Abstract
Soybean isoflavones play diverse roles in human health, including cancers, osteoporosis, heart disease, menopausal symptoms and pabulums. The objective of this study was to identify the quantitative trait loci (QTL) associated with the isoflavones daidzein (DC), genistein (GeC), glycitein (GlC) and total isoflavone contents (TIC) in soybean seeds. A population of 184 F21:0 recombinant inbred lines derived from a ‘Xiaoheidou’ בGR8836’ cross was planted in pot and field conditions to evaluate soybean isoflavones. Twenty-one QTL were detected by composite interval mapping. Several QTL were associated with the traits for DC, GeC, GlC and TIC only. QDGeGlTIC4_1 and QDGlTIC12_1 are reported first in this study and were associated with the DC, GeC, GlC and TIC traits simultaneously. The QTL identified have potential value for marker-assisted selection to develop soybean varieties with desirable isoflavone content.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soybean (Glycine max L. Merr.) is one of the world’s most important oilseed crops and comprises ∼20% oil and 40% protein. Soybean seeds have received considerable attention for their high isoflavone concentrations (1.0–3.0 μg·mg−1) (Wang and Murphy 1994; Cardinal et al. 2007). The three main isoflavone components, dadzein (DC) genistein (GeC) and glycitein (GlC) made up ∼95% of total isoflavones in soybean seeds (Kudou et al. 1991; Latunde-Dada et al. 2001). Isoflavone have pharmacological activities in preventing ovarian, breast, colon and prostate cancers as well as osteoporosis and cardiovascular diseases (Naim et al. 1976; Weidenborner et al. 1990; Aedin et al. 2000; Tikkanen and Adlerereutz 2000; Watanable et al. 2002; Lo et al. 2007). Manipulation of crop characteristics using molecular biology is now possible. Previous studies have demonstrated that the main isoflavone compounds are synthesized in a reaction catalysed by several key enzymes from a branch of the general phenylpropanoid pathway, and that isoflavone synthase (IFS), phenylalanine ammonialyase, chalcone synthase (CHS), chalcone isomerase and certain CytP450s are involved in the GeC and DC biosynthetic pathways (Akashi et al. 1999; Jung et al. 2000; Yu et al. 2003). Compared with transgenic technology, molecular markers may be easier to utilize in a breeding programme. The two genetic IFS loci polymorphisms (IFS1 and IFS2) have been mapped and associated with isoflavone concentration (Cheng et al. 2008) and several quantitative trait loci (QTL) for isoflavones are located on the same linkage group as the CHS family genes (Kassem et al. 2004; Matsumura et al. 2005; Primomo et al. 2005; Sangeeta et al., 2007). For example, the QTL for GlC share the same chromosome (Chr) 5, 9, 1 and 11 as CHS2, CHS6, CHS7 and CHS8, respectively (Matsumura et al. 2005; Sangeeta et al. 2007). Thus, these QTL might be important.
Other QTL among various mapping populations have been mapped for DC, GeC, GlC and TIC (Njiti et al. 1999; Meksem et al. 2001; Kassem et al. 2004, 2006; Primomo et al. 2005; Sangeeta et al. 2007; Yang et al. 2008, 2011; Gutierrez-Gonzalez et al. 2009; Liang et al. 2009; Murphy et al. 2009; Zeng et al. 2009, Zhanget al. 2012). In total, 18 major QTL for DC have been mapped on Chr1, Chr3, Chr5, Chr7, Chr8, Chr13 and Chr14, and these explain 3.4% to 50.2% of the phenotypic variation. Twenty-one QTL for GlC are located on Chr1, Chr3, Chr5, Chr7, Chr11 and Chr18; these explain 3.4–50.2% of the genetic variation. Eighteen QTL for GeC are present on Chr1, Chr3, Chr5, Chr7, Chr11, Chr13 and Chr18 and these explaine 9.3% to 29.5% of the phenotypic variation, respectively. Twenty major QTL for TIC are mapped on Chr5, Chr6, Chr7, Chr8, Chr12 and Chr13; these explain 4.1–32.3% of the genetic variation.
Soybean isoflavone content is a quantitative trait (Hoeck et al. 2000) and is regulated by environmental factors, including crop year, planting date, fertilizer, water and temperature (Tsukamoto et al. 1995; Bennett et al. 2004; Caldwell et al. 2005; Lozovaya et al. 2005; Murphy et al. 2009). The effects of genotype, environment, and the genotype × environment interaction are significant (Wang and Murphy 1994; Hoeck et al. 2000; Lee et al. 2003; Primomo et al. 2005; Murphy et al. 2009). The genetic basis of regulation of the amount of isoflavones is not well-understood, due to the tremendous variability in isoflavone content in seeds harvested from different environments (Njiti et al. 1999; Meksem et al. 2001, Kassem et al. 2004, 2006; Primomo et al. 2005; Zeng et al. 2009; Gutierrez-Gonzalez et al. 2009; Yang et al. 2011; Meng et al. 2011). Several QTL have been detected across different environments (Primomo et al. 2005; Zeng et al. 2009); however, they are insufficient for use in a breeding programme. Thus, it is important to identify soybean isoflavone QTL in different environments and populations.
The main objective of this study was to identify QTL associated with loci-conditioned variations in DC, GlC, GeC and TIC used a recombinant inbred lines (RILs) population derived from ‘Xiaoheidou’ and ‘GR8836’ grown in field-cultured and pot-cultured environments. The parents were derived from varieties that displayed wide genetic diversity in terms of isoflavone content.
Materials and methods
Plant materials
The mapping population contained 184 RILs of F210 advanced by single-seed-descent and derived from a ‘Xiaoheidou’ × ‘GR8836’ cross. The parents, Xiaoheidou (♀, with low individual as well as total isoflavone content in seeds: DC, 1.85 μg·mg−1; GeC, 0.21 μg·mg−1; GlC, 1.26 μg·mg−1; TIC, 3.32 μg·mg−1) is a Chinese landrace and GR8836 (PI534647, maturity group III, ♂, with high individual and total isoflavone content in seeds: DC, 2.04 μg·mg−1; GeC, 0.44 μg·mg−1; GlC, 1.95 μg·mg−1; TIC, 4.43 μg·mg−1) was introduced from USA.
Field and greenhouse experiments
A set of 184 RILs were grown together with parents in a field-culture environment at the Jilin University Experimental Station, Changchun, China (43\(^{\circ }54^{\prime }\)N, 125\(^{\circ }19^{\prime }\)E), following a randomized complete block design with three replicates. The frost-free period was 145 days. The farming soil style was phaeozem which contained 120.19 μg·mg−1 rapidly available nitrogen, 17.33 μg·mg−1 rapidly available phosphorus and 153.74 μg·mg−1 rapidly available potassium. The previous crop was corn. The population was sown on 2 May 2010 in a field containing 5-m long rows with a row width of 0.65 m and was thinned to a uniform density with a space of 12.5 cm between plants two weeks after emergence. Weeds and pests were controlled routinely. The same RIL population was sown with three replicates on 20 April 2010 in plastic barrels (30 cm in diametre) contained ∼6 kg local soil, each pot contained three plants. Management of plants was similar to field-culture environment, except that irrigation was used as needed for the pot-cultured condition. Seed samples were harvested from single plants until complete ripeness. No symptoms of fertilizer deficiency were identified in plants under two conditions.
Isoflavone extraction and determination
Sample preparation:
Soybean seed powder (100 mg) was dissolved in 4 mL of 80% methanol (Dingguo, Beijing, China) in distilled water and stirred in an ultrasonic cleaning bath (Kunshan KQ3200DE, Jiangsu, China) for 30 min at 80∘C and then left overnight at room temperature. The supernatant was filtered through a 0.45- μm filter and transferred to a 5-mL high-performance liquid chromatography (HPLC) volumetric flask. A 20 μL aliquot of the filtrate was subjected to HPLC analysis.
Chromatographic conditions:
A C18 column (Shimadzu LC-20A, Tokyo, Japan; 150 × 4.6 mm, 5.0 μm) was used for all separations at a column temperature of 40∘C. The linear gradient system consisted of solvent A (HPLC-grade methanol) and solvent B (0.4% orthophosphoric acid in distilled water). The solvent flow rate was 1.0 mL/min−1 and UV absorption was measured at 254 nm.
Genetic analysis:
Young trifoliate leaves of the parents and each recombinant inbred line were collected from seedlings in the pot-cultured condition. Total DNA was purified as described by Yuan et al. (2002). SSR primers developed by Song et al. (2004) were screened from Soybase (2005). Polymerase chain reaction (PCR) amplification was performed as described by Zeng et al. (2009). The PCR products were separated in 6% (w/v) denaturing polyacrylamide gels and visualized by silver staining (Trigizano and Caetano-Anolles 1998).
Frequency distribution and statistical parameters for the parental and RIL populations were analysed by using the SPSS 13.0 (SPSS, Chicago, USA) and Excel 2003 (Microsoft, Redmond, USA) software. A linkage map contained 87 SSR markers was constructed using Mapmaker/ Exp 3.0b (Lander et al. 1987) and the Kosambi mapping function. The commands ‘group’, ‘map’, ‘try’, and ‘compare’ were used to build the linkage groups. The type I error detection ratio was set to 5%. The Haldane mapping function (Haldane 1919) was used with a minimum LOD score of 2.5 and a maximum distance of 50 cM (Promomo et al. 2005). QTL were identified by composite interval mapping (Zeng 1993, 1994).
Results
Phenotypic analysis of individual and total isoflavone contents
Ranges, means, standard deviations, coefficients of variation (CV), skewness, kurtosis, and broad-sense heritability for seed isoflavone contents of the parents and RIL population across the two environments (field-cultured and pot-cultured) are presented in table 1. ‘GR8836’ had significantly higher values than those of ‘Xiaoheidou’ for all isoflavone contents across both environments, indicated that the two parents differed in the genes controlling individual and total isoflavone contents. In fact, isoflavone content was significantly higher in pot-cultured than in the field-cultured environment, and the reason may be consistent with the large environmental interaction generally associated with isoflavone content in soybean seeds (Wang and Murphy 1994, Hoeck et al. 2000, Lee et al. 2003, Primomo et al. 2005; Murphy et al. 2009). Frequency distribution of field cultured and pot cultured conditions for the RILs population was determined for the samples and both displayed a continuous distribution (table 1; figure 1). DC was higher than GeC and GlC was the lowest in soybean seeds. Widely transgressive segregations were detected in all research environments.
Frequency distribution of daidzein, genistein, glycitein and total isoflavone contents in soybeans seeds among 184 F210 RIL derived from a cross between the cultivars ‘Xiaoheidou’ and ‘GR8836’ in field-cultured environments (A) and pot-cultured environments (B). Values next to the x-axis are the upper limit of each category. Parental values are indicated for ‘Xiaoheidou’ and ‘GR8836’.
The CV values that were not significant were < 0.5 for the isoflavone content means. Broad-sense heritability estimates for DC, GeC, GlC and TIC across field-cultured and pot-cultured environments were 0.54, 0.66, 0.61, 0.52, 0.44, 0.49, 0.56, and 0.69 (table 1), individually. Broad-sense heritability estimates for the isoflavone content were similar to those reported previously by Primomo et al. (2005) and Zeng et al. (2009), which ranged from 0.35 to 0.57. However, heritability estimates reported by Meksem et al. (2001) and Yang et al. (2011) were higher than the values obtained here.
Both skewness and kurtosis values for relative traits were < 1.0, and all isoflavone contents were normally distributed. Positive values for skewness (table 1; figure 1) indicated that all distributions were skewed towards ‘GR8836’. An analysis of variance of isoflavone content indicated a significant (P < 0.001) genotypic variation for DC, GeC, GlC and TIC among RILs. Significant variation was also detected between the different environments (P < 0.001).
Linkage analysis
A total of 667 SSR markers were screened between the two parents and 232 SSR primers had polymorphisms. A genetic linkage map that covered 20 chromosomes and contained 87 SSR markers was constructed (Lander et al. 1987). Total length of the map was 1733 cM with an average distance between markers of 25.1 cM. The markers were initially grouped and anchored based on a consensus map (Song et al. 2004).
Isoflavone QTL mapping
Twenty-one QTL associated with DC, GeC, GlC and TIC, including many novel regions, were identified in the field-cultured and pot-cultured environments (table 2; figure 2). The QTL were located on eight chromosomes (Chr3, Chr4, Chr7, Chr9, Chr10, Chr12, Chr13 and Chr17). The total explained phenotypic variation for specific isoflavone content was 4.48–8.83%. Nine of the 10 QTL effects were positive that was contributed by ‘GR8836’ and only one QTL named QDGlTIC3_1 had a negative effect that was donated by ‘Xiaoheidou’.
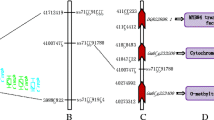
Locations of some of the chromosomes with major QTL for DC, GeC, GlC and TIC in field-cultured and pot-cultured environments in Changchun in 2010. \(\bigstar \) Daidzein content, \(\blacktriangle \) glycitein content, • genistein content and \(\blacklozenge \) total isoflavone content. Chromosome number is indicated at the top of the linkage group diagram. QTL names, marker names and distances for the interval are given. Genetic distances are from the RIL function of Mapmaker/EXP 3.0b (Lander et al. 1987). Linkage groups were named using the consensus map (Song et al. 2004) and coincided with the chromosome number on Soybase (http://www.soybase.org/)
Five QTL (table 2) associated with DC were mapped on Chr3, Chr4, Chr9 and Chr12. Of them, two QTL for DC, QDGeGlTIC4_1 and QDTIC4_2, both located on Chr4, explained 6.70% and 7.73% of the phenotypic variation. Four QTL for GeC were mapped on Chr4, Chr7, Chr10 and Chr13. Six QTL for GlC were detected on Chr3, Chr4, Chr12 and Chr17. Of them, two QTL, QGlC4_1 and QDGeGlTIC4_1 both located on Chr4, explained 5.53% and 4.87% of the phenotypic variation in the two environments, respectively. One QTL, QGlC17_1 for GlC located on Chr17, was identified in both field-cultured and pod-cultured conditions and was associated with Satt488. Six QTL (table 2) for TIC were detected on Chr3, Chr4, Chr9 and Chr12, which could explain 4.48–6.63% of the phenotypic variations. Of them, two QTL, QDGeGlTIC4_1 and QDTIC4_2 for TIC both located on Chr4, explained 6.63% and 5.09% of the genetic variation, individually.
Most of the QTL were clustered in genomic regions, particularly on Chr3, Chr4, Chr9 and Chr12 (table 2; figure 3). One novel QTL, QDGeGlTIC4_1 associated with Satt524, was identified for DC, GeC, GlC, and TIC, simultaneously. Two QTL, QDGlTIC12_1 associated with Satt353 and QDGlTIC3_1 with Satt009, were identified for DC, GlC and TIC, respectively. Another two QTL, QDTIC4_2 with Sat_140 and QDTIC9_1 with Sat_319, were detected across only one environment for DC and TIC, respectively. Two major QTL, QGlC17_1 for GlC and QDGlTIC12_1 for TIC were detected in both culture environments. One QTL, QGlC17_1 for GlC, explained 6.7% of the phenotypic variation in both environments. Another QTL, QDGlTIC12_1 for TIC, was also detected in both environments and it explained 4.87% of the phenotypic variation in the field-cultured environment and 4.48% in the pot-cultured environment, individually.
Discussion
Individual and total isoflavone contents in the pot-cultured environment were markedly higher than those in the field-cultured environment. The reason might be the interaction between genetic and multiple environmental factors (Tsukamoto et al. 1995; Bennett et al. 2004; Caldwell et al. 2005; Lozovaya et al. 2005; Murphy et al 2009), as we detected a significant environmental effect (P < 0.001). Differences between the two environments were water availability and early sowing, which have been demonstrated to benefit the accumulation of individual and total isoflavone contents (Bennett et al. 2004; Caldwell et al. 2005; Lozovaya et al. 2005). Thus, it could be inferred that irrigation and/or an early sowing date should be considered to produce high isoflavone content soybeans.
Notably, two QTL, QDGeGlTIC4_1 located on Chr4 associated with Satt524 and QDGlTIC12_1 located on Chr12 with Satt353, are first reported here. The two QTL were associated with the DC, GeC, GlC and TIC traits simultaneously, suggesting that these QTL are linked to the same gene or represent the action of clustered genes. If so, it may be inferred that these QTL are linked to the upstream genes in the isoflavone biosynthetic and regulatory pathways. Genes linked to new intervals were identified on the SoyBase website. QDGeGlTIC4_1 might associate with Glyma04g42110 which was noted as a R2R3-MYB transcription factor. Previous studies had proved that the R2R3- MYB transcription factor gene family could increased the expression abundance of some key enzyme genes in flavonid biosynthesis in transgenic Arabidopsis plants (Stracke et al. 2001). It can be inferred that Glyma04g42110 is a candidate gene for QDGeGlTIC4_1. No candidate gene associate with QDGlTIC12_1 was found to relate to the synthesis of isoflavone in the adjacent region of Satt353.
Stability of the QTL across environments and genetic backgrounds is key to determine if they can be used in a breeding programme (Brummer et al. 1997). In this study, three QTL were consistent with previously mapped results. The QTLQDGlTIC3_1 for DC, GeC and TIC associated with Satt009 was first mapped by Liang et al. (2009). QGeC7_1 for GeC associated with Satt323 detected in the present study was ∼20 cM away from the QTL loci associated with Satt540 detected by Zeng et al. (2009) and Primomo et al. (2005). QDTIC4_2 corresponded to Sat_140 and was similar to the QTL QGC4 for GeC detected by Yang et al. (2011). In addition, three QTL of QGlC4_1, QGlC17_1, and QDGlTIC12_1 were detected simultaneously in pot cultured and field cultured conditions and were significantly different (P < 0.001) in the present study. However, these QTL were not detected in other study populations, suggested that they may be unique to the ‘Xiaoheidou’ and ‘GRRR36’ parents, which have a distant genetic relationship. All these stable QTL should be considered to narrow down the genomic regions and identify related genes for future research.
Previous studies have shown that genomic regions associated with soybean isoflavone content are always linked to other agronomic (seed yield, weight, maturity, lodging and height) and quality (oil and protein content) traits (Wang et al. 2000; Meksem et al. 2001; Kassem et al. 2004; Primomo et al. 2005). In this study, the location of QDGlTIC3_1 was shared by oil content, iron efficiency, reaction to sclerotinia sclerotiorum, and flower number; the location ofQDGeGlTIC4_1 was shared by oil content and protein content; and the locus of QDGlTIC12_1 was shared by somatic embryos per explant and protein content (Qi et al. 2011; Lin et al. 1997; Zhang et al. 2010; Song et al. 2010). Such linkages should be considered in a breeding programme.
Several QTL located on the same chromosome as key enzymes or transcription factor genes have been reported (Cheng et al. 2008; Kassem et al. 2004; Primomo et al. 2005; Matsumura et al. 2005). In the present study, three QTL, named QDGlTIC12_1, QDTIC9_1 and QDGlTIC3_1, were matched with the candidate genes, CHS,bHLH and those of the DFR2family, which are related to isoflavone accumulation. Although these assumptions should be confirmed by further studies, these QTL may facilitate improvement of soybean lines in terms of their isoflavone content.
References
Aedin C., Bryn H. and Rosa M. 2000 Isoflavones, lignans and stilbenes – origins, metabolism and potential importance to human health. J. Sci. Food. Agric. 80, 1044–1062.
Akashi T., Aoki T. and Ayabe S. 1999 Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involed in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 121, 821–828.
Bennett J. O., Yu O., Heatherly L. G. and Krishnan B. 2004 Accumulation of genistein and daidzein, soybean isoflavones implicated in promoting human health, is significantly elevated by irrigation. J. Agric. Food Chem. 52, 7574–7579.
Brummer E. C., Graef G. L., Orf J., Wilcox J. R. and Shoemaker R. C. 1997 Mapping QTL for seed protein and oil content in eight soybean populations. Crop Sci. 37, 304–310.
Caldwell C. R., Britz S. J. and Mirecki R. M. 2005 Effect of temperature, elevated carbon dioxide, and drought during seed development on the isoflavone content of dwarf soybean (Glycine max (L.) Merrill) grown in controlled environments. J. Agric. Food Chem. 53, 1125–1129.
Cardinal A. J., Burton J. W., Camacho-Roger A. M., Yang J. H., Wilson R. F. and Dewey R. E. 2007 Molecular analysis of soybean lines with low palmitic acid content in the seed oil. Crop Sci. 47, 304–310.
Cheng H., Yu O. and Yu D. Y. 2008 Polymorphisms of IFS1 and IFS2 gene are associated with isoflavone concentrations in soybean seeds. Plant Sci. 175, 505–512.
Gutierrez-Gonzalez J. J., Wu X., Zhang J., Lee J. D., Ellersieck M., Shannon J. G., Yu O., Nguyen H. T. and Sleper D. A. 2009 Genetic control of soybean seed isoflavone content: importance of statistical model and epistasis in complex traits. Theor. Appl. Genet. 119, 1069–1083.
Haldane J. B. S. 1919 The combination of linkage values and the calculation of distances between the loci of linked factors. J. Genet. 8, 299–309.
Hoeck J. A., Fehr W. R., Murphy P. A. and Welke G. A. 2000 Influence of genotype and environment on isoflavone contents of soybean. Crop Sci. 40, 48–51.
Jung W., Yu O., Lau S. M., O’Keefe D. P., Odell J., Fader G. and McGonigle B. 2000 Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotwchnol. 18, 208–212.
Kassem M. A., Meksem K., Iqbal M. J., Njiti V. N., Banz W. J., Winters T. A., et al. 2004 Definition of soybean genomic regions that control seed phytoestrogen amounts. J. Biomed. Biotechnol. 1, 52–60.
Kassem M. A., Shultz J., Meksem K., Cho Y., Wood A., Iqbal M. J. and Lightfood D. A. 2006 An updated ‘Essex’ by ‘Forrest’ linkage map and first composite interval map of QTL underlying six soybean traits. Theor. Appl. Genet. 113, 1015–1026.
Kosambi D. D. 1944 The estimation of map distances from recombination values. Ann. Eugen 12, 172–175.
Kudou S., Fleury Y., Welti D., Magnolato D., Uchida T., Keisuke K. and Okubo K. 1991 Malonyl isoflavone glycosides in soybean seeds (Glycinem max. Merrill.) Agric. Biol. Chem. 55, 2227–2233.
Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., Lincoln S. E. and Newburg L. 1987 Mapmaker: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181.
Latunde-Dada A. O., Cabello-Hurtado F., Czittrich N., Didierjean L., Schopfer C., Hertkorn N. et al. 2001 Flavonoid 6-hydroxylase from soybean (Glycine max L.), a novel plant P-450 monooxygenase. Biol. Chem. 276, 1688–1695.
Lee S. J., Yan W., Ahn J. K. and Chung I. M. 2003 Effects of year, site, genotype and their interactions on various soybean isoflavones. Field Crops Res. 81, 181–192.
Liang H., Wang S., Yu Y., Lian Y., Wang T., Wei Y., Gong P., Liu X. and Fang X. 2009 QTL mapping of isoflavone, oil and protein content in soybean. Sci. Agric. Sin 42, 2652–2660.
Lin S., Cianzio S. R. and Shoemaker R. C. 1997 Mapping genetic loci for iron deficiency chlorosis in soybean. Mol. Breed. 3, 219–229.
Lo F. H., Mak N. K. and Leung K. N. 2007 Studies on the anti-tumor activities of the soy isoflavone daidzein on murine neuroblastoma cells. Biomed. Pharmaeother. 61, 591–595.
Lozovaya V. V., Lygin A.V., Ulanov A. V., Nelson R. L., Dayde J. and Widholm J. M. 2005 Effect of temperature and soil moisture status during seed development on soybean seed isoflavone concentration and composition. Crop Sci 45, 1934–1940.
Matsumura H., Watanabe S., Harada K., Senda M., Akada S., Kawasaki S. et al. 2005 Molecular linkage mapping and phylogeny of the chalcone synthase multigene family in soybean. Theor. Appl. Genet. 110, 1203–1209.
Meksem K., Njiti V. N., Banz W. J., Iqbal M. J., Kassem M. M., Hyten D. L., Yuang J., Winters T. A. and Lightfood D. A. 2001 Genomic regions that underlie soybean seed isoflavone content. J. Biomed. Biotechnol. 1, 38–44.
Meng F., Han Y., TengW., Li Y. and LiW. 2011 QTL underlying the resistance to soybean aphid (Aphis glycines Matsumura) through isoflavone-mediated antibiosis in soybean cultivar ‘Zhongdou isoflavone-mediated antibiosis in soybean cultivar ‘Zhongdou. Theor. Appl. Genet 123, 1459–1465.
Murphy S. E., Lee E. A., Woodrow L., Seguin P., Kumar J., Rajcan I. and Ablett G. R. 2009 Genotype × environment interaction and stability for isoflavone content in soybean. Crop Sci. 49, 1313–1321.
Naim M., Gestetner B., Bondi A. and Birk Y. 1976 Antioxidative and antihemolytic activity of soybean isoflavone. J. Agric. Food Chem. 22, 806–811.
Njiti V., Meksem K., Lightfood D. A., Banz W. J. and Winters T. A. 1999 Molecular markers of phytoestrogen content in soybeans. J. Med. Food 2, 165–167.
Primomo V. S., Poysa V., Ablett G. R., Jackson C. J., Gijzen M. and Rajcan I. 2005 Mapping QTL for individual and total isoflavone content in soybean seeds. Crop Sci. 45, 2454–2464.
Qi Z., Wu Q., Han X., Sun Y., Du X., Liu C., Jiang H., Guo H. and Chen Q. 2011 Soybean oil content QTL mapping and integrating with meta-analysis method for mining genes. Euphytica 179, 499–514.
Sangeeta D., Mark G., Pat M. and Mana F. 2007 Transcriptome analysis reveals a critical role of CHS7 and CHS8 genes for isoflavonoid synthesis in soybean seeds. Plant Physiol. 143, 326–338.
Song Q. J., Marek L. F., Shoemaker R. C., Lark K. G., Concibido V. C., Delannay X. et al. 2004 A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109, 122–128.
Song X., Han Y., Teng W., Sun G. and Li W. 2010 Identification of QTL underlying somatic embryogenesis capacity of immature embryos in soybean (Glycine max (L.) Merr.) Plant Cell Rep. 29, 125–131.
Stracke R., Werber M. and Weisshaar B. 2001 The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456.
Tikkanen M. J. and Adlerereutz H. 2000 Dietary soy-derived isoflavone phytoestrogens: Could they have a role in coronary heart disease prevention? Biochem. Pharmacol. 60, l–5.
Trigizano R. N. and Caetano-Anolles G. 1998 Laboratory exercises on DNA amplification fingerprinting for evaluating the molecular diversity of horticultural species. Hort. Technol. 8, 413–423.
Tsukamoto C., Shimada S., Igita K., Kudou S., Kokubun M., Okubo K. and Kitamura K. 1995 Factors affecting isoflavone content in soybean seeds: Changes in isoflavones, saponins, and composition of fatty acids at different temperatures during seed development. J. Agric. Food Chem. 43, 1184–1192.
Wang C., Sherrard M., Pagadala S., Wixon R. and Scott R. A. 2000 Isoflavone content among maturity group 0 to II soybeans. J. Am. Oil Chem. Soc. 77, 483–487.
Wang H. and Murphy P. 1994 Isoflavone composition of American and Japanese soybeans in lowa: Efferents of variety, crop year, and location. J. Agric. Food Chem. 42, 1674–1677.
Watanabe S., Uesugi S. and Kikuchi Y. 2002 Isoflavones for prevention of cancer, cardiovascular diseases, gynecological problems and possible immune potentiation. Biomed Pharmacother 56, 302–312.
Weidenborner M., Hindorf H., Jha H. C., Tsotsonos P. and Egge H. 1990 Antifungal activity of isoflavones in different reduced stages on Rhizoctonia solani and Sclerotium rolfsii. Phytochemistry 29, 801–803.
Yang K., Moon J. K., Jeong N., Back K., Kim H. M. and Jeong S. C. 2008 Genome structure in soybean revealed by a genomewide genetic map constructed from a single population. Genomics 9, 52–59.
Yang K., Moon J. K., Jeong N., Chun H. K., Kang S. T., Back K. and Jeong S. C. 2011 Novel major quantitative trait loci regulating the content of isoflavone in soybean seeds. Genes Genomics 33, 685–692.
Yu O., Shi J., Hession A. O., Maxwell C. A., Mcgonigle B. and Odell C. A. 2003 Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63, 753–763.
Yuan J., Njiti V. N., Meksem K., Iqbal M. J., Triwitayakorn K., Kassem M. A. et al. 2002 Quantitative trait loci in two soybean recombinant inbred line populations segregating for yield and disease resistance. Crop Sci. 42, 271–277.
Zeng G., Li D., Han Y., Teng W., Wang J., Qiu L. and Li W. 2009 Identification of QTL underlying isoflavone contents in soybean seeds among multiple environments. Theor. Appl. Genet. 118, 1455–1463.
Zeng Z. B. 1993 Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90, 10972–10976.
Zeng Z. B. 1994 Precision mapping of quantitative trait loci. Genetics 136, 1457–1468.
Zhang D., Cheng H., Wang H., Zhang H. Y., Liu C. Y. and Yu D. Y. 2010 Identification of genomic regions determining flower and pod numbers development in soybean (Glycine max L.) J. Genet. Genomics 37, 545–556.
Zhang J., Ge Y., Sun J., Han F., Yu F., Yan S. and Yang H. 2012 Identification of QTL for isoflavone components among multiple environment in soybean seeds. Sci. Agric. Sin 45, 3909–3920.
Acknowledgements
The financial support for this research was provided by the National Natural Science Foundation of China (no. 31000717), Specialized Research Fund for the Doctoral Programme of Higher Education (20090061120002), the Fundamental Research Funds for the Central Universities, and the 211 Project of Jilin University.
Author information
Authors and Affiliations
Corresponding authors
Additional information
[Zhang H. J., Li J.W., Liu Y. J., Jiang W. Z., Du X. L., Li L., Li X.W., Su L. T.,Wang Q. Y. and Wang Y. 2014 Quantitative trait loci analysis of individual and total isoflavone contents in soybean seeds. J. Genet. 93, xx–xx]
Hai Jun Zhang and Jing Wen Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
ZHANG, H.J., LI, J.W., LIU, Y.J. et al. Quantitative trait loci analysis of individual and total isoflavone contents in soybean seeds. J Genet 93, 331–338 (2014). https://doi.org/10.1007/s12041-014-0371-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-014-0371-2