) Physiological and molecular basis of extreme radioresistance in Deinococcus radiodurans. Curr Sci 104:194–206
Modi KM, Tewari R, Misra HS (2014) FtsZDr, a tubulin homologue in radioresistant bacterium deinococcus radiodurans is characterized as a GTPase exhibiting polymerization/depolymerization dynamics in vitro and FtsZ ring formation in vivo. Int J Biochem Cell Biol 50:38–46. https://doi.org/10.1016/j.biocel.2014.01.015
Patel A, Shuman S, Mondragón A (2006) Crystal structure of a bacterial type IB DNA topoisomerase reveals a preassembled active site in the absence of DNA. J Biol Chem 281(9):6030–6037. https://doi.org/10.1074/jbc.M512332200. (Epub 2005 Dec 19 PMID: 16368685)
Patel A, Yakovleva L, Shuman S, Mondragón A (2010) Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure 18(6):725–733. https://doi.org/10.1016/j.str.2010.03.007.PMID:20541510;PMCID:PMC2886027
Pommier Y, Nussenzweig A, Takeda S et al (2022) Human topoisomerases and their roles in genome stability and organization. Nat Rev Mol Cell Biol 23:407–427
Ptacin JL, Shapiro L (2013) Chromosome architecture is a key element of bacterial cellular organization. Cell Microbiol. 1:45–52. https://doi.org/10.1111/cmi.12049. (Epub 2012, PMID: 23078580; PMCID: PMC3660146)
Schäfer M, Schmitz C, Facius R, Horneck G, Milow B, Funken KH, Ornery J (2000) Systematic study of parameters influencing the action of Rose Bengal with visible light on bacterial cells: comparison between the biological effect and singletoxygen production. Photochem Photobiol 71(2000):514–523
Slade D, Radman M (2011) Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133–191
Slade D, Lindner AB, Paul G, Radman M (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136(6):1044–1055. https://doi.org/10.1016/j.cell.2009.01.018. (PMID: 19303848)
Soren BC, Dasari JB, Ottaviani A, Iacovelli F, Fiorani P (2020) Topoisomerase IB: a relaxing enzyme for stressed DNA. Cancer Drug Resist 3(1):18–25. https://doi.org/10.20517/cdr.2019.106.PMID:35582040;PMCID:PMC9094055
Stracy M, Wollman AJM, Kaja E, Gapinski J, Lee J-E, Leek VA, McKie SJ, Mitchenall LA, Maxwell A, Sherratt DJ, Leake MC, Zawadzki P (2019) Single-molecule imaging of DNA gyrase activity in living Escherichia coli. Nucleic Acids Res 47(1):210–220. https://doi.org/10.1093/nar/gky1143
Tadesse S, Graumann PL (2006) Differential and dynamic localization of topoisomerases in Bacillus subtilis. J Bacteriol 188(8):3002–3011. https://doi.org/10.1128/JB.188.8.3002-3011.2006.PMID:16585761;PMCID:PMC1446999
Takahashi DT, Gadelle D, Agama K, Kiselev E, Zhang H, Yab E, Petrella S, Forterre P, Pommier Y, Mayer C (2022) Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states. Nat Commun 13(1):59. https://doi.org/10.1038/s41467-021-27686-7.PMID:35013228;PMCID:PMC8748870
White, O., Eisen, J.A., Heidelberg, J.F., Hickey, E.K., Peterson, J.D., Dodson, R.J., et al. (1999). Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 2861571–1577.
Wuqiang Huang, Zhiping Liu, Yikang S Rong. (2021) Dynamic localization of DNA topoisomerase I and its functional relevance during Drosophila development. G3 Genes Genomes Genetics https://doi.org/10.1093/g3journal/jkab202
Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443(7111):569–573. https://doi.org/10.1038/nature05160. (Epub 2006 Sep 27 PMID: 17006450)
Zawadzki P, Stracy M, Ginda K, Zawadzka K, Lesterlin C, Kapanidis AN, Sherratt DJ (2015) The localization and action of topoisomerase IV in escherichia coli chromosome segregation is coordinated by the SMC complex. MukBEF. Cell Rep. 13(11):2587–2596. https://doi.org/10.1016/j.celrep.2015.11.034
Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem 275(11):8103–8113. https://doi.org/10.1074/jbc.275.11.8103. (PMID: 10713132)
Acknowledgements
We sincerely thank Dr. Hema Rajaram, Bhabha Atomic Research Centre, Mumbai, for her suggestions and support while preparing the manuscript. Miss Himani Tewari is grateful to the Department of Atomic Energy, Government of India, for the research fellowship.
Author information
Authors and Affiliations
Contributions
SM performed the experiments, analyzed the results, and prepared the manuscript. HT performed the experiments and manuscript preparation. RC conducted the experiments and manuscript preparation. HSM analyzed the data and manuscript preparation. SK conceived the idea, planned and conducted the experiments, analyzed the data, wrote the manuscript, and communicated for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Communicated by Atomi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
792_2023_1323_MOESM1_ESM.pdf
Supplementary file1 Fig S1: Growth studies of D. radiodurans cells expressing RFP fusions of TopoIB and DNAgyrA episomally. Different dilutions of exponentially growing cells of D. radiodurans (R1) expressing TopoIB-RFP and DNAgyrA-RFP under unirradiated (UIrr) and post-irradiation (Irr) were spotted on TYG agar plates. Fig S2: Time-lapse microscopy of a population of D. radiodurans cells expressing TopoIB-RFP. Microscopic images were taken in differential inference contrast (DIC) and TRITC (TopoIB-RFP- red) channels. The scale bar is 1 μm. Fig S3: Time-lapse microscopy of a population of D. radiodurans cells expressing DNAgyrA-RFP. Microscopic images were taken in differential inference contrast (DIC) and TRITC (DNAgyrA-RFP-red) channels. The scale bar is 1 μm. Fig S4: Growth studies of D. radiodurans (R1) and topo:nptII mutant (Δtopo) under unirradiated (UIrr), post-irradiated (Irr) conditions, post-hydroxyurea (HU) treatment (HU-UIrr) and post-HU treatment followed by irradiation (HU-Irr). Different dilutions of exponentially growing cells were spotted on TYG agar plates. Fig S5: Microscopic images of D. radiodurans cells expressing drDNAgyrA-RFP after hydroxyurea (HU) treatment. Cells were grown in TYG broth and monitored under the microscope at specific time intervals (t=hrs) post-HU treatment. Fig S6: DNA binding motif analysis of D. radiodurans DNA Gyrase A. Sequence alignment of D. radiodurans DNA Gyrase A C- terminal domain (CTD) with M. tuberculosis DNA Gyrase A CTD was done using PROMALS3D (a). A schematic representation of the conserved DNA binding motif GyrA-box and a unique GyrA-box-I at the GyrA-CTD is given (b). Boundaries of the motifs are marked as a black box for GyrA-box and a red box for GyrA-box-I. (PDF 1088 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, S., Tewari, H., Chaudhary, R. et al. Differential cellular localization of DNA gyrase and topoisomerase IB in response to DNA damage in Deinococcus radiodurans. Extremophiles 28, 7 (2024). https://doi.org/10.1007/s00792-023-01323-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00792-023-01323-1