Abstract
The application of DNA intercalator 9-aminoacridine allowed us to increase the resolution of chromosome C-banding and DAPI-banding patterns and to investigate chromosomal polymorphism in karyotypes of seven spring and six winter rape varieties. It was shown that the pericentromeric and intercalary C-bands of most of the chromosomes in spring rape were smaller in size and less polymorphic than those of winter rape. More 26S and 5S rDNA sites were found in the winter rape karyotypes than the spring varieties. Separate or colocalized 26S and 5S rDNA sites were revealed on chromosomes 4, 5, 6, 8, 10, 14, 15, 16 and 18. Intervarietal and intravarietal polymorphism of the number and chromosomal localization of rDNA sites were detected. The generalized idiogram of chromosomes of 13 Brassica napus varieties with account of all possibilities of C-banding patterns as well as localization of 26S and 5S rDNA sites were constructed. Polymorphism of the examined molecular and cytogenetic markers as well as the heterozygosis level of FAE1.1 gene controlling erucic acid synthesis in rapeseed was higher in the winter varieties than in the spring ones. The obtained data were in a satisfactory agreement with increased tolerance to environmental stress conditions of winter rape.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oilseed rape (Brassica napusL. AACC, 2n = 38) is an allopolyploid species formed by spontaneous hybridization of B. oleracea L. (CC, 2n = 18) and B. rapa L. (AA, 2n = 20) followed by diploidization (Morinaga 1928; Nagaharu 1935). The rapeseed has spring (B. napus oleifera annuaMetzg.) and winter (B. napus oleifera biennisMetzg.) varieties. Nowadays the rapeseed genome (1127 Mb/1C) (Johnston et al. 2005) is being extensively investigated. A number of genes have been mapped for B. napus (Foisset et al. 1996; Qiu et al. 2006).
Oilseed rape chromosomes are small in size (1.53–3.3 μm) (Hasterok and Maluszynska 2000a). C-banding patterns of this species were shown to be simple and uniform, and they were represented mostly by pericentromeric and telomeric heterochromatic bands (Olin-Fatih and Heneen 1992; Olin-Fatih 1996). Many polymorphic rDNA sites were localized by fluorescence in situ hybridization (FISH) in the proximal and/or terminal chromosome regions (Kamisugi et al. 1998; Schrader et al. 2000; Kulak et al. 2002; Snowdon et al. 2002; Hasterok et al. 2001). In addition, some efforts (with different degrees of accuracy and precision) were undertaken to identify rapeseed chromosomes by molecular markers using FISH (Xiong and Pires 2011) and to discriminate the chromosomes of A and C subgenomes by GISH (Snowdon et al. 1997; Howell et al. 2008). However, there is still no unified classification of oilseed rape chromosomes that would take into account morphological characters of chromosomes, their subgenomic affiliations and the results of chromosomal marking. But the application of modern methods and techniques for analysing rapeseed chromosomes is problematic as they are small in size. Therefore, the intraspecific polymorphism of chromosome markers and C-banding patterns or chromosomal distribution of rDNA sites in the winter and spring rape are poorly investigated (Olin-Fatih 1996; Hasterok et al. 2006).
Rapeseed oil is used for industrial purposes and also in food (Grundy 1986; McDonald 1995). The content of erucic acid in rapeseeds is one of the important factor of its food or industrial application. Nowadays, canola or ‘00’ rapeseed varieties low in erucic acid and glucosinolates are the obligatory standard in the countries where oilseed rape is cultivated. The erucic acid content in rapeseeds is known to be under additive control of alleles of FAE1.1 and FAE1.2 genes that encode the 3-ketoacyl-CoA synthase, the enzyme of erucic acid synthesis from the oleoyl-CoA (Harvey and Downey 1964). It was revealed that loss of functions of FAE1.2 (in C subgenome) and FAE1.1 genes (in A subgenome) lead to ‘00’ rape plant formation (Fourmann et al. 1998; Katavic et al. 2002). The development of genome-specific DNA markers, which reveal homozygous and heterozygous state of FAE1.1 locus, gives an opportunity to evaluate the genetic polymorphism of rapeseed varieties and to accelerate the breeding of new canola varieties with desired properties. Some other specific molecular markers, that could be applied to select the forms with desired properties, were revealed in A and C subgenomes of oilseed rape. In particular, the molecular markers linked to the genes, that control the content of oleic and linolenic acids, can be used for breeding of high oleic or low linolenic oilseed rape genotypes (Hu et al. 1995; Jourdren et al. 1996). At the same time, there are no data on the correlation of molecular and chromosome markers of A and C subgenomes of oilseed rape that are specific to winter or spring varieties and/or associated with agronomic characters.
Apparently the investigation of oilseed rape genetic polymorphism by molecular and chromosomal markers is important for carrying out the analysis of intraspecific diversity of this useful plant. Further, such research stimulates intensive study of the peculiarities of genome organization of winter and spring rape varieties. The approach based on application of DNA intercalator 9-aminoacridine (9-AMA), that increased resolution of chromosome banding of small-sized plant chromosomes (Muravenko et al. 2003), had previously been described. Using this approach, the present paper examines chromosomal and genetic polymorphism of 13 winter and spring oilseed rape varieties of Russian and Belarusian breeding by studying C-banding and DAPI-banding, 26S and 5S rDNA sites localization on chromosomes, and DNA markers specific to FAE1.1 gene controlling the erucic acid synthesis.
Materials and methods
Plant material
The list of studied 13 rape varieties and their origins are given in table 1. The seeds were obtained from the germplasm collections of the Scientific and Practical Centre of NAS of Belarus for Arable Farming, Zhodino, Belarus and of All-Russian Williams Fodder Research Institute of RAAS, Moscow, Russia.
Chromosome spreads
Oilseed rape root tips were incubated for 12–16 h at 0°C in 5 mM solution of DNA intercalator 9-AMA to inhibit chromosome condensation process and to accumulate prometaphase chromosomes (Muravenko et al. 2003). Then the roots were fixed in 96% ethanol:glacial acetic acid fixative (3:1) which was changed twice every 30 min. The roots were stored in the fixative at –20°C.
For FISH and DAPI banding, before chromosome spread preparation, the roots were placed into 1% acetocarmine solution in 45% acetic acid for half an hour, the tip caps with root meristem were cut on the object plate, the meristem was macerated in a drop of 45% acetic acid, and then squashed chromosome preparations were made. The cover slips were removed after freezing, and the preparations were then dehydrated and stored in 96% ethanol.
For C-banding procedure, before chromosome spread preparation, the fixative was removed with six washes in distilled water (10 min each wash), the root tips were immersed in cold 0.2 N HCl for 15 min, and then incubated in 0.2 N HCl at 60°C for 5 min. To stop the hydrolysis, the root tips were placed into chilled distilled water and kept in the ice bath for 10 min. Then the root tips were washed once more in chilled distilled water and kept on the ice bath for another 30 min before being placed into 1.5 mL microtubes to which enzyme mixture (0.1% (w/v) Onozuka R10 cellulase (Serva, Heidelberg, Germany) and 0.1% (w/v) pectinase (Sigma, St Louis, USA) in citrate buffer (10 mM, pH 4)) was added. The root tips were digested for 16–20 h at room temperature. Then they were washed in distilled water, and the meristem was dissected in 45% (v/v) aqueous acetic acid solution and squashed under a coverslip. The cover slips were removed by freezing, and the preparations were then dehydrated and stored in 96% ethanol.
C-banding
C-banding procedure was carried out according to the technique developed for small-sized chromosome plants as previously described (Muravenko et al. 2001).
Fluorescence in situ hybridization (FISH)
FISH with 26S (isolated from Linum usitatissimum) and 5S (isolated from common wheat) rDNA probes was conducted as previously described (Semenova et al. 2006). Biotin labelled 26S rDNA probe was detected with FITC-conjugated avidin (Vector laboratories, Peterborough, UK). Digoxigenin labelled 5S rDNA probe was visualized with anti-digoxigenin-rhodamine, Fab fragments (Roche Biochemicals, Sussex, UK).
DAPI-banding
After FISH procedure, chromosome slides were stained with 0.1 μg/mL DAPI (4′,6-diamidino-2-phenylindole) in Vectashield medium (Vector laboratories, Peterborough, UK).
Chromosomal analysis
The slides were examined using Olympus BX61 fluorescence microscope (Tokyo, Japan), and images captured using monochrome CCD camera (Cool Snap, Roper Scientific, USA). The obtained images were processed with Adobe Photoshop 6.0 (Adobe, Birmingham, USA) and VideoTest-Kario 1.5 (Ista-VideoTest, St Petersburg, Russia) software. At least 10 metaphase plates were investigated for each variety.
DNA extraction and PCR
Total genomic DNA was extracted from 100 mg of young leaves of individual plants by Fermentas Genomic DNA purification kit (Fermentas, Vilnius, Lithuania). Fifteen individual plants of every variety were used to estimate the genetic heterogeneity.
The technique of FAE1.1 gene identification consisted of two stages: genome-specific and allele-specific PCR of FAE1.1gene fragment. Genome-specific primers to the promoter site of FAE1.1 gene allowed us to amplify selectively the fragment of the A genome of B. napus containing the target gene FAE1.1. PCR amplification was performed in a volume of 25 μL, containing 10 mM of Tris-HCl (pH 8.3), 50 mM of KCl, 1.5 mM of MgCl 2, 1.5 units of Taqpolymerase, 0.2 mM of dNTP, 0.25 μM of each primer (table 2), and 50–100 ng of template DNA. After the initial denaturation step at 94°C for 45 s, 35 PCR cycles at 94°C for 45 s, 60°C for 30 s and 72°C for 90 s were performed followed by a final extension at 72°C for 7 min.
The obtained PCR product of 980 bp was used as template DNA for the allele-specific amplification step with derived cleaved amplified polymorphic sequences-primers (dCAPS). Primer SAT_R to a mutant allele of FAE1.1gene was designed in such a way that an artificial restriction site for TaqI endonuclease appeared in case of SNP substitution (table 3).
PCR amplification was performed in a volume of 25 μL, containing 10 mM of Tris-HCl (pH 8.3), 50 mM of KCl, 1.5 mM of MgCl 2, 1.5 units of Taqpolymerase, 0.2 mM of dNTP, 0.25 μM of each primer (table 3), and 2 μL of PCR-mixture of rapeseed DNA amplification with Er_A primers. After the initial denaturation step at 94°C for 3 min, 35 PCR cycles at 94°C for 30 s, 60°C for 30 s and 72°C for 30 s were performed, followed by the final extension at 72°C for 7 min.
The obtained PCR product of 135 bp was digested at 65°C for 3 h. The reaction conditions: 1 × buffer TaqI: 10 mM Tris-HCl (pH 8.0), 5 mM MgCl 2, 100 mM NaCl and 0.1 mg/mL BSA, and 2 units of TaqI endonuclease in the incubation buffer. The resulting markers were electrophoresed in nondenaturing 8% polyacrylamide gel and visualized by ethidium bromide (Laemmli 1970).
The mean of expected heterozygosity forFAE1.1locus was estimated as a ratio of the number of heterozygotes H to the total amount of the analysed genotypes of the studied variety or hybrid N (Makarieva 2001).
Results
C-banding analysis of karyotypes
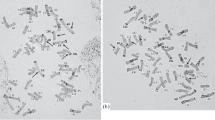
C-banding analysis of karyotypes of all studied rapeseed varieties showed that large C-bands were found in the pericentromeric regions of chromosomes, while small C-bands were revealed in the telomeric and intercalary regions (figure 1).
We subdivided chromosomes in the karyotypes into three groups based on their centromeric position and morphology as it had been suggested earlier (Olin-Fatih and Heneen 1992). The first group comprised seven metacentric chromosomes, the second group comprised six submetacentric chromosomes, and the third group comprised six acrocentric chromosomes. The chromosomes within each group were identified according to their C-banding patterns.
Visual analysis of C-banding patterns in karyotypes of the examined varieties revealed C-band polymorphism. In Dobrodei, Mayak, VIK-2 and Garant karyotypes the greater amount of heterochromatin was observed than in the rest of the varieties.
In karyotypes of winter varieties pericentromeric bands of larger sizes were observed. In contrast, in karyotypes of spring varieties there were more intercalary C-bands. Telomeric C-bands were highly polymorphic in karyotypes of every studied rape variety. In addition, a large C-band was found on the short arm of chromosome 19.
Secondary constrictions of satellite chromosomes 14 and 15 were revealed in the distal regions of the short arms, therefore the chromosomes were included with the group of acrocentric chromosomes. On chromosome 14, small polymorphic C-bands were revealed at the end of the satellite and in the region adjacent to the nucleolus organizer region (NOR). The satellite size as well as the length of the well-defined satellite thread was polymorphic. In addition, a polymorphic intercalary C-band was revealed in the pericentromeric region of the long arm of this chromosome. On chromosome 15, a satellite thread was ill-defined, and at the end of the satellite a small polymorphic C-band was detected. In karyotype of every variety, with the exception of Gedimin, a small polymorphic intercalary C-band was located in the proximal region of the long arm of chromosome 15. In this region of Gedimin chromosome 15, there were two intercalary C-bands.
Fluorescence in situ hybridization with 26S and 5S rDNA probes
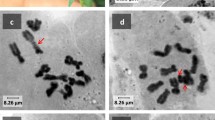
We conducted FISH on metaphase spreads of spring and winter rape using 26S and 5S rDNA probes (figure 2). Chromosomes were identified based on their morphology, centromeric positions and localization of DAPI bands which was mainly similar to C-banding pattern. The signals of hybridization of 26S and/or 5S rDNA probes were detected on chromosomes 4, 5, 6, 8, 10, 14, 15, 16 and 18.
A separate 26S rDNA signal was revealed in a number of varieties on two pairs of chromosomes, in the proximal regions of the long arm of chromosome 4 and/or in the subtelomeric regions of the short arm of chromosome 16. Only one signal on chromosome 16 was revealed in Garant, Severyanin, Dobrodei, Martyn, Mayak, Podmoskovnyi and Germes varieties, while a separate signal on chromosome 4 was detected in Pramen′. Separate 26S rDNA signals on chromosomes 4 and 16 were detected in Novik and Gedemin. There were no separate 26S rDNA signals in Grant, Lugovskoi and VIK-2 varieties.
In every studied variety we observed separate 5S rDNA signals in the pericentromeric and proximal regions of the long arm of chromosome 8 as well as in the subtelomeric regions of the short arm of chromosome 18.
Colocalized 26S and 5S rDNA sites were revealed in the secondary constriction region on the satellite chromosomes 14 and 15, and also in the proximal regions of long arm of chromosomes 4, 5, 6 and 10 (table 4).
As table 4 shows, in every variety there were colocalized 26S and 5S rDNA signals on chromosomes 5, 6, 14 and 15 as well as separate 5S rDNA signals on chromosomes 8 and 18.
In all the examined winter varieties we detected separate 5S rDNA sites on chromosomes 8 and 18. Colocalized 26S and 5S rDNA sites were observed on chromosomes 4, 5, 6, 14, 15 and on chromosome 10 in Severyanin, and also in the subtelomeric region of the short arm of chromosome 16 in VIK-2. The rest of the winter varieties possessed no signals on chromosome 10, but in the subtelomeric region of the short arm of chromosome 16 we detected a separate large 26S rDNA signal except VIK-2.
In all the examined spring varieties we observed separate 5S rDNA signals on chromosomes 8 and 18. Colocalized 26S and 5S rDNA sites were detected on chromosomes 4, 5, 6, 10, 14 and 15. In Novik, Gedimin and Pramen′ there were no 5S rDNA signals on chromosome 4, but 26S rDNA sites were found. In Novik, Podmoskovnyi, Gedimin and Germes we detected a separate 26S rDNA signal in subtelomeric region of the short arm of chromosome 16.
Thus, in the karyotypes of spring Russian rape we observed five to seven chromosome pairs bearing 26S rDNA sites and six to eight chromosome pairs with 5S rDNA sites, while in spring Belorusian rape we detected six to seven chromosome pairs with 26S rDNA sites and seven to eight chromosome pairs with 5S rDNA sites. Winter Russian rape karyotypes possessed six to seven chromosome pairs bearing 26S rDNA sites and seven to eight chromosome pairs with 5S rDNA sites. In winter Belorusian rape karyotypes revealed six to seven chromosome pairs with 26S rDNA sites and seven chromosome pairs with 5S rDNA sites (table 4).
In Novik and Severyanin we found intravarietal polymorphism in a number of rDNA sites. Both varieties possessed polymorphic colocalized 26S and 5S rDNA signals in the proximal regions of long arms of chromosome 10. In Novik we also detected a polymorphic 26S rDNA signal in the subtelomeric region of the short arm of chromosome 16.
Based on the chromosome morphology, C-banding patterns, DAPI-bands localization and also results of FISH with 26S and 5S rDNA probes, every chromosome in the karyotypes of the studied varieties was identified. A chromosomal passport of the species–a generalized idiogram of chromosomes of 13 B. napus varieties with account of all possibilities of C-banding patterns as well as 26S and 5S rDNA localization, was constructed (figure 3).
FAE1.1 gene identification by dCAPS-markers
Analysis of spring and winter rape varieties by using the developed dCAPS-markers make it possible to recognize single nucleotide polymorphism (SNP) in FAE1.1gene which results in disability to synthesize erucic acid. In addition, the analysis allows for efficient discrimination of mutant and wild-type alleles as well as estimation of heterozygous state of the FAE1.1locus (figure 4). The analysis of Russian and Belarusian spring and winter rape varieties revealed their heterogeneity in FAE1.1locus (table 5).
dCAPS products of Dobrodei variety. PCR products were amplified by allele-specific FAE1.1 primers and digested with TaqI. Patterns 2, 4–6, 8–14: homozygous genotypes free from erucic acid; patterns 1, 3, 7, 15: heterozygous genotypes with intermediate content of erucic acid; M, pUC19 DNA/MspI ladder (Fermentas, Vilnius, Lithuania). Right side arrows indicate the dCAPS products sizes.
The identification (screening for) of FAE1.1genes polymorphism in spring and winter rape revealed a number of FAE1.1heterozygotes among the individual plants. The mean of expected heterozygosity of FAE1.1 gene in the winter varieties (0.243) was nearly twice of spring ones (0.12).
Discussion
The rapeseed karyotype was shown to consist of 38 small-sized chromosomes (1.53–3.3 μm) (Hasterok and Maluszynska 2000a). Several classifications subdivided the chromosomes into three to four groups based on the centromeric position and chromosome morphology (Olin-Fatih and Heneen 1992; Hasterok and Maluszynska 2000a; Kulak et al. 2002). Additionally, an attempt to identify rapeseed chromosomes by C-banding showed that their C-banding patterns were rather poor and mostly represented by large pericentromeric and small telomeric bands (Olin-Fatih and Heneen 1992).
The application of DNA intercalator 9-AMA, which slowed down the process of chromosome condensation, allowed us to obtain longer rapeseed chromosomes in the spreads (2.1–4.7 μm) and to reveal more intercalary C-bands on the chromosomes. We subdivided rapeseed chromosomes into three groups according to the previously described classification (Olin-Fatih and Heneen 1992), but within each group we were able to identify every chromosome as C-banding patterns in our study were more informative (chromosomes were less condensed and contained more intercalary C-bands).
The analysis of C-banding chromosome polymorphism, that is a variation in size and number of C-bands, promotes effective studying of intraspecific chromosome variability (distinctions between subspecies, ecotypes and varieties) of large-size and small-size chromosome species. In addition, the polymorphism level correlates with the environmental conditions of the area of plant cultivation (Badaeva et al. 1990; Samatadze et al. 1997; Guerra 2000; Morales et al. 2007; Rachinskaya et al. 2011).
We found very few published studies on rapeseed C-heterochromatin analysis which had shown that C-banding patterns of B. napus,B. oleracea and B. rapa were not informative as C-bands were located mostly in pericentromeric and terminal regions of chromosomes (Olin-Fatih and Heneen 1992). Moreover, intraspecific C-band polymorphism of rapeseed chromosomes is still poorly studied (Olin-Fatih 1996). Application of DNA 9-AMA allowed us to obtain less condensed chromosomes containing more intercalary C-bands (figure 1). Accordingly, we were able to identify chromosomes in karyotypes of the examined varieties, to investigate intraspecific C-band polymorphism and also to construct an idiogram of the rapeseed genome with account of polymorphism of chromosome C-banding patterns (figure 3). Comparative analysis of rapeseed chromosomes revealed C-band polymorphism in the examined varieties. In particular, karyotypes of winter varieties were more heterochromatic than those of spring ones. All the varieties demonstrated high variability in length of the satellite thread of chromosome 14 as well as in C-bands adjacent to the secondary construction. It was probably associated with different functional status of NORs on chromosomes (Muravenko et al. 1991; Samatadze et al. 1997; Muravenko et al. 2009).
After FISH, chromosome staining with DAPI is shown to permit characterization of heterochromatic regions as AT-rich (DAPI +ve) or AT-poor (DAPI –ve) (Tagashira et al. 2009; Barros e Silva and Guerra 2010; Lavania et al. 2010). Additionally, DAPI-bands in the karyotypes of higher plants mostly coincide with C-bands (Schweizer 1980; Heng and Tsui 1993; Ali et al. 2000; Guerra 2000) although the mechanisms of C-specific and DNA-specific fluorochrome bandings are different (Schweizer 1980; Pignone et al. 1995; Peterson et al. 1999).
C-banding pattern of the plants with small chromosomes is known to be mainly represented by large heterochromatic bands found in the pericentromeric regions and small bands detected in the telomeric and/or intercalary regions of the chromosomes (Guerra 2000; Nakamura et al. 2001; Koo et al. 2002). The combination of C-banding, DAPI bandings and localization of rDNA loci could be an effective tool to refine karyotypes of the plants with small chromosomes (Nakamura et al. 2001; Koo et al. 2002; Muravenkoet al. 2009). In our study we followed such approach, and analysis of DAPI-banding patterns in karyotypes of the examined rapeseed varieties allowed us to identify the chromosomes, as they were considerably, although not completely, similar to the C-banding patterns.
Comparative analysis of the results, reported in previous studies, revealed the existence of intervarietal and intravarietal polymorphism of the number and chromosomal distribution of rDNA sites in karyotypes of foreign rapeseed cultivars (Kamisugi et al. 1998; Schrader et al. 2000; Hasteroket al. 2001; Kulak et al. 2002; Snowdon et al. 2002; Hasteroket al. 2006; Xiong and Pires 2011). In our study we also detected intervarietal and intravarietal polymorphism of the number and chromosomal localization of rDNA sites in seven winter and six spring varieties of Russian and Belorusian selection.
There were several rapeseed chromosome classifications in use, and it was difficult to compare our results of chromosome rDNA sites distribution (with the exception of satellite chromosomes) with the data of other investigators. Even so, it was possible to match the number of revealed 26S and 5S rDNA in a karyotype. In the examined varieties we observed five to seven pairs of 26S rDNA sites (table 4), and it was one to three pairs more than the number of pairs described earlier (Kamisugi et al. 1998; Schrader et al. 2000; Snowdonet al. 2000; Hasterok et al. 2006; Xiong and Pires 2011). But only 26S rRNA genes located on the satellite chromosomes were shown to be transcriptionally active (Hasterok and Maluszynska 2000b).
According to our results, the number of chromosome pairs containing 5S rDNA was also one to three more in karyotypes of Russian and Belorusian varieties than those of foreign rapeseed cultivars that had been analysed in the above mentioned studies. In addition, we revealed separate 5S rDNA sites localized on two chromosomes (table 4), that was consistent with the experimental data of some researchers (Kamisugi et al. 1998; Schrader et al. 2000, Hasterok et al. 2006), but it was not in line with the results of Snowdon et al. (2000) who had revealed separate 5S rDNA sites on four chromosomes.
In all the varieties, karyotypes comprised the set of four to six chromosome pairs with colocalized 26S and 5S rDNA sites. However, in all the spring varieties one of the colocalized rDNA sites was revealed on chromosome 10, and in all the winter varieties, on chromosome 4. Besides, we found the intravarietal polymorphism of localization of rDNA sites in two varieties. In the winter-hardy variety Severyanin a colocalized 26S and 5S rDNA site was sometimes revealed on chromosome 10. In Novik variety we detected two polymorphic sites: a colocalized 26S and 5S rDNA site on chromosome 10 and a separate 26S rDNA site on chromosome 16. In addition, we found colocalized 26S and 5S rDNA sites on the satellite chromosomes of both A and C subgenomes. In contrast, the investigation on foreign rapeseed cultivars revealed colocalization of 26S and 5S rDNA sites only on satellite chromosomes of A subgenome (Hasterok et al. 2006; Xiong and Pires 2011) or they were not detected on the satellite chromosomes at all (Snowdon et al. 2000). The total number of chromosomes containing colocalized 26S and 5S rDNA sites in the varieties of Russian and Belorusian origin was 1.5–2.0 times more than in foreign rapeseed cultivars.
A correlation between chromosome C-heterochromatin polymorphism in different populations of species and their adaptive characteristics was previously described (Vosa 1973; Greihuber and Speta 1978; Muravenko et al. 1991; Samatadze et al. 1998; Muravenko et al. 2001; Samatadze et al. 2012). Intraspecific variability of the number, sizes and localization of ribosomal genes was detected in different plant species (Hanson et al. 1996; Shishido et al. 2000; Rachinskaya et al. 2011) including Brassica species (Maluszynska and Heslop-Harrison 1993; Fukui et al. 1998; Hasterok et al. 2001, 2006). In addition, the number of ribosomal genes as well as their functional activity might vary under the influence of environmental and climatic conditions (Cullis 1979; Sobol’ 2001; Boulon et al. 2010; Goryachkina et al. 2013).
Our result showed that karyotypes of winter varieties possessed more C-bands as well as rDNA sites, than those in the spring rape. Winter rape is known to be more frost hardy (–8–10°C), and it is also more productive and oil-bearing than spring rape. In contrast, spring rape is less demanding of growth conditions (Allard 1999). Perhaps, the distinctions of molecular-cytogenetic markers between winter and spring rape, revealed in our study, were associated with the differences in adaptive characteristics of these varieties. This suggestion was confirmed indirectly by the fact that in karyotypes of foreign rapeseed cultivars grown under mild climatic conditions fewer rDNA sites were revealed than in Russian and Belorusian varieties.
The differences in adaptive characteristics of the studied rapeseed varieties were also revealed while we were analysing the heterozygosity level of the FAE1.1.gene. Association ofFAE1.1gene function and erucic acid synthesis was revealed by genetic transformation of the varieties low in erucic acid (Lassner et al. 1996). Both ofFAE1.1gene homologs (Bn-FAE1.1 and Bn-FAE1.2) of A and C genomes were defined, and they were showed to display 99.4% nucleotide sequence affinity (Katavic et al. 2002). Genome-specific amplification was performed by using of gene flanking sequences with high polymorphism level (Rahman et al. 2008). Based on comparison of these sequences, we developed several primers for obtaining fragments of A and C genomes of rapeseed with FAE1.1genes. Using the developed primers, we found that heterozygosity level of theFAE1.1gene, which controlled the erucic acid synthesis of rapeseed A subgenome, in the winter varieties was higher than that in the spring ones. It was probably caused by their origin as well as their raised tolerance to environmental stress conditions. High erucic acid containing rapeseed varieties were found to be more frost resistant and more winter hardy than canola (Rapacz and Markowski 1999). It may be suggested that, in spite of permanent selection of low erucic acid-containing genotypes in rapeseed, natural selection taking place at the background of lower temperatures supports FAE1.1gene heterozygous genotypes in winter rape population.
Our experimental data indicated the need of using chromosomal and molecular markers in the process of rapeseed breeding because classical approaches as well as oil biochemical analysis are inadequate for the equalization of varieties according to the erucic acid content in seed oil. Based on molecular and cytogenetic analysis of rapeseed, the suggested approach would be useful for further breeding of appreciable varieties and/or offering practical guidelines for cultivation of varieties at different environmental and climate conditions.
References
Ali H. B. M., Meister A. and Schubert I. 2000 DNA content, rDNA loci and DAPI bands reflect the phylogenetic distance between Lathyrus species. Genome 43, 1027–1032.
Allard R. W. 1999 Principals of plant breeding, 2nd edition. Wiley, New York, USA.
Badaeva E. D., Boguslavsky R. L., Badaev N. S. and Zelenin A.V. 1990 Intraspecific chromosomal polymorphism of Triticum araraticum (Poaceae) detected by C-banding technique. Plant Syst. Evol. 169, 13–24.
Barros e Silva A. E. and Guerra M. 2010 The meaning of DAPI bands observed after C-banding and FISH procedures. Biotech. Histoche. 85, 115–125.
Boulon S., Westman B. J., Hutten S., Boisver F-M. and Lamond A. I. 2010 The nucleolus under stress. Mol. Cell. 40, 216–227.
Cullis C. A. 1979 Quantitative variations of ribosomal RNA genes in flax genothrophs. Heredity 42, 237–246.
Foisset N., Delourme R., Barret P., Hubert N., Landry B. S. and Renard M. 1996 Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled-haploid progeny. Theor. Appl. Genet. 93, 1017–1025.
Fourmann M., Barret P., Renard M., Pelletier G., Delourme R. and Brunel D. 1998 The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor. Appl. Genet. 96, 852–858.
Fukui K., Nakayama S., Ohmido N., Yoshiaki H. and Yamabe M. 1998 Quantitative karyotyping of three diploid Brassica species by imaging methods and localization of 45S rDNA loci on the identified chromosomes. Theor. Appl. Genet. 96, 325–330.
Goryachkina O. V., Badaeva E. D., Muratova E. N. and Zelenin A. V. 2013 Molecular cytogenetic analysis of Siberian Larix species by fluorescence in situ hybridization. Plant Syst. Evol. 299, 471–479.
Greihuber J. and Speta F. 1978 Quantitative analysis of C-banded karyotypes and systematics in the cultivated species of the Schillasiberica group (Liliaceae). Plant Syst. Evol. 129, 63–109.
Grundy S. M. 1986 Comparison of monounsaturated fatty acids and carbohydrates for lowering plasma cholesterol. New Engl. J. Med. 314, 745–748.
Guerra M. 2000 Patterns of heterochromatin distribution in plant chromosomes. Genet. Mol. Biol. 23, 1029–1041.
Hanson R. E., Islam-Faridi M. N., Percival E. A., Crane C. F., Ji Y., McKnight T. D. et al. 1996 Distribution of 5S and 18S–25S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105, 55–61.
Harvey B. L. and Downey R. K. 1964 The inheritance of erucic acid content in rapeseed (Brassica napus L.)Can. J. Plant Sci. 44, 104–111.
Hasterok R. and Maluszynska J. (2000a) Cytogenetic markers of Brassica napus chromosomes. J. Appl. Genet. 41, 1–9.
Hasterok R. and Maluszynska J. (2000b) Nucleolar dominance does not occur in root tip cells of allotetraploid Brassica species. Genome 43, 574–579.
Hasterok R., Jenkins G., Langdon T., Jones R. N. and Maluszynska J. 2001 Ribosomal DNA is an effective marker of Brassica chromosomes. Theor. Appl. Genet. 103, 486–490.
Hasterok R., Wolny E., Hosiawa M., Kowalczyk M., Kulak-Ksiazczyk S., Ksiazczyk T. et al. 2006 Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann. Bot. 97, 205–216.
Heng H. H. Q. and Tsui L.-C. 1993 Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma 102, 325–352.
Howell E. C., Kearsey M. J., Jones G. H., King G. J. and Armstrong S. J. 2008 A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180, 1849–1857.
Hu J., Quiros C., Arus P., Strass D. and Robbelen G. 1995 Mapping of a gene determining linolenic acid concentration in rapeseed with DNA based markers. Theor. Appl. Genet. 90, 258–262.
Johnston J. S., Pepper A. E., Hall A. E., Chen Z. J., Hodnett G., Drabek J. et al. 2005 Evolution of genome size in Brassicaceae. Ann. Bot. 95, 229–235.
Jourdren C., Barret P., Brunel P., Delourme R. and Renard M. 1996 Specific molecular marker of the genes controlling linolenic acid content in rapeseed. Theor. Appl. Genet. 93, 512–518.
Kamisugi Y., Nakayama S., O’Neil C. M., Mathias R. J., Trick M. and Fukui K. 1998 Visualization of the Brassica self-incompatibility S-locus on identified oilseed rape chromosomes. Plant Mol. Biol. 38, 1081–1087.
Katavic V., Mietkiewska E., Barton D. L., Giblin E. M., Reed D. W. and Taylor D. C. 2002 Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. Eur. J. Biochem. 269, 5625–5631.
Koo D. H., Hur Y., Jin D. C. and Bang J. W. 2002 Karyotype analysis of a Korean cucumber cultivar (Cucumis sativus L. cv. Winter Long) using C-banding and bicolor fluorescence in situ hybridization. Mol. Cells 13, 413–418.
Kulak S., Hasterok R. and Maluszynska J. 2002 Karyotyping of Brassica amphidiploids using 5S and 25S rDNA as chromosome markers. Hereditas 136, 144–150.
Laemmli U. K. 1970 Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685.
Lassner M., Lardizabal K. and Metz J. 1996 A jojoba - ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutationin transgenic plants. Plant Cell 8, 281–292.
Lavania U. C., Kushwaha J., Lavania S. and Basu S. 2010 Chromosomal localization of rDNA and DAPI bands in solanaceous medicinal plant Hyoscyamus niger L. J. Genet. 89, 493–496.
Makarieva A. M. 2001 Variance of protein heterozygosity in different species of mammals with respect to the number of loci studied. Heredity 87, 41–51.
Maluszynska J. and Heslop-Harrison J. S. 1993 Physical mapping of rDNA loci in Brassica species. Genome 36, 774–781.
McDonald B. E. 1995 Oil properties of importance in human nutrition. In Brassica oilseeds – production and utilization (ed. D. S. Kimber and D. I. McGregor), pp. 291–299. CAB, Wallingford, UK.
Morales A. P., Filho W. and Guerra M. 2007 Karyotype diversity and the origin of grapefruit. Chromosome Res. 15, 115–121.
Morinaga T. 1928 Preliminary note on interspecific hybridization in Brassica. Proc. Imp. Acad. 4, 620–622.
Muravenko O. V., Badaev N. S., Borisov Yu. M., Borisova Z. Z., Surin N. A. and Zelenin A. V. 1991 Formation of karyotype in related barley varieties. Genetika 27, 2109–2118.
Muravenko O. V., Samatadze T. E., Popov K. V., Amosova A. V. and Zelenin A. V. 2001 Comparative study of genomes of two species of flax by C-banding of chromosomes. Genetika 37, 332–335.
Muravenko O. V., Amosova A. V., Samatadze T. E., Popov K. V., Poletaev A. I. and Zelenin A. V. 2003 9-aminoacridin- an efficient reagent to improve human and plant chromosome banding patterns and to standardize chromosome image analysis. Cytometry A. 51, 52–57.
Muravenko O. V., Yurkevich O. Yu., Bolsheva N. L., Samatadze T. E., Nosova I. V., Zelenina D. A. et al. 2009 Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica 135, 245–255.
Nakamura R., Kitamura S., Inoue M., Ohmido N. and Fukui K. 2001 Karyotype analysis of Nicotiana kawakamii Y. Ohashi using DAPI banding and rDNA FISH. Theor. Appl. Genet. 102, 810–814.
Olin-Fatih M. 1996 The morphology, cytology and C-banded karyotypes of Brassica campestris, B. oleracea and B. napus plants regenerated from protoplasts. Theor. Appl. Genet. 93, 414–420.
Olin-Fatih M. and Heneen W. K. 1992 C-banded karyotypes of Brassica campestris, B. oleracea and B. napus. Genome 35, 583–589.
Qiu D., Morgan C., Shi J., Long Y., Liu J., Li R. et al. 2006 A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 114, 67–80.
Peterson D. G., Lapitan N. L. V. and Stack S. M. 1999 Localization of single- and low copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152, 427–439.
Pignone D., Galasso I., Rossino R. and Mezzanotte R. 1995 Characterization of Dasypyrum villosum (L.) Candargy chromosomal chromatin by means of in situ restriction endonucleases, fluorochromes, silver staining and C-banding. Chromosome Res. 3, 109–114.
Rachinskaya O. A., Lemesh V. A., Muravenko O. V., Yurkevich O. Yu., Guzenko E. V., Bol’sheva N. L. et al. 2011 Genetic polymorphism of flax Linum usitatissimum based on use of molecular cytogenetic markers. Genetika 47, 65–75.
Rahman M., Sun Z., McVetty P. B. E. and Li G. 2008 High throughput genome-specific and gene-specific molecular markers for erucic acid genes in Brassica napus (L.) for marker-assisted selection in plant breeding. Theor. Appl. Genet 117, 895–904.
Rapacz M. and Markowski A. 1999 Winter hardiness, frost resistance and vernalization requirement of European winter oilseed rape (Brassica napus var. oleifera) cultivars within the last 20 years. J. Agron. Crop Sci. 183, 243–253.
Samatadze T. E., Muravenko O. V., Klimakhin G. I. and Zelenin A. V. 1997 Intraspecific chromosome polymorphism in Matricaria chamomilla L. (syn. M. recutita L.) studied by the C-banding technique. Genetika 33, 111–113.
Samatadze T. E., Muravenko O. V. and Zelenin A. V. 1998 Comparison of C-banded chromosomes in karyotypes of three species of the genus Matricaria L. Genetika 34, 1720–1724.
Samatadze T. E., Zelenin A. V., Suslina S. N., Amosova A. V., Popov K. V., Zagumennikova T. N. et al. 2012 Comparative cytogenetic study of the forms of Macleaya cordata (Willd.) R Br. from different localities. Genetika 48, 72–79.
Schrader O., Budahn H. and Ahne R. 2000 Detection of 5S and 25S rRNA genes in Sinapis alba, Raphanus sativus and Brassica napus by double fluorescence in situ hybridization. Theor. Appl. Genet. 100, 665–669.
Semenova O. Yu., Samatadze T. E., Zelenin A. V. and Muravenko O. V. 2006 The comparative genome study of the species of Adenolinum and Stellerolinum sections by means of FISH technique. Biologicheskie Membrany 23, 453–460.
Shishido R., Sano Y. and Fukui K. 2000 Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol. Gen. Genet. 263, 586–591.
Snowdon R. J., Kohler W., Friedt W. and Kohler A. 1997 Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids. Theor. Appl. Genet. 95, 1320–1324.
Snowdon R. J., Friedt W., Kohler A. and Kohler W. 2000 Molecular cytogenetic localization and characterization of 5S and 25S rDNA loci for chromosome identification in oilseed rape (Brassica napus L.)Ann. Bot. 86, 201–204.
Snowdon R. J., Friedrich T., Friedt W. and Kohler W. 2002 Identifying the chromosomes of the A- and C-genome diploid Brassica species B. rapa (syn. campestris) and B. oleracea in their amphidiploid B. napus. Theor. Appl. Genet. 104, 533–538.
Sobol’ M. A. 2001 Role of the nucleolus in plant cell response to environmental physical factors. Tsitol. Genet 35, 72–84.
Schweizer D. 1980 Fluorescent chromosome banding in plants; applications, mechanisms and implications for chromosome structure. In The plant genome (ed. D. R. Davids and D. A. Hopwood), pp. 61–71. The John Innes Institute, Norwich, UK.
Tagashira N., Hoshi Y., Yagi K., Pl der W. and Malepszy S. 2009 Cytogenetic comparison among three cultivars of cucumber (Cucumis sativus L.) by using post-heated DAPI band, 45S and 5S rDNA sites. Chromosome Bot. 4, 19–23.
U N. 1935 Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452.
Vosa C. G. 1973 Heterochromatin recognition and analysis of chromosome variation in Schillacibirica. Chromosoma 43, 269–278.
Xiong Z. and Pires J. C. 2011 Karyotype and identification of all homoelogous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187, 37–49.
Acknowledgements
This work was financially supported by RFBR grants no. 11-08-00716, 12-04-90046, B10P-081; subprogramme 1 ‘Innovative Biotechnologies in Belarus’ of EAEC Interstate special-purpose programme ‘Innovative Biotechnologies’; Fundamental Research Programme of the RAS ‘Dynamics of Plant, Animal and Human Genofonds’ of Presidium of the RAS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Amosova A. V., Zemtsova L. V., Grushetskaya Z. E., Samatadze T. E., Mozgova G. V., Pilyuk Y. E., Volovik V. T., Melnikova N. V., Zelenin A. V., Lemesh V. A. and Muravenko O. V. 2014 Intraspecific chromosomal and genetic polymorphism in Brassica napus L. detected by cytogenetic and molecular markers. J. Genet. 93, xx-xx
A. V. Amosova and L. V. Zemtsova contributed equally to this work
Rights and permissions
About this article
Cite this article
AMOSOVA, A.V., ZEMTSOVA, L.V., GRUSHETSKAYA, Z.E. et al. Intraspecific chromosomal and genetic polymorphism in Brassica napus L. detected by cytogenetic and molecular markers. J Genet 93, 133–143 (2014). https://doi.org/10.1007/s12041-014-0351-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-014-0351-6