Abstract
A single base change in the Bn-FAE1.1 gene in the A genome and a two-base deletion in the Bn-FAE1.2 gene in the C genome produce the nearly zero content of erucic acid observed in canola. A BAC clone anchoring Bn-FAE1.1 from a B. rapa BAC library and a BAC clone anchoring Bn-FAE1.2 from a B. oleracea BAC library were used in this research. After sequencing the gene flanking regions, it was found that the dissimilarity of the flanking sequences of these two FAE1 homologs facilitated the design of genome-specific primers that could amplify the corresponding genome in allotetraploid B. napus. The two-base deletion in the C genome gene was detected as a sequence-characterized amplified region (SCAR) marker. To increase the throughput, one genome-specific primer was labeled with four fluorescence dyes and combined with 20 different primers to produce PCR products with different fragment sizes. Eventually, a super pool of 80 samples was detected simultaneously. This dramatically reduces the cost of marker detection. The single base change in the Bn-FAE1.1 gene was detected as single nucleotide polymorphic (SNP) marker with an ABI SNaPshot kit. A multiplexing primer set was designed by adding a polyT to the 5′ primer end to increase SNP detection throughput through sample pooling. Furthermore, the Bn-FAE1.1 and Bn-FAE1.2 were integrated into the N8 and N13 linkage groups of our previously reported high-density sequence-related amplified polymorphism (SRAP) map, respectively. There were 124 SRAP markers in a N8 bin in which the Bn-FAE1.1 gene-specific SCAR marker was located and 46 SRAP markers in a N13 bin into which the Bn-FAE1.2 SNP marker was integrated. These three kinds of high throughput molecular markers have been successfully implemented in our canola/rapeseed breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Canola/rapeseed (Brassica napus L.) is a major oilseed crop in Canada, Europe, Australia, China and the Indian subcontinent. The quality of canola/rapeseed oil is determined primarily by its constituent fatty acids. The major fatty acid constituents of Brassica oil are palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), arachidic acid (C20:0), eicosenoic acid (C20:1) and erucic acid (C22:1). Erucic acid is one of the main fatty acids in rapeseed oil. Low erucic acid in rapeseed improves the quality of the oil, because high erucic acid is relatively low in digestibility and has been associated with health problems (Beare et al. 1963). On the other hand, high erucic acid rapeseed (HEAR) has several potential applications in the oleo-chemical industry for the production of high-temperature lubricants, nylon, plastics, slip and coating agents, soaps, painting inks and surfactants (Töpfer et al. 1995).
In B. napus, additive alleles at two gene loci control the erucic acid content in seeds (Harvey and Downey 1964), and these two genes (E1 and E2) are mapped in rapeseed (Ecke et al. 1995; Jourdren et al. 1996; Thormann et al. 1996). Development of low or high erucic acid content B. napus cultivars requires a long process of crossing, backcrossing and selfing of the segregating generations followed by identification of low or high erucic acid content lines from fatty acid profiles of seed lots from individual seeds by gas chromatography (GC). However, DNA molecular markers, which are tightly linked to the erucic acid genes or inside the candidate genes, can be applied with high efficiency in marker-assisted selection (MAS) for rapid transfer of a character into an otherwise desirable genotype efficiently and effectively. For instance, 15 of 16 plants in the F2 generation of canola/rapeseed crosses could be discarded at the seedling stage by selecting homozygous Bn-FAE1.1 and Bn-FAE1.2 genotypes with high or low erucic acid content instead of growing plants to full maturity and then doing seed quality analysis and selection.
The pathway for erucic acid biosynthesis and the major reactions involved in this pathway has been well characterized in Arabidopsis. Oleic acid is the main precursor for erucic acid biosynthesis via an elongation process in the developing embryos of B. napus (Bao et al. 1998). In A. thaliana seeds, it was reported that fatty acid elongase 1 (FAE1) is the candidate gene and that the gene product was essential for elongation of C18:1 (oleic acid) to C22:1 (erucic acid) (Lemieux et al. 1990; Kunst et al. 1992). The elongation process has four different steps. The first step is the condensation of oleoyl-CoA to malonyl-CoA to form a 3-ketoacyl-CoA. The second step is the reduction of the 3-ketoacyl-CoA to produce 3-hydroxyacyl-CoA. The third step is the dehydration of the 3-hydroxyacyl-CoA to form trans-(2,3)-enoyl-CoA. The final step is the further reduction of the trans-(2,3)-enoyl-CoA. These reactions are catalyzed by four different enzymes 3-ketoacyl-CoA synthase, 3-ketoacyl-CoA reductase, 3-hydroxyacyl-CoA dehydratase and trans-(2,3)-enoyl-CoA reductase, respectively (Fehling and Mukherjee 1991). The role of the FAE1 gene in producing erucic acid was genetically ascertained by genetic transformation of a low erucic acid content rapeseed (Lassner et al. 1996). In rapeseed, the two loci E1 and E2 of FAE1 homologs encode the rapeseed 3-ketoacyl-CoA synthases for the elongation process to generate erucic acid from oleoyl-CoA (Barret et al. 1998; Fourmann et al. 1998). In B. napus, these two homologs of the FAE1 gene (Bn-FAE1.1 and Bn-FAE1.2) have been characterized. These two homologs show 99.4% nucleotide identity and a two-base deletion results in a functional loss of Bn-FAE1.2 gene in the C genome (Fourmann et al. 1998). Katavic et al. (2002) reported that the amino acid serine at position 282 in high erucic acid content genotypes is substituted by phenylalanine in low erucic acid content genotypes due to single base change in the Bn-FAE1.1 gene in the A genome. In this report, BAC clones containing Bn-FAE1.1 and Bn-FAE1.2 genes from the A and C genome libraries were used to extend the sequence adjacent to these two genes to develop genome-specific high throughput molecular markers. These markers will considerably facilitate the selection of the four different erucic acid content control alleles in canola/rapeseed breeding programs.
Materials and methods
Plant materials
The pure breeding high erucic acid content cultivar ‘MillenniUM 03’ (~55% erucic acid) was crossed with the pure breeding canola line ‘SRYS-3’ (0% erucic acid) and the F1 was backcrossed to SRYS-3. The F1, F2, F3 and BC1 generations were grown in the greenhouse. Self-pollinated seeds of 279 F2 and 159 BC1 plants were examined for erucic acid content by gas chromatography (GC). Another 58 cultivars or breeding lines with high or low erucic acid content (determined by GC) in different Brassica species that were collected from various countries of the world were used to check the FAE1 genes markers (Table 3). Fifty-eight DH lines for constructing a B. napus high density map (Sun et al. 2007) were analyzed for erucic acid content and used for marker integration.
DNA extraction and sequencing of flanking sequence of the FAE1 genes
DNA was extracted using a modified version of the CTAB method according to Li and Quiros (2001) from the flower buds of B. rapa, B. oleracea and B. napus. The broccoli BAC library and library screening strategy, as described by Quiros et al. (2001), was used to pick up the BAC clone anchoring the Bo-FAE1.2 gene. The B. rapa BAC library, constructed from a male sterile line (kindly provided by Dr. Quiros, University of California, Davis), was used to select the BAC clone containing the Br-FAE1.1 gene. BAC DNA was prepared from the A-genome library and the C-genome library of pooled cultures of all clones from each 384-well plate. Targeted positive clones from positive plates were identified using a column and row (24 × 16) pooling strategy. The length of the FAE1 gene is approximately 1.6 kb with a single exon, and the primers were designed according to the FAE1 sequence in the GenBank data (u50771, AF009563, Y14974, Y14975, Y14980). The flanking sequences of the FAE1 gene in the A-genome and C-genome were sequenced by primer walking with the selected BAC clones. With the extended flanking sequences, the polymorphic regions between the A-genome and C-genome were found and used to design genome-specific primers.
Multiplexing SCAR markers by targeting the two-base deletion in the Bn-FAE1.2 gene
The two-base deletion in the Bn-FAE1.2 gene is located at the end of this gene. With the extended 3′-end flanking sequence of Bn-FAE1.1 and Bn-FAE1.2, a polymorphic region was found and a genome-specific primer FE42A targeting the Bn-FAE1.2 in the C genome was designed and another 20 primers were located inside the gene to form 20 primer pairs for multiplexing detection (Table 1). The genome-specific primer was labeled with one of the four fluorescent dyes (6-FAM, VIC, NED and PET, ABI, California) and the PCR products in a range of 200–500 bp were separated with an ABI 3100 DNA analyzer (ABI, California).
SNaPshot detection of the Bn-FAE1.1 alleles
SNP markers were developed through targeting the single base change in the Bn-FAE1.1 gene in the A genome. A genome-specific primers, ‘FEAG4 and FEAS1,’ were designed on the basis of the extended gene flanking sequence to amplify the Bn-FAE1.1 gene in the A genome. For multiplexing, another 22 primers were designed to detect the single nucleotide polymorphism (SNP) in the Bn-FAE1.1 by adding polyT at the 5′ end of the primers (Table 1).
The first step for SNP detection was to produce genome-specific PCR products containing the SNP position. PCR reactions were performed in a 10 μl volume containing 60 ng of genomic DNA, 100 μM of each dNTP, 0.15 μM of each primer, 1× PCR buffer, 1.5 mM MgCl2 and 1 Unit of Taq polymerase. The PCR program was as follows: 94°C for 3 min, followed by 35 cycles of 94°C for 1.0 min, 55°C for 1.0 min, 72°C for 1.0 min and final extension at 72°C for 10 min. SNP detection was performed using the single nucleotide primer extension method in the ABI 3100 Genetic Analyzer using a SNaPshot multiplex kit (ABI, California), following the instructions in the kit.
The SNaPshot products were pooled first and 2 μl pooled DNA was mixed with 8 μl formamide containing GeneScanTM 120 LIZTM size standard (ABI, California). After the DNA was denatured for 5 min at 95°C, the DNA fragments were analyzed with an ABI 3100 Genetic Analyzer. Genotypes were scored manually, using peak color verification.
Marker testing with different Brassica species and marker integration on a high-density map
DNA samples from 25 cultivars or breeding lines in different Brassica species and 58 DH lines used for the high-density SRAP map (Sun et al. 2007) was prepared as in the aforementioned protocol and were analyzed with the SNP marker for the Bn-FAE1.1 gene and the SCAR marker for the Bn-FAE1.2 gene.
For integrating these two Bn-FAE1 genes on the available high-density SRAP map, new data for the SNP and SCAR markers were put together with the data for the B. napus map construction and run with MAPMAKER v2.0 on a Macintosh computer to find the integrated linkage groups and the corresponding bins (Sun et al. 2007).
Results
Study on erucic acid content of B. napus
Self-pollinated seeds of 279 F2 and 159 BC1 plants from a cross of ‘MillenniUM03’ and ‘SRYS-3’ were examined for erucic acid content. The parental lines, ‘MillenniUM03’ and ‘SRYS-3’, have 56 and 0% erucic acid in seed, respectively. Twenty out of 279 plants produced low erucic content in F2, and 39 out of 159 BC1 plants showed low erucic acid content in seed. These results were consistent with a digenic (15:1 for F2 and 3:1 for BC1) segregation ratio indicating that the two genes are responsible for erucic acid content (χ 2 test, P = 0.52 and 0.87 in F2 and BC1).
Gene-specific SNP markers for Bn-FAE1.1 in the A genome in B. napus
Gene-specific markers for the A-genome (Bn-FAE1.1 gene) were developed by targeting a transition type base substitution at position 846 (C to T) from the starting codon in low erucic acid content lines. Because of the high level of sequence conservation between Bn-FAE1.1 and Bn-FAE1.2, some primers targeting the sequence dissimilarities between the Bn-FAE1.1 and Bn-FAE1.2 coding regions were found to be unreliable to produce genome-specific PCR products. To design reliable primers for genome-specific PCR products, chromosome walking 2.5 kb upstream and 1.5 kb downstream of the flanking sequence of the Bn-FAE1.2 gene from the broccoli BAC clone, and 750 bp upstream and 700 bp downstream flanking sequences of the Bn-FAE1.1 gene from the B. rapa BAC clone was conducted. After Clustalw analysis (http://www.ebi.ac.uk/clustalw), dissimilarities between the upstream and downstream flanking sequences of Bn-FAE1.1 and Bn-FAE1.2 genes were found (Fig. 1). On the basis of these dissimilar sequences, one primer FEAG4 in the upstream flanking region and another primer FEAF1 inside the gene were designed to amplify a 1.2 kb A genome-specific fragment containing the single nucleotide polymorphic (SNP) position. The SNP was detected by using the single base extension method with an ABI SNaPshot Multiplex kit. The SNaPshot data were analyzed by GeneScan software and peak information was transformed manually for each loci. The polymorphic position containing homozygous ‘C’ for high erucic acid content exhibited a black peak, homozygous ‘T’ for low erucic acid content, a red peak, and ‘C/T’ for heterozygous plants, both a black peak and a red peak (Fig. 2). Genotypes were assigned accordingly with E1E1 for the black peak, e1e1 for the red peak and E1e1 for both the black peak and the red peak. One hundred and seventy-four F2 plants were tested for the Bn-FAE1.1 gene using SNaPshot. In this F2 population, 43 plants exhibited the E1E1 genotype, 81 showed the E1e1 genotype and 50 showed the e1e1 genotype, a good fit to a 1:2:1 genotypic ratio for single locus segregation for the Bn-FAE1.1 gene (χ 2 = 1.385, P = 0.5–0.7). Similarly, in the BC1 population, 49 plants were found to be the E1e1 genotype and 59 were the e1e1 genotype, consistent with the 1:1 expected genotypic segregation for a single locus inheritance for Bn-FAE1.1 gene (χ 2 = 0.926, P = 0.3–0.5).
SNP detection and multiplexed with various length of SNP primers using GeneScan software in ABI 3100 genetic analyzer to analyze the SNaPshot data. The peak information has been transformed manually for each loci (e.g., black for ‘C’ and the genotype is E1E1; red for ‘T’ and genotype is e1e1; black/red for ‘C/T’ and genotype E1e1)
Gene-specific SCAR markers for the Bn-FAE1.2 gene in the C genome in B. napus
Gene-specific markers for the Bn-FAE1.2 gene in the C genome were developed after the genome-specific primers were designed. The two-base deletion is located at 1,425–1,426 bp position for low erucic acid content lines, which result in a truncated protein due to an early stop codon (Fourmann et al. 1998). Fortunately, this deletion was close to the downstream dissimilarity region, where a C genome-specific primer FE42A was designed. Twenty different primers within the gene coding region from the upstream of the polymorphic region (CAAAT/C–AT) were also designed to amplify various fragments ranging from 270 to 484 bp in size. The DNA fragment specific for high erucic acid content lines consistently produced two-base larger fragments than those of low erucic acid content lines. This small difference was easily detected by the ABI 3100 Genetic Analyzer when the primer FE42A was labeled with a fluorescent dye. Therefore, the Bn-FAE1.2 gene for homozygous high erucic acid content lines generated two-base larger fragments, the low erucic acid content lines had two-base smaller fragments and the heterozygous lines had both larger and smaller fragments (Fig. 3a). Since these twenty different primer combinations generated fragments that were different in size, it was possible to pool the PCR products from 20 different primer combinations to achieve multiplexed, high throughput markers. With one fluorescent dye color, 320 DNA samples (16 × 20) were separated in one run in 30 min with the ABI 3100 DNA analyzer (Fig. 3b).
a Detection of C genome-specific High ER, F1, F1′ and low ER lines using C15 genome-specific primers FE42A/FE42F. The upper band is responsible for high ER 16 (E2E2), lower band is for low ER (e2e2) and double band is for intermediate ER (E2e2). b Identification of different erucic acid contents segregating populations using 20 labeled FE42A primer with twenty different upstream unlabelled primers to produce 21 various DNA fragments from 270 to 484 bp. The upper band is two bases larger than 22 the lower band for all pairs
A total of 174 F2 plants were analyzed for the Bn-FAE1.2 gene in the C genome. Of these, 46 were E2E2 genotype (single upper band), 45 were e2e2 genotype (single lower band) and 83 were E2e2 genotype (both upper and lower bands). The F2 population fits a 1:2:1 genotypic ratio for single locus inheritance for Bn-FAE1.2 for high, intermediate and low erucic acid content alleles (χ 2 = 0.378, P = 0.7–0.9). Similarly, 108 BC1 plants segregated into two groups, 51 for the E2e2 genotype and 57 for the e2e2 genotype, which also fit a 1:1 genotypic ratio for single locus segregation for Bn-FAE1.2 (χ 2 = 0.334, P = 0.5–0.7).
Association of FAE1 molecular markers with erucic acid content
Erucic acid content in the F2 and BC1 populations were associated with the two FAE1 homologs present in the A genome and the C genome. On the basis of SNP and SCAR markers, the F2 plants were classified into nine different genotypes: E1E1E2E2, E1E1E2e2, E1E1e2e2, E1e1E2E2, E1e1E2e2, E1e1e2e2, e1e1E2E2, e1e1E2e2 and e1e1e2e2 for Bn-FAE1.1 and BN-FAE1.2 genes in the A genome and the C genome, respectively (Fig. 2, 3b, and Table 2). A total of 170 F2 plants were tested for both genes, of which 10 were E1E1E2E2, 22 E1E1E2e2, 11 E1E1e2e2, 23 E1e1E2E2, 37 E1e1E2e2, 19 E1e1e2e2, 11 e1e1E2E2, 24 e1e1E2e2 and 13 e1e1e2e2 (Table 2). The segregation ratio was consistent with a 1:2:1:2:4:2:1:2:1 genotypic segregation ratio for two genes (χ 2 = 2.358, P ≥ 0.95). All genotypic classes were tested for erucic acid content in seeds and it was found that all four dominant allele genotypes had the highest erucic acid content with an average of 41.2%, three dominant allele genotypes had an average of 34.4% (E1E1E2 average = 34.5% and E1E2E2 average = 34.3%), two dominant alleles contributed an average of 23.7% (E1E1 = 24.2%, E1E2 = 24.1% and E2E2 = 22.9%), one dominant allele genotypes had an average of 12.5% (E1 = 12.0% and E2 = 13.0%), while e1e1e2e2 genotype plants had an average 0.26% erucic acid content (Table 2). The results indicated that all four dominant alleles had almost identical contribution to erucic acid content in seed with an average of 11.2% erucic acid per allele.
A further study was conducted to test the association of the two genes with erucic acid content in 108 BC1 plants developed from a cross of ‘MillenniUM03’ and ‘SRYS-3,’ with ‘SRYS-3’ (low ER) as recurrent parent. The genotyping was done on the basis of SNP detection and two base deletion detection in ABI 3100 genetic analyzer with four groups obtained from the BC1 population as E1e1E2e2, E1e1e2e2, e1e1E2e2 and e1e1e2e2 (Fig. 2, 3b, and Table 3). Of the 108 BC1 plants, 26 were E1e1E2e2 with an average 19.8% erucic acid content, 23 E1e1e2e2 with an average 10.5% erucic acid content, 24 e1e1E2e2 with an average 10.4% erucic acid content and 35 e1e1e2e2 with an average 0.18% erucic acid content. The four different genotypic classes fit a 1:1:1:1 backcross segregation ratio for two genes (χ 2 = 3.332, P = 0.3–0.5). The contributions of each allele for erucic acid content in seed in the BC1 population were similar to each other at 10.2%.
Applicability of the markers in different Brassica species
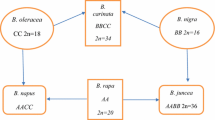
Fifty-eight cultivars or breeding lines with high or low erucic acid content in different Brassica species that were collected from Australia, Bangladesh, Canada, China, Denmark, Ethiopia, Germany, India, Japan, Netherlands, Poland, Sweden, Thailand, United Kingdom and USA were tested with the Bn-FAE1.1 and the Bn-FAE1.2 gene markers. Among the cultivars or breeding lines, forty-one were in B. napus, nine in B. rapa, two in B. oleracea and six in B. juncea. All the cultivars or breeding lines had the SNP and SCAR markers for the FAE1 genes perfectly matched with their erucic acid content in seeds (Table 3). In B. napus, all eighteen canola cultivars showed both mutations in the A and C genomes and all rapeseed cultivars did not, demonstrating that the genome-specific SNP and SCAR markers were reliable in the canola cultivars collected worldwide. As expected, the primers for the A genome-specific SNP marker did not amplify the FAE1 homolog in the C genome of B. oleracea, and the primers for the C genome SCAR marker did not produce bands in B. rapa and B. juncea. The genome specification of the SNP and SCAR markers were demonstrated with the species such as B. rapa, B. oleracea and B. juncea.
Integration of the SNP and SCAR markers into a high-density SRAP map
All 58 DH lines that were used for constructing the high-density SRAP map in B. napus were checked with GC for their erucic acid content. The A genome-specific SNP marker and the C genome-specific SCAR markers showed exactly the expected alleles corresponding to the erucic acid content (data not shown). The marker data were put together with the data that were previously used for the SRAP map construction to find the linkage groups that the SNP and SCAR markers were integrated into. As expected, the A genome-specific SNP marker was mapped into the N8 linkage group in the A genome and the C genome-specific SCAR marker into the N13 linkage group in the C genome. The N8 position integrated with the SNP marker was the N8-28 bin that contains 124 SRAP markers and the bin for the SCAR marker was the N13-70 that has 48 SRAP markers (Sun et al. 2007). Since most SRAP markers on the high-density B. napus map are genome-specific, they can be used directly for marker-assisted selection of erucic acid content in canola and rapeseed breeding.
Discussion
In rapeseed, the wild-type genotypes produce high erucic acid content in the seed. The first and only naturally occurring low erucic acid rapeseed line ‘Liho’ was discovered by using gas chromotography analysis in 1963 at the University of Manitoba. It had a mutation in the gene for erucic acid biosynthesis in B. napus (Stefansson and Hougen 1963). This is the only source of the low erucic acid B. napus that is currently used in canola breeding in the world. Low erucic acid content genotypes possess mutations affecting both E1 and E2 loci controlling erucic acid content in the seed (Harvey and Downey 1964). The fatty acid elongation 1 (FAE1) encoding 3-ketoacyl-CoA synthase has been characterized in Arabidopsis thaliana (James et al. 1995) and jojoba (Lassner et al. 1996). Barret et al. (1998) identified the FAE1 gene in B. napus with two homologous sequences (Bn-FAE1.1 and Bn-FAE1.2) from the embryo. The Bn-FAE1.1 gene has been shown to be linked to the E1 locus for B. rapa, and Bn-FAE1.2 was assigned to the E2 locus in B. oleracea. Only three nucleotide changes, a single base change in Bn-FAE1.1 and a two-base deletion in Bn-FAE1.2 account for the fatty acid difference of canola and rapeseed (Barret et al. 1998; Fourmann et al. 1998; Katavic et al. 2002). By targeting these three base changes, high throughput gene-specific markers for both Bn-FAE1.1 and Bn-FAE1.2 were developed. These markers are perfect matches with erucic acid content and no recombination occurs between the markers and trait. As the source of the low erucic acid content line is common worldwide, the markers showed the expected pattern in different high and low erucic acid cultivars and species of Brassica collected from various countries of the world, which confirms the acceptability of the markers for MAS in plant breeding.
Sequence similarity among the homologous genes in the A and C genes in B. napus is very high for most genes even in intergenic regions, which is an obstacle for developing gene-specific markers. The Bn-FAE1.1 gene in the A genome and the Bn-FAE1.2 gene in the C genome have similar sequence similarities. Since the sequence similarity of these two genes is very high, it is difficult to amplify the SNP position for low erucic acid in the A genome, because the same gene sequence is present in the C genome for high erucic acid content. Similarly, it is difficult to separately amplify the two-base deletion mutation for low erucic content genotypes in the C genome, because the same gene sequence is present in the A genome for high erucic acid, which hinders the identification of genotypes with homozygous deletion alleles. To overcome these problems, the extended sequence from the flanking regions of Bn-FAE1.1 in the A genome and Bn-FAE1.2 in the C genome were determined. Sequence dissimilarities outside the gene’s regions in the A and C genomes were large enough for genome-specific primers to amplify the A genome-specific Bn-FAE1.1 containing a SNP position and the C genome-specific Bn-FAE1.2 containing a two-base deletion position. This is the first reported approach to amplify the A and C genome-specific FAE1 genes in B. napus for precise tagging of the high and low erucic acid content genotypes in B. napus.
Sequencing the BAC clones from the B. rapa and B. oleracea BAC libraries allowed the accurate identification of the sequences from the A genome and the C genome in B. napus. Han et al. (2001) cloned the Bn-FAE1.1 promoter through inverse PCR in B. napus (GenBank accession number AF275254). After ClustalW alignment of AF275254 and our promoter sequences, we found that the AF275254 shared 99.8% identity to the C genome promoter and only 62.1% identity to the A genome with multiple deletions (S1). Therefore, the promoter in Han’s analysis is not a Bn-FAE1.1 promoter in the A genome, but it is a Bn-FAE1.2 promoter in the C genome.
Increasing throughput and reducing the cost for molecular marker detection is critical for large-scale application of MAS in canola/rapeseed breeding programs, since large number of plants have to be screened in a short time. In this report, multiplexed SNP detection and multiplexed SCAR markers were demonstrated using a series of SNP detection primers varying in size by adding polyT at the primer 5′ end. Twenty-two SNP detection primers allow a mixture of 22 reactions in one well. In theory, 17,600 samples (22 × 16 × 50) can be done with a medium throughput DNA analyzer, such as ABI 3100 DNA analyzer, within 24 h. Similarly, with an ABI 3100 Genetic analyzer, over sixty thousand (4 × 20 × 16 × 48) samples can be detected with the SCAR markers for Bn-FAE1.2 within 24 h, since the genome-specific primer labeled with four fluorescent dye colors can be used in our laboratory. Using this approach, it is possible to pool 80 PCR products for detection in an ABI genetic analyzer, which could reduce running time and cost by approximately 80 times.
The ABI 3100 genetic analyzer can be used with high efficiency to detect two-base difference markers. The two base deletion in the C genome could also be detected using SNaPshot, but using a SCAR marker is much cheaper than using SNaPshot, since only a small amount of specific PCR product is required for detection. Additionally, it is a one-step PCR with a short PCR amplification program, with about 15–20 cycles. Using a PCR pooling technique, approximately 60,000 samples could be analyzed with a medium throughput DNA analyzer and about three million samples could be analyzed with a high throughput DNA analyzer, such as ABI 3700 genetic analyzer, within 24 h, with a per sample cost of just a few cents instead of more than one dollar.
References
Bao X, Pollard M, Ohlrogge J (1998) The biosynthesis of erucic acid in developing embryos of B. rapa. Plant Physiol 118:183–190
Barret P, Delourme R, Renard M, Domergue F, Lessire R, Delseny M, Roscoe TJ (1998) A rapeseed FAE1 gene is linked to the E1 locus associated with variation in the content of erucic acid. Theor Appl Genet 96:177–186
Beare JL, Campbell JA, Youngs CG, Craig BM (1963) Effects of saturated fat in rats fed rapeseed oil. Can J Biochem Physiol 41:605–612
Ecke W, Uzunova M, Weißleder K (1995) Mapping the genome of rapeseed (Brassica napus L.). II. Localization of genes controlling erucic acid synthesis and seed oil content. Theor Appl Genet 91:972–977
Fehling E, Mukherjee KD (1991) Acyl-CoA elongase from higher plant (Lunaria annua): metabolic intermediates of very-longchain acyl-CoA products and substrate specificity. Biochim Biophys Acta 1082:239–246
Fourmann M, Barret P, Renard M, Pelletier G, Delourme R, Brunel D (1998) The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor Appl Genet 96:852–858
Han J, Luhs W, Sonntag K, Zahringer U, Borchardt DS, Wolter FP, Heinz E, Frentzen M (2001) Functional characterization of β-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol Biol 46:229–239
Harvey BL, Downey RK (1964) The inheritance of erucic acid content in rapeseed (Brassica napus). Can J Plant Sci 44:104–111
James DW Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Directed tagging of the Arabidopsis Fatty Acid Elongation 1 (FAE1) gene with the maize transposon Activator. Plant Cell 7:309–319
Jourdren C, Barret P, Horvais R, Foisset N, Delourme R, Renard M (1996) Identification of RAPD markers linked to the loci controlling erucic acid level in rapeseed. Mol Breed 2:61–71
Katavic V, Mietkiewska E, Barton DL, Giblin EM, Reed DW, Taylor DC (2002) Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. Eur J Biochem 269:5625–5631
Kunst L, Taylor DC, Underhill EW (1992) Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem 30:425–434
Lassner MW, Lardizabal K, Metz JG (1996) A jojoba b-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8:281–292
Lemieux B, Miquel M, Somerville C, Browse J (1990) Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet 80:234–240
Li G, Quiros CF (2001) Sequence related amplified polymorphism (SRAP) a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Quiros CF, Grellet F, Sadowski J, Suzuki T, Li G, Wroblewski T (2001) Arabidopsis and Brassica comparative genomics: sequence, structure and gene content in the ABI-Rps2-Ck1 chromosomal segment and related regions. Genetics 157(3):1321–1330
Stefansson BR, Hougen FW (1963) Selection of rape plants (Brassica napus) with seed oil practically free from erucic acid. Can J Plant Sci 44:359–364
Sun Z, Wang Z, Tu J, Zhang J, Yu F, McVetty PBE, Li G (2007) An ultradense genetic recombination map for Brassica napus, consisting of 13551 SRAP markers. Theor Appl Genet 114:1305–1317
Thormann CE, Romero J, Mantet J, Osborn TC (1996) Mapping loci controlling the concentrations of erucic and linolenic acids in seed oil of Brassica napus L. Theor Appl Genet 93:282–286
Töpfer R, Nartini N, Schell J (1995) Modification of plant lipid synthesis. Science 268:681–686
Acknowledgments
The research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), Bunge Canada and the Manitoba Canola Growers Association in an Industrial Research Chair program. Mr. Rahman was supported by a University of Manitoba Graduate Fellowship, a Manitoba Graduate Scholarship and an NSERC Scholarship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. L. Goldman.
Rights and permissions
About this article
Cite this article
Rahman, M., Sun, Z., McVetty, P.B.E. et al. High throughput genome-specific and gene-specific molecular markers for erucic acid genes in Brassica napus (L.) for marker-assisted selection in plant breeding. Theor Appl Genet 117, 895–904 (2008). https://doi.org/10.1007/s00122-008-0829-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0829-9