Abstract
A new mixed-ligand binuclear copper(II) complex [Cu(MS)(bpy)]2.(ClO4)2, built of 5-methylsalicylaldehyde and 2,2′-bipyridyl has been synthesized and characterized by using elemental analysis, IR and UV-Vis spectroscopy. Crystal structure of the complex shows that copper(II) ion lies in a square pyramidal coordination environment. The structure consists of two symmetrical half units in which the copper(II) ion of one half unit connected with the phenolate oxygen atom of other half unit along with one perchlorate anion in the crystal lattice as free molecule. Presence of uncoordinated perchlorate anion was also confirmed by IR spectroscopy. Absorption spectroscopy exhibits d-d transition at 628 nm, which further supports the square pyramidal geometry around the copper(II) ions. EPR spectrum of the copper(II) complex at room temperature shows a broad signal without any splitting pattern at g || = 2.26,g ⊥ = 2.03 and the magnetic moment (μ eff= 1.31 BM) obtained at room temperature indicate an antiferromagnetic interaction between the two copper(II) ions through phenoxide-bridge. Binding studies reveal that the complex possesses good binding propensity (K b = 5.2 ± 1.7 × 104 M−1) and bind to nitrogenous bases of DNA through intercalation. Nuclease activity of the complex with pBR322 DNA shows that the effect of hydrolytic cleavage is dose-dependent and the oxidative cleavage indicates the involvement of hydroxyl radical and singlet-oxygen as reactive oxygen species.

Mixed-ligand binuclear copper(II) complex was isolated and characterized. The binding studies of the complex suggest the intercalation mode of binding with CTDNA. The nuclease activity with pBR322 DNA shows that the hydrolytic cleavage is dose-dependent and the oxidative mechanism implies the involvement of hydroxyl radical and singlet-oxygen as reactive oxygen species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Recent literature on mixed-ligand complexes shows a wide range of applications in industrial, environmental and medicinal fields such as pharmacology, toxicology and biochemistry.[1–3] Transition metal ions are essential for the normal functioning of living organisms. Therefore, it is not surprising that transition metal complexes are of great interest as effective drugs and these complexes with tunable coordination environments and versatile spectral and electrochemical properties offer a great scope for design of new species that are suitable for DNA interactions.[4–6) DNA which is the storage and carrier molecule of genetic information is a major target for drug interaction due to the ability of DNA-bound drugs to interfere with transcription and/or replication, an essential step in cell growth and division. Transition metal complexes of mixed-ligands have attracted much attention because they are found to strongly bind and cleave DNA.[7,8] Bipyridyl (bpy) is an important chelating bidentate ligand for transition metal ions in the development of coordination chemistry.[9] It is a rigid planar, hydrophobic, electron-poor hetero aromatic system whose nitrogen atoms are appropriately placed to act cooperatively in cation binding. Copper(II) complexes of mixed-ligands are of great interest since they exhibit numerous biological activities. Copper is a bio-impact element with two biologically accessible oxidation states, and its mixed-ligand complexes have been synthesized because they bring about not only oxidative cleavage of DNA but also hydrolytic photo-induced and electrolytic cleavage of DNA.[10]
The impact of mixed-ligand complexes allows the development of methods with increasing selectivity and highly useful in the field of biological and environmental chemistry. In view of all these observations, we herein report the synthesis, characterization, crystal structure, DNA binding and nuclease activities of mixed-ligand binuclear copper(II) complex of 5-methylsalicylaldehyde and 2,2′-bipyridyl.
2 Experimental
2.1 Materials
All the chemicals are commercially available in analytical grade and used as received. Solvents are dried and purified before being used according to the standard procedure and stored over molecular sieves. Tetra(n-butyl)ammonium perchlorate (TBAP) was purchased from Fluka (Switzerland) and recrystallized from hot methanol. CT DNA and supercoiled pBR322 DNA was purchased from Bangalore Genei (India). 5 mM Tris-HCl/50 mM NaCl buffer solution was prepared from Tris base and NaCl dissolved in aqueous solution in which the pH was adjusted (till pH 7.2) with HCl/NaOH solution.
2.2 Physical measurements
A micro-analysis (% CHN) was performed using Carlo Erba model 1106 elemental analyser. FT-IR spectrum of the complex was recorded using KBr pellets in the range of 4000–400 cm−1 on a JASCO FT/IR-4100 spectrometer. UV-Vis spectrum of the complex was recorded in spectral grade DMF at 25∘C, on Agilent-8453 spectrophotometer in the range of 200–1100 nm. Cyclic voltammograms were obtained on CHI 602D (CH Instruments Co., USA) electrochemical analyser. Measurements were carried out under oxygen-free conditions using a three-electrode cell in which a glassy carbon, saturated Ag/AgCl and platinum wire are used as the working, reference and auxiliary electrodes, respectively. A ferrocene/ferrocenium (Fc/Fc+) couple was used as an internal standard. TBAP (0.1 M) was used as the supporting electrolyte. Molar conductivity was measured with an Elico digital conductivity bridge model CM-88, using a freshly prepared DMF solution of the complex. X-band EPR spectrum of the binuclear complex was recorded at room temperature on a Varian EPR-E 112 spectrometer. DPPH (2,2′-diphenyl-1-picrylhydrazyl) with ‘g’ = 2.0023 was used as the standard ‘g’ marker. The room temperature magnetic moment was measured on a PAR (Model 155) vibrating sample magnetometer.
Caution! perchlorate salts of metal complexes with organic ligands should be handled with care as they can cause explosion.
2.3 Synthesis of mixed-ligand complex [Cu(MS)(bpy)]2.(ClO4)2
The mixed-ligand copper(II) complex was prepared by adding a methanolic solution (10 mL) of 5-methylsalicylaldehyde (0.136 g, 1 mmol) and bipyridyl (0.156 g, 1 mmol) followed by an equimolar amount of triethylamine for deprotonating the former reactant, to an aqueous solution (10 mL) of copper(II) perchlorate (0.37 g, 1 mmol) with constant stirring and refluxed for 2 h. After cooling the reaction mixture to room temperature, the resulting solution was kept aside for slow evaporation. The title compound was obtained as dark green crystals and washed with cold methanol and dried in vacuo. Yield: 0.384 g (84%). Anal. Calc. for C36 H 30Cl2Cu2 N 4 O 12 (FW: 908.64): C, 47.59; H, 3.33; N, 6.17%. Found: C, 47.53; H, 3.52; N, 6.21%. FT-IR (KBr, cm−1): 2929–2600 v(C–H); 1664 ν(C =O); 1607 ν(C =N) of bpy; 1448 ν(C =C); 1097 & 625 [ ν(ClO4)− uncoordinated]. UV-Vis (DMF) λ/nm (ε/M−1 cm −1): 268 (16,825, π→ π*), 306 (6680, n →π*), 628 (835, d →d transition). Conductance Λ M (DMF, Ω−1 cm2 mol−1): 217. EPR at 298 K: g ||= 2.26, g ⊥= 2.03. μ eff= 1.31 BM at 298 K.
2.4 Crystal structure determination
Suitable single crystals were obtained for XRD measurements by slow evaporation of the mixed-ligand copper(II) complex [Cu(MS)(bpy)]2.(ClO4)2, in methanol at room temperature. The obtained dark green crystals were sorted using polarizing microscope (Leica DMLSP). Crystals having good morphology were chosen for three-dimensional intensity data collection. A crystal with dimensions of 0.30 × 0.25 × 0.20 mm was mounted on a glass fibre for diffraction experiment. X-ray single crystal data were collected on a Kappa Apex2 CCD diffractometer equipped with a fine-focus sealed tube X-ray source and graphite monochromated MoK α radiation in the wavelength (λ) of 0.71073 Å at room temperature (293 ± 2 K). Intensity data were collected using ω and φ scans with frame width of 0.5∘. Frame integration and data reduction were performed using Bruker SAINT-plus (Version 7.06a) software. Empirical absorption corrections were applied for the complex, using SADABS program.[11] The structure was solved by using SIR92[12] and the full-matrix least-squares refinement on F 2 was performed using SHELXL-97 program.[13] The scattering factors incorporated in SHELXL97 were used. After several cycles of refinement, positions of hydrogen atoms were calculated and added to the refinement process. Molecular graphics and packing figure was generated by using the softwares ORTEP 3.0[14] and Mercury 3.0,[15] respectively. Final R-value of the complex was 0.0275, and the final electrodensity map contains maximum and minimum peak heights of 0.405 and –0.285 e Å−3, respectively.
2.5 DNA binding studies
Application of electronic absorption spectra in DNA interaction is one of the most effective methods to examine the binding mode and strength of metal complex with DNA. A solution of calf thymus DNA in Tris-HCl/NaCl buffer gave a ratio of UV absorbance at 260 and 280 nm of about 1.89:1, indicating that the DNA was sufficiently free from proteins.[16] Stock solution of CT DNA was prepared in Tris-HCl/NaCl buffer and stored at 4∘C for less than 4 days. DNA concentration per nucleotide was determined by absorption spectroscopy using molar absorption coefficient 6600 M−1 cm−1 at 260 nm.[17] Stock solution of the complex was prepared by using 5% DMF of Tris-HCl/NaCl buffer and diluting suitably with the corresponding buffer to the required concentration for the experiments. Absorption titrations were carried out by varying nucleic acid concentration (0–500 μM) and maintaining the complex concentration constant (50 μM). Complex-DNA solutions were allowed to incubate for 30 min at room temperature before measurements were taken. While measuring absorption spectra, equal amounts of DNA was added to both complex solutions and the reference solution to eliminate absorbance of DNA itself. Titration curve was constructed from fractional change in absorption intensity as a function of DNA concentration.[18,19] The intrinsic binding constant K b, can be obtained by the following equation:
where [DNA] – DNA concentration in M (nucleotide), ε a – absorption coefficient observed at a given DNA concentration, ε f – absorption coefficient of the complex in the absence of DNA, ε b – absorption coefficient of the complex when fully bound to DNA, and K b – intrinsic binding constant in M−1. Each set of data was fitted to the above equation, and the plot of [DNA]/(ε b – ε f) versus [DNA] gave a slope and the y-intercept which are equal to 1/(ε b– ε f) and 1/K b(ε b– ε f), respectively. The intrinsic binding constant K bwas obtained from the ratio of the slope to the intercept.
Viscometric titrations were performed with an Ostwald micro viscometer (2 mL capacity) immersed in a water bath maintained at 25 ± 0.2∘C. Desired concentrations of DNA (100 μM) and complex (0–100 μM) were prepared by using Tris-HCl/NaCl buffer (pH = 7.2). Mixing of the solution was achieved by purging the N2 gas through viscometer. Flow time was measured by using digital stopwatch and, the experiment was repeated in triplicate to obtain the concurrent values. Data were presented as (η/ η 0)1/3 versus binding ratio (1/R) [complex]/[DNA],[20,21] where η and η 0 are the specific viscosity of DNA in the presence and absence of the complex, respectively. Values of η and η 0 were calculated from the relation of η = (t – t b)/t b,[22] where t b is flow time of buffer alone and t is observed flow time for DNA in the absence and presence of complex. Relative viscosities for DNA were obtained from the relation, η/ η 0.
2.6 Nuclease activity
The DNA cleavage efficacy of the mixed-ligand copper(II) complex on supercoiled pBR322 plasmid DNA was monitored by determining its ability to convert supercoiled DNA (Form I) to nicked circular (Form II) and linear forms (Form III) using agarose gel electrophoresis technique. In the cleavage reaction, plasmid DNA (33.3 μM) was treated with the complex (50–500 μM) and hydrogen peroxide (100 μM, co-reactant) in Tris-HCl/NaCl buffer (pH 7.2). To test the involvement of reactive oxygen species (ROS) during strand scission, the ROS scavengers sodium azide (NaN3, 50 mM), dimethylsulphoxide (DMSO, 10 mM) and catalase (0.5 μL) were added alternatively to the reaction mixture. The mixture was incubated for 1 h at 37∘C. A loading buffer containing 0.25% bromophenol blue, 0.25% xylene cyanol, 30% glycerol (3 μL) was added and the electrophoresis of the DNA cleavage products was performed on 0.8% agarose gel containing ethidium bromide (1 μg/mL). The gel was run at 50 V for 1 h in TAE buffer (40 mM Tris base, 20 mM acetic acid, 1 mM EDTA, pH 8.3). The bands were viewed by placing the gel on UV illuminator and photographed. The DNA ligation experiments were conducted as follows: The cleavage product was purified by a DNA gel extraction kit. After purification, the nicked circular (NC) DNA (2 μL) was incubated for 12 h at 16∘C with 1.5 μL of 10X ligation buffer, 1 μL of T4 ligase (4 units) and 2.5 μL of 1 mM ATP. Afterwards, the ligation products were stained with EtBr, electrophoresed and imaged.
3 Results and discussion
The synthesized mixed-ligand binuclear copper(II) complex, [Cu(MS)(bpy)]2.(ClO4)2, has been isolated in good yield. It is non-hygroscopic in nature and, soluble in Tris-HCl/NaCl buffer, DMF and DMSO. Authenticity of the complex was ascertained by elemental analysis, IR, UV-Vis spectroscopy and cyclic voltammetry. Analytical data for the complex is in good agreement with the corresponding molecular formula, and it was confirmed by single crystal X-ray diffraction technique.
3.1 Description of crystal structure
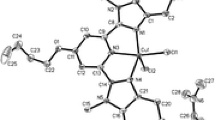
Slow evaporation of the reaction mixture offered dark green crystals with adequate quality to obtain good interpretable reflections. The single crystal structure of [Cu(MS)(bpy)]2.(ClO4)2 has been solved and the crystal data and structure refinement details are provided in table 1. The solved crystal structure of the complex contains two half units which are equivalent; one of them is in a general position and another unit is placed in a centre of inversion. Each unit of the mixed-ligand copper(II) complex contains [Cu(MS)(bpy)] and a perchlorate anion to balance the charge. Perchlorate anions found in the crystal are not coordinated to the copper(II) ion and occupy the crystal lattice as free molecules. The ORTEP diagram of the complex, along with atom numbering scheme is shown in figure 1, and the selected bond distances and bond angles relevant to the copper coordination sphere are provided in table 2. The copper(II) ions in the complex is five-coordinated and the geometry around the copper nuclei is best described as a perfect square pyramidal with a large contribution from the value of distortion constant (or) structural index parameter τ = 0.001 (τ = (β−α)/60, where β and α (in∘) are the two largest L–M–L angles O(6)–Cu(1)–N(2) = 173.30(7)∘ and O(5)–Cu(1)–N(1) = 173.23(7)∘, a regular trigonal-bipyramid (TBP) and square pyramidal (SP) have τ values of 1 and 0, respectively).[23–25] The five coordination sites of copper(II) metal centre can be described as below: Two imine nitrogen atoms (N1), (N2) of bpy, an oxygen atom of phenolate (O5) and aldehydic (O6) group of 5-methylsalicylaldehyde were coordinated at a distance of N(1)–Cu(1) = 1.9831(18), N(2)–Cu(1) = 1.9874(18), O(5)–Cu(1) = 1.8972(15), O(6)–Cu(1) = 1.9433(15) Å, respectively. The fifth coordination site of the copper(II) ion has been observed with the phenolate oxygen (O5#) atom of other half unit at a distance of O(5#)–Cu(1) 2.694(15) Å. The uncoordinated perchlorate anions are placed in the crystal lattice as free molecules with a distance of Cu(1)...Cl(1) 3.574(6) Å, from the copper nuclei.
The phenolic oxygen atom of 5-methylsalicylaldehyde fulfills a chelating bridging function, linking the two half monomeric complexes in dimeric [Cu(MS)(bpy)]2 associates with Cu(1)...Cu(1#) distance of 3.462(3) Å. Intermolecular Cu–O and Cu...Cu distances of the heteroleptic (mixed-ligand) complex [Cu(MS)(bpy)]2.(ClO4)2 are 2.694 and 3.462 Å, respectively. The bridged metal–oxygen and metal–metal distances are longer, when compared to the recently reported heteroleptic complex.[26] Non-covalent interactions such as π-stacking interactions with aryl hydrogen and hydrogen bonding network play a vital role in supramolecular chemistry and crystal engineering.[27] The molecular packing diagram is shown in figure S1, and it shows no hydrogen bonding in the crystal structure.
3.2 FT-IR, electronic and electrochemical properties
IR spectrum of the complex shows medium intensity bands in the region of 2929–2600 cm−1, assigned to C–H stretching. Sharp bands at 1664 and 1607 cm−1 are attributed to coordinated carbonyl group and C =N stretching of bpy ligand, respectively and, C =C stretching of aromatic ring was observed at 1448 cm−1. Presence of perchlorate anions in the complex is inferred from the bands at 1097 cm−1 (v 3-antisymmetric stretching) and 625 cm−1 (v 4-antisymmetric bending). No splitting pattern at 1097 cm−1 and the absence of band around 930 cm−1 (v 2-symmetric stretching) indicate that perchlorate anions are uncoordinated in the complex.[28] In order to gain further structural information about the mixed-ligand copper(II) complex, the electronic spectrum was recorded in DMF medium at room temperature. Three absorption bands with varied intensities were observed. The intense band at 268 nm is assigned to intra-ligand π– π* transition, while the medium intense band at 306 nm corresponds to charge transfer transition. Besides, a broad band observed at 628 nm can be unambiguously attributed to the d–d transition of copper(II) ions in a square pyramidal environment.1 Spectroscopic data of the complex is consistent with the results of the X-ray crystal structure analysis.
Electrochemical properties of the redox-active metal complexes have been extensively studied because the redox properties provide information for its further application in electrochemical, catalytic and biological reactions. Conductivity measurement of the binuclear copper(II) complex in dimethylformamide resulted in Λ M value at 217 Ω−1 cm2 mol−1 which indicates that the complex is of the 1:2 electrolyte type.[29] Generally, electrochemical properties of the complexes depend on a number of factors such as chelation, axial ligation, degree and distribution of unsaturation and substitution pattern.[30,31) Phenolic metal complexes usually undergo reduction at cathodic potentials, because of the negative influence and hard nature of phenoxide atom in the ligand.[32] In the cathodic region, copper(II) complex shows two reduction waves at \(E_{\text {pc}}^{1}= -\)0.805 and \(E_{\text {pc}}^{2} =-\)1.628 V (figure S2). Based on the observations, reduction process for Cu(II) complex is said to be irreversible in nature. The following steps may be involved in the reduction process:
3.3 Magnetic studies
Magnetic studies of the binuclear copper(II) complex has been studied to evaluate magnetic interactions between the two copper ions. The EPR spectrum of the binuclear copper(II) complex was recorded on X-band at a frequency of 9.4 GHz under magnetic field strength of 3200 G at 298 K. The spectra show a broad signal without any hyperfine splitting, and exhibit axially symmetric g-tensor parameters with g ∥ > g ⊥>2.0023 revealing that the unpaired electron is localized in dx 2 −y 2 orbital of the Cu(II) ions and an appreciable covalency with the orbital. The broad centred EPR spectrum (figure 2) in the region g ∥=2.26,g ⊥=2.03 observed for the binuclear copper(II) complex is attributed to magnetic interaction between two copper(II) ions,[33] through the phenoxide bridge. Inspite of the Cu–Cu separation, an antiferromagnetic interaction is observed due to the favourable relative orientation of the magnetic orbitals. The probe room temperature magnetic studies of the binuclear copper(II) complex resulted in magnetic moment value of 1.31 BM, which suggests the presence of an antiferromagnetic interaction between two copper(II) ions. Relatively speaking, g ∥ > 2.3 is characteristic of anionic environment and g ∥ < 2.3 of a covalent environment in M–L bonding.[34] The observed g ∥ value for the complex was less than 2.3 in agreement with the covalent nature of the metal–ligand bond.
3.4 DNA binding studies
3.4.1 Absorption spectral titration
The binding mode and strength of the drugs to CT DNA have been usually studied by various spectroscopic techniques. Monitoring the effect of increasing amounts of DNA on the absorption spectrum of a drug is one of the most widely used methods for determining overall binding constants. Transition metal complexes can bind to DNA via both covalent and/or non-covalent interactions. In the case of covalent binding, the labile ligand of the complex can be replaced by a nitrogen base of DNA such as guanine (N7), while the non-covalent interactions include intercalative, electrostatic and groove or surface binding (major/minor groove). In general, an intercalative binding mode of molecule to DNA is accompanied by hypochromism with significant red (bathochromism) or blue (hypsochromism) shift due to the strong π– π stacking interaction between the DNA base pairs and aromatic chromophore of the ligand with the extent of hypochromism commonly consistent with the strength of the intercalation.[8]
Absorption spectra of the complex in the absence and presence of CT DNA are provided in figure 3. With increasing concentration of DNA, absorption of the complex was affected, resulting in the tendency towards hypochromism with blue shift. Observed hypochromism (28%) and blue shift (2 nm) for the complex at a ratio (R) of [DNA]/[complex] = 10, unambiguously reveal non-covalent interaction (intercalative binding mode) of the complex with CT DNA.[35,36] Hypochromism suggests stacking interaction between an aromatic ring of the ligand and the base pairs of DNA. After interaction of the complex with the base pairs of DNA, π* orbital of the intercalated ligand can couple with π orbitals of the base pairs, thus decreasing the π– π* transition probabilities and concomitantly resulting in hypochromism.[37] Binding constant K b of the complex to CT DNA was determined by monitoring the changes of absorbances with increasing concentration of DNA. The calculated binding constant K b of complex is 5.2 ± 1.7 × 104 M−1. Binding strength of the complex is lower than that of the reported classical intercalators (EtBr, in Tris-HCl/NaCl buffer (25:40), pH 7.9 and [Ru(phen)2(dppz)]2+), in which the binding constants have been found to be in the order of 106–107 M−1.[38] Structurally, aromatic moiety of the complex should provide to overlap with the stacking base pairs of the DNA helix by intercalation, which results in hypochromism.
Absorption spectra of binuclear copper(II) complex (50 μM) in Tris-HCl/NaCl (pH 7.2) buffer upon addition of CT DNA (0–500 μM). Arrow shows that absorption decreases upon increase in concentration of DNA. Inset: Plot of [DNA]/(ε b– ε f) versus [DNA] for absorption titration of CT DNA with the complex.
3.4.2 Hydrodynamic (viscosity) titration
Viscometric measurement plays a crucial role in clarifying the binding mode of complex with DNA. To apprehend the nature of DNA binding of the complex, we have carried out viscosity measurements on CT DNA by varying the concentration of the complex. Values of relative specific viscosity (η/ η 0), where η and η 0 are specific viscosities of DNA in the presence and absence of the complex, are plotted against 1/R ([Complex]/[DNA] = 0.1–1). In classical intercalation, the DNA helix lengthens as base pairs are separated to accommodate the binding ligand leading to increase in DNA viscosity, whereas a partial, non-classical intercalation of ligand causes a bend (or kink) in DNA helix reducing its effective length and thereby its viscosity. Therefore, viscosity measurements, which are sensitive to the changes in the length of DNA molecule, are regarded as the least ambiguous and most critical means of studying the binding mode of complex with DNA in solution and provide stronger arguments for an intercalation binding mode.[22,39] Ethidium bromide is a well-known DNA intercalator which increases relative viscosity for the lengthening of DNA helix. Relative viscosity of DNA is steadily increased upon adding increasing amount of complex, similar to that observed for the proven intercalator EB (figure 4). These results suggest the binding of complex between the base pairs of DNA through classical intercalation. Thus, the complex shows an increase in relative viscosity of CT DNA but less compared to that of classical intercalator ethidium bromide.[40] This behaviour is similar to that of previously reported mixed-ligand copper(II) complexes.[26,41]
3.5 Nuclease activity
Cleavage of plasmid DNA by the transition metal complexes has been of interest to researchers. Nuclease activity of binuclear copper(II) complex has been studied using pBR322 DNA (33.3 μM) in Tris-HCl/NaCl (pH 7.2) buffer at 37∘C. This can be determined by gel electrophoresis of the plasmid. Migration of DNA in gel electrophoresis occurs under the influence of electric potential. DNA is negatively charged species, and when it is placed under electric field, it will migrate towards anode and this migration depends on the DNA size, electric field, gel density and the buffer nature. When circular plasmid DNA is run in horizontal using electrophoresis, relatively fast migration will be observed for the supercoiled (SC) form (Form I). If scission occurs, the supercoiled form will relax to generate a slower-moving nicked circular (NC) form (Form II), and if both strands are cleaved, a linear form (Form III) will be generated that migrates between Form I and Form II.
3.5.1 Nuclease activity in the absence of co-reactant
Hydrolytic cleavage of plasmid DNA by the copper(II) complex has been studied at room temperature. Transition metal complexes are the primary criteria for catalysing hydrolytic cleavage of DNA, i.e., coordination of the phosphate moiety of DNA to the metal(II) centre of complex, its DNA cleaving ability has been investigated with different concentrations of the complex. Nuclease activity of the complex was observed in absence of external agents and the increasing intensity of NC form was found with increase in concentrations of the complex (50–500 μM) without formation of linear form of DNA. Electrophoretogram (figure 5) of the complex shows prominent cleavage to give nicked circular (NC) form (lanes 2–7) and the control experiment does not show any considerable cleavage (lane 1). At higher concentrations, the SC form is completely converted to NC form. Presence of more aromatic moieties and hard Lewis acid properties of the complex plays a vital role in the DNA scission process.[24] The following interaction was concluded between complex and phosphate moieties of DNA: the copper(II) ions may recognize and bind with the phospho-diester bond of DNA through the coordinate linkage and/or electrostatic interaction, and the metal ions activate the central phosphorus atom of DNA, and this activated phosphorus atom is attacked nucleophilically by the metal ions due to their Lewis acidity via charge neutralization, and finally one of the P–O ester bonds of the nucleic acid is cleaved. The observation made during the cleavage experiment are reminiscent of those reported earlier for mixed-ligand[41] and bis(phen)[42] copper(II) complexes under identical conditions. To ascertain the hydrolytic nature of the scission process, an additional cleavage experiment was conducted using T4 ligase enzyme. The obtained cleavage product of SC DNA was reacted with T4 ligase enzyme and we have observed the conversion of 30% NC DNA to its original SC DNA (figure S3) which supports the hydrolytic cleavage.
3.5.2 Nuclease activity in the presence of co-reactant
Degradation of pBR322 DNA is also dependent on the co-reactant used. Cleavage of plasmid DNA induced by the binuclear copper(II) complex was investigated and clarified in the presence of H2 O 2 as a co-reactant. Control experiments with DNA alone (lane 1) and DNA + H2 O 2 (lane 7) were also carried out and did not show any considerable scission. At 25 μM concentration, the complex converts SC DNA into NC form (lane 2) revealing that the complex exhibits nuclease activity in the presence of H2 O 2 (figure 6). A mechanism was proposed for oxidative cleavage of DNA by the copper(II) complex as follows: interaction of the redox-active complex with DNA in the presence of a co-reactant is believed to produce different oxygen intermediates, depending on the specific complex and conditions. A non-diffusible metal-peroxo intermediate has been invoked in some cleavage reactions while in others, Fenton-like chemistry, which invokes release of freely diffusible hydroxyl (OH∙) or hydroperoxyl (HO\(_{\mathrm {2}}^{\mathrm {\bullet }})\) radical, has been assumed. Metal in the presence of H2 O 2 may generate reactive oxygen species (ROS) that can damage the deoxyribose ring (C3′), or alternatively a metal-peroxo species may participate directly in oxidation of the deoxyribose ring.
Electrophoretogram shows nuclease activity of the binuclear copper(II) complex (25 μM) on pBR322 DNA (33.3 μM) in the presence of H2 O 2 in Tris-HCl/NaCl buffer (pH 7.2) at room temperature. Lane 1, DNA control; lane 2, DNA + complex; lane 3, DNA + complex+H2 O 2 (40 μM); lane 4, DNA + complex+H2 O 2 + NaN3 (50 mM); lane 5, DNA + complex+H2 O 2 + DMSO (10 mM); lane 6, DNA + complex+H2 O 2 + catalase (0.5 μL); lane 7, DNA + H2 O 2.
In order to obtain a good knowledge of the reactive chemical species that was responsible for the DNA scission process for the complex, we further investigated the influence of different potentially inhibiting agents including singlet-oxygen quencher (NaN3), hydroxyl radical (DMSO) and hydrogen peroxide (catalase) scavengers. There is no appreciable inhibition of cleavage reaction in the presence of catalase (lane 6), which ruled out the possibility of DNA damage by hydrogen peroxide radical. Cleavage of DNA mediated by the complex in the presence of H2 O 2 was diminished by adding NaN3 (lane 4) and DMSO (lane 5), which conclusively indicates the involvement of singlet-oxygen and hydroxyl radical as the reactive oxygen species in the DNA scission process. In addition, coordination environment of the central metal ions and geometry of the complexes not only governs the DNA binding but also determines the nucleolytic action.
4 Conclusions
The principal findings and conclusions of this study are as follows: A new binuclear mixed-ligand copper(II) complex [Cu(MS)(bpy)]2.(ClO4)2 has been successfully synthesized from 5-methylsalicylaldehyde and 2,2′-bipyridyl and characterized by elemental analysis, IR and UV-Vis spectroscopy. Crystal structure of the complex has been determined by XRD method, which shows that the complex exists as square pyramidal coordination geometry. EPR study of the complex shows a broad spectrum without any hyperfine splitting, suggesting a covalent nature between the metal ions and donor atoms of the ligands. Absorption spectra and viscometric titration experiments suggest that the complex interacts with calf thymus DNA through the nitrogenous bases of DNA viaintercalation. Interestingly, the complex exhibits pronounced nuclease activity on plasmid DNA by the hydrolytic pathway. Oxidative pathway reveals that the underlying mechanism involves singlet-oxygen species and hydroxyl radical as reactive oxygen species.
References
Barve A, Kumbhar A, Bhat M, Joshi B, Butcher R, Sonawane U and Joshi R 2009 Inorg. Chem. 48 9120
Hui J K H, Yu Z and MacLachlan M J 2007 Angew. Chem. Int. Ed. 119 8126
Loganathan R, Ramakrishnan S, Suresh E, Riyasdeen A, Akbarsha M A and Palaniandavar M 2012 Inorg. Chem. 51 5512
Ramakrishnan S and Palaniandavar M 2005 J. Chem. Sci. 117 179
Roy M, Pathak B, Patra A K, Jemmis E D, Nethaji M and Chakravarty A R 2007 Inorg. Chem. 46 11122
Kumar A, Mitra A, Ajay A K, Bhat M K and Rao C P 2012 J. Chem. Sci. 124 1217
Ng C H, Kong K C, Von S T, Balraj P, Jensen P, Thirthagiri E, Hamada H and Chikira M 2008 Dalton Trans. 447
Ramakrishnan S, Rajendiran V, Palaniandavar M, Periasamy V S, Srinag B S, Krishnamurthy H and Akbarsha M A 2009 Inorg. Chem. 48 1309
Sammes P G and Yahioglu G 1994 Chem. Soc. Rev. 23 327
Chen G J, Qiao X, Qiao P Q, Xu G J, Xu J Y, Tian J L, Gu W, Liu X and Yan S P 2011 J. Inorg. Biochem. 105 119
Bruker-Nonius 2004APEX-II and SAINT-plus (Version 7.06a), Bruker AXS Inc., Madison, Wisconsin, USA
Altomare A, Cascarano G, Giacovazzo C and Guagliardi A 1993 J. Appl. Crystallogr. 26 343
Sheldrick G M 2008 Acta Crystallogr. A 64 112
Farrugia L J 1997 J. Appl. Crystallogr. 30 565
Macrae C F, Edgington P R, McCabe P, Pidcock E, Shields G P, Taylor R, Towler M and van de Streek J 2006 J. Appl. Crystallogr. 39 453
Marmur J 1961 J. Mol. Biol. 3 208
Reichmann M E, Rice S A, Thomas C A and Doty P 1954 J. Am. Chem. Soc. 76 3047
Carter M T, Rodriguez M and Bard A J 1989 J. Am. Chem. Soc. 111 8901
Smith S R, Neyhart G A, Kalsbeck W A and Thorp H H 1994 New J. Chem. 18 397
Friedman A E, Chambron J C, Sauvage J P, Turro N J and Barton J K 1990 J. Am. Chem. Soc. 112 4960
Cohen G and Eisenberg H 1969 Biopolymers 8 45
Satyanarayana S, Dabrowiak J C and Chaires J B 1992 Biochemistry 31 9319
Addison A W, Rao T N, Reedijk J, van Rijn J and Verschoor G C 1984 J. Chem. Soc. Dalton Trans. 1349
Dhara P K, Pramanik S, Lu T H, Drew M G B and Chattopadhyay P 2004 Polyhedron 23 2457
Weyhermüller T, Wagner R and Chaudhuri P 2011 Eur. J. Inorg. Chem. 2011 2547
Fei B L, Li W, Xu W S, Li Y G, Long J Y, Liu Q B, Shao K Z, Su Z M and Sun W Y 2013 J. Photochem. Photobiol. B 125 32
Desiraju G R and Steiner T 1999 The weak hydrogen bond in structural chemistry and biology. Oxford University PressNew York
Bharathi K S, Sreedaran S, Rahiman A K and Narayanan V 2013 Spectrochim. Acta A 105 245
Geary W J 1971 Coord. Chem. Rev. 7 81
Barefield E K, Freeman G M and Derveer D G V 1986 Inorg. Chem. 25 552
Connick P A and Macor K A 1991 Inorg. Chem. 30 4654
Benzekri A, Dubourdeaux P, Latour J M, Rey P and Laugier J 1991J. Chem. Soc. Dalton. Trans. 3359
Tolman W B, Rardin R L and Lippard S J 1989 J. Am. Chem. Soc. 111 4532
Guzar S H and Qin-han J I N 2008 J. Appl. Sci. 8 2480
Dougherty G and Pigram W J 1982 Crit. Rev. Biochem. 12 103
Bejune S A, Shelton A H and McMillin D R 2003 Inorg. Chem. 42 8465
Raja D S, Bhuvanesh N S P and Natarajan K 2011 Eur. J. Med. Chem. 46 4584
Cory M, Mckee D D, Kagan J, Henry D W and Miller J A 1985 J. Am. Chem. Soc. 107 2528
Fin L and Yang P 1997 J. Inorg. Biochem. 68 79
Gabbay E J, Scofield R E and Baxter C S 1973 J. Am. Chem. Soc. 95 7850
Ramakrishnan S, Shakthipriya D, Suresh E, Periasamy V S, Akbarsha M A and Palaniandavar M 2011 Inorg. Chem. 50 6458
Anbu S, Kandaswamy M, Kamalraj S, Muthumarry J and Varghese B 2011 Dalton Trans. 40 7310
Acknowledgements
The authors thank Sophisticated Analytical Instruments Facility (SAIF), Indian Institute Technology Madras (IIT-M), Chennai 600 025, for solving the crystal structure of the complex.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary information
CCDC-924217 contains supplementary crystallographic data of this article. These data files can be obtained free of charge from The Cambridge Crystallographic Data Centre via http://www.ccdc.cam.ac.uk/data_request/cif). Figures , and show the packing diagram, cyclic voltammogram and T4 ligation experiment of the complex, respectively. For details, see www.ias.ac.in/chemsci.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
GURUMOORTHY, P., RAVICHANDRAN, J. & RAHIMAN, A.K. Mixed-ligand binuclear copper(II) complex of 5-methylsalicylaldehyde and 2,2′-bipyridyl: Synthesis, crystal structure, DNA binding and nuclease activity. J Chem Sci 126, 783–792 (2014). https://doi.org/10.1007/s12039-014-0607-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12039-014-0607-y