Abstract
Accumulating evidence suggest that microRNAs play crucial roles in the development and progression of bladder cancer (BC). Here, we found that miR-212-3p was significantly down-regulated and negatively correlated with nuclear factor IA (NFIA) in human BC tissues. Bioinformatics analysis predicted that NFIA was a target gene of miR-212-3p. Then BC cell lines, T24 and J82 cells were transfected with miR-212-3p mimics or siNFIA to obtain miR-212-3p overexpression or NFIA knockdown cell lines, respectively. Quantitative real-time PCR was used to determine the expression of miR-212-3p and NFIA. Western blot analysis was utilized to detect NFIA expression. MTT assay showed either miR-212-3 overexpression or NFIA knockdown significantly inhibited the BC cell proliferation. Double staining with Annexin V-APC and 7-AAD showed the total number of apoptotic BC cells were remarkably increased after miR-212-3p overexpression or NFIA knockdown. Collectively, our results indicated that miR-212-3p targeting NFIA might serve as a promising target for BC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Bladder cancer (BC) is the ninth most frequently diagnosed cancer around the world (Al-Ahmadie et al. 2016), with higher incidence in Southern and Western Europe, North America, Northern Africa and Western Asia (Antoni et al. 2017). Despite a dramatic decline in mortality rates, BC still ranks the eighth in cancer-related death in European Union (Matsushita et al. 2015). Until now, patients with BC are usually treated by radical cystectomy, systemic cisplatin-based chemotherapy and immunotherapy. However, the five-year survival rate still remains poor (Kamat et al. 2016). Therefore, it is urgently needed to well understand the pathological molecular mechanisms underlying the development and progression of BC.
There is substantial empirical literature showing that dysregulation of microRNAs (miRNAs) links many malignancies leading to oncogenesis and metastasis (Lin and Gregory 2015). As an abundant class of small non-coding RNAs (~19–24 nt long), miRNAs can recognize the special motif of their target mRNAs and modulate gene expression via post-transcriptional, RNA interference and other pathways (Chen et al. 2015). Till now, several miRNA family members were found to contribute to BC physiological and pathological processes, such as miR-144-5p (Matsushita et al. 2015), miR-23b/27b (Chiyomaru et al. 2015), miR-145 (Matsushita et al. 2016) and miR-411 (Jin et al. 2018). The pre-miR-212 is processed by Dicer, leading to production of yield sense (miR-212-3p) and antisense strand (miR-212-5p) (Hansen et al. 2016). Hyperexpression of miR-212-3p decreases glioblastoma multiforme cells viability via pair to 3′UTRs of SGK3 (Liu et al. 2015). It has been reported that emergence of miR-212-3p binding motif mutation in CD80 is associated with the occurrence of gastric cancer in a Chinese population (Wu et al. 2014). However, the role of miR-212-3p is as yet undefined in BC.
The nuclear factor I (NFI) is a family of transcription factors that consists of four members, NFIA, NFIB, NFIC and NFIX (Messina et al. 2010). This family of site-specific (5′-TTGGC(N)5GCCAA-3′) DNA-binding proteins has been implicated in viral replication and activating or suppressing gene expression (Lee et al. 2017). NFIA is located in the 1p31 locus. In recent years, NFIA has emerged as a critical regulator in cancer development (Bernard et al. 2009). Zhao et al. (2017) showed the miR-191/NFIA signal transduction pathway in non-small cell lung cancer. A previous study identified that long non-coding RNA RP5-833A20.1 inhibition of NFIA as an important intrinsic mechanism suppressing carcinogenesis (Kang et al. 2016). In addition, NFIA plays important roles in glial development, including maintaining the glial progenitor cell status, controlling specification of neuronal-glial fate, and governing terminal differentiation (Lee et al. 2017). It also promotes glioma cell proliferation, survival and growth via negative modulation of p53, p21 and PAI1 (Lee et al. 2014).
In this study, we found that miR-212-3p expression was inversely correlated with NFIA in BC tissues. Bioinformatics analysis revealed that miR-212-3p could potentially target NFIA. Therefore, we decided to focus our study on the crosstalk of miR-212-3p and NFIA in the development and progression of BC. This study may be helpful to deepen our understanding of the molecular basis of BC.
2 Materials and methods
2.1 Tissue samples
A total of 32 pairs of fresh BC tissues and adjacent normal tissues were collected at the Department of Urology, the First Hospital of Jiaxing (Zhejiang, China) and immediately snap-frozen in liquid nitrogen until RNA extraction. All patients signed the informed consent and their clinicopathologic characteristics were summarized in table 1. This study was approved by the Ethics Committee of the First Hospital of Jiaxing (Zhejiang, China).
2.2 Cell lines and culture
Human BC cell lines, T24 and J82 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Human embryonic kidney cell line 293T (HEK293T) were purchased from the Cell Bank of Shanghai Institute of Cell Biology (Chinese Academy of Sciences, Shanghai, China). All cell lines were cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and maintained in a humidified incubator containing 5% CO2 at 37°C.
2.3 Quantitative real-time PCR
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For detection of miR-212-3p, cDNA was synthesized using a miRNA Reverse Kit (TaKaRa, Dalian, China) using the SYBR Premix Ex Taq kits (TaKaRa, Dalian, China). For detection of the NFIA mRNA level, cDNA was synthesized using the PrimeScript RT Reagent kit (TaKaRa). Quantitative real-time PCR was performed on ABI Prism 7500 HUT (ABI, MA, USA). The primers used in the experiments were as follows: miR-212-3p forward, 5′-GGTAACAGTCTCCAGTCA-3′ and reverse, 5′-GCAATTGCACTGGATACG-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; NFIA forward, 5′-TGGCCAAGTTACGGAAAGAT-3′ and reverse, 5′-GCGCTCGCCATCAGTACT-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. The relative expression levels of miR-212-3p and NFIA were normalized, respectively, to U6 and GAPDH by using the 2−ΔΔCt method (Livak and Schmittgen 2001).
2.4 Gene transfection
Human miR-212-3p mimic (miR-212-3p) and the corresponding negative control mimic (miR-NC), as well as NFIA siRNA (siNFIA) and its negative control siRNA (siNC), were purchased from RiboBio Co., Ltd. (Guangzhou, China). For gene transfection, T24 and J82 cells were plated into six-well plates and transfected with 100 nM miR-212-3p or siNFIA as well as their corresponding negative control using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol.
2.5 Cell proliferation assay
The cell proliferation was evaluated using the MTT assay. Briefly, the transfected cells at a density of 2.5 × 103 cells/well were seeded into 96-well plates and cultured for consecutive 5 days. Subsequently, cells in each well were incubated with 100 μL MTT (10 mg/mL, Sigma) at 37°C for 2 h. Next, 200 μL of DMSO was added to each well after the cell supernatant was removed. Finally, the optical density (OD) value at a wavelength of 595 nm was measured using a microplate spectrophotometer (BioTek Instruments, Inc. Winooski, VT, USA).
2.6 Cell apoptosis analysis
Flow cytometry was performed to determine cell apoptosis after 48 h transfection. Briefly, BC cells were harvested with Trypsin-EDTA, washed twice with PBS and underwent dual stained with an Annexin V-APC and 7-AAD Apoptosis Detection kit (Keygen, Nanjing, China) according to the manufacturer’s instruction. The percentage of early and late apoptotic cells was quantified between different groups using FlowJo software (TreeStar, San Carlos, CA, USA).
2.7 Luciferase reporter assay
Possible miR-212-3p binding sites were obtained from a miRNA database (http://www.targetscan.org/vert_71/). Of which, NFIA was predicted and selected as a potential target gene of miR-212-3p. Subsequently, luciferase reporter assay was performed to confirm this prediction. In brief, the wild-type (WT) of NFIA mRNA and its homologous mutant (MUT) sites were amplified and cloned into psiCHECK-2 vector (Promega Corp., Madison, WI, USA) to obtain WT NFIA and MUT NFIA plasmids, respectively, according to the manufacturer’s instructions. For luciferase assays, approximately 1 × 105 HEK293T cells were seeded into 24-well plates and co-transfected with 100 ng luciferase reporter plasmids and 100 nM miR-212-3p or miR-NC using Lipofectamine 2000 reagent (Invitrogen). At 48 h post-transfection, the relative luciferase activities were detected with the Dual-Luciferase Reporter Assay kit (Promega Corp.) according to the manufacturer’s instructions.
2.8 Western blot analysis
Total cellular protein was extracted using ice-cold RIPA lysis buffer (Roche Diagnostics, Basel, Switzerland) and the protein concentration was measured using A BCA protein assay kit (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer’s protocol. Then 30 µg of protein samples were separated with 10% Tris-SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, MA, USA). Membranes were blocked with 5% non-fat milk at room temperature for 2 h and incubated with primary antibodies against NFIA (Abcam, Cambridge, MA) and GAPDH (ProteinTech, IL, USA) at 4°C overnight. After washing with TBST three times, membranes were incubated with horseradish peroxidase labeled secondary antibodies for 2 h at room temperature. The protein signal was detected by enhanced chemiluminescence detection kit (Beijing Solarbio Science & Technology Co., Ltd.).
2.9 Statistical analysis
All statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). The experimental data were expressed as the mean ± standard deviation (SD) from three independent experiments. The quantitative data were statistically analyzed using the Student’s t-test between two groups or one-way analysis of variance followed by Tukey’s test among multiple groups. The association of miR-212-3p with NFIA expression was statistically analyzed using Spearman’s correlation test. The p value of less than 0.05 was regarded to be a statistically significant difference.
3 Results
3.1 MiR-212-3p was down-regulated and negatively correlated with NFIA expression in BC tissues
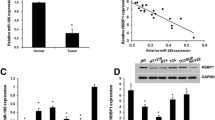
To investigate the functional role of miR-212-3p and NFIA in BC, we first determined their expression levels in BC tissues using quantitative real-time PCR. As shown in figure 1A, the endogenous miR-212-3p expression level was significantly down-regulated in BC tissues compared with adjacent normal tissues (p<0.001). In contrast, the expression level of NFIA was obviously higher in BC tissues than adjacent normal tissues (figure 1B, p<0.00). We next analyzed whether there was any relationship between their expression levels in BC tissues. As expected, Spearman’s correlation test showed that the expression of miR-212-3p was inversely correlated with NFIA expression in the same BC tissues (figure 1C, p = 0.0059, r = −0.4760).
Endogenous expression level of miR-212-3p and NFIA is negatively correlated in BC tissues. Quantitative real-time PCR analyzed endogenous (A) miR-212-3p and (B) NFIA expression level in human BC tissues and adjacent normal tissues. (C) Negative linear correlation between miR-212-3p and NFIA in human BC tissues was determined by Spearman’s correlation test. ***p<0.001 vs adjacent tissues.
3.2 Identification of NFIA as a target gene of miR-212-3p
Previous results have indicated miR-212-3p and NFIA were negatively correlated in BC tissues. Using target prediction tool TargetScan, miR-212-3p was predicted to directly bind to 3′UTR of NFIA (figure 2A). We then performed luciferase reporter assays to further validate their regulatory relationship. As shown in figure 2B, co-transfection with WT NFIA and miR-212-3p markedly decreased the luciferase activity in 293T cells (p<0.05), while there was no significant change in luciferase activity after co-transfection with MUT NFIA and miR-212-3p. Moreover, mRNA and protein expression level of NFIA were examined by quantitative real-time PCR and western blot, respectively. The results showed that miR-212-3p overexpression significantly suppressed the expression of NFIA at mRNA (figure 2C, p<0.01) and protein levels (figure 2D) in both T24 and J82 cells. These data suggest miR-212-3p could down-regulate NFIA expression through directly binding to its 3′-UTR.
MiR-212-3p modulates BC cells through targeting NFIA. (A) The direct binding sites of miR-212-3p with NFIA were predicted by TargetScan. The NFIA 3′-UTRs containing the wild- or mutant-type miR-212-3p targeting sequence were inserted downstream of the luciferase reporter vector. (B) Luciferase reporter gene assay were used to detect interaction between miR-212-3p and direct binding and mutation sites of NFIA in HEK293T cells. (C) Quantitative real-time PCR and (D) western blot analysis were used to measure NFIA mRNA and protein level in BC cell with transfection of miR-212-3p or miR-NC. Data are expressed as means ± SD of three independent experiments. *p<0.05, **p<0.01 vs miR-NC.
3.3 MiR-212-3p inhibits proliferation and induced apoptosis of BC cells
Since miR-212-3p has been shown to be lower expressed in BC tissues, we thus speculated that it might be a tumor suppressor in BC. To confirm this hypothesis, T24 and J82 cells were transfected with miR-212-3p. As shown in figure 3A, miR-212-3p transfection significantly increased the expression of miR-212-3p in both T24 and J82 cells (p<0.001). MTT assay showed that miR-212-3p overexpression inhibited the cell proliferation at different time points, especially at day 4th and 5th in T24 and J82 cells (figure 3B, p<0.001). Furthermore, flow cytometry analysis revealed that miR-212-3p overexpression remarkably increased the total number of apoptotic cells from 7.18 ± 0.30% to 18.28 ± 0.72% in T24 cells and 11.11 ± 0.52% to 22.06 ± 0.69% in J82 cells (figure 3C, p<0.001). These results demonstrated that miR-212-3p negatively regulated cell growth and proliferation in BC cells.
MiR-212-3p inhibits proliferation and induced apoptosis of BC cells. T24 and J82 cells were transfected with miR-212-3p or miR-NC, respectively. (A) Quantitative real-time PCR was used to determine the expression of miR-212-3p in T24 and J82 cells. (B) Cell proliferation was determined using the MTT assay. (C) Flow cytometry with double staining with Annexin V/7-AAD was utilized to analyze cell apoptosis in T24 and J82 cells. Data are expressed as means ± SD of three independent experiments. **p<0.01 vs miR-NC.
3.4 Knockdown of NFIA imitated the effects of miR-212-3p on cell proliferation and apoptosis in BC cells
As NFIA expression was significantly up-regulated and inversely correlated with miR-212-3p, we thus performed loss-of-function assays in BC cells to investigate the biological function of NFIA. First, T24 and J82 cells were transfected with siNFIA and underwent NFIA expression determination. The results showed that the expression of NFIA was significantly down-regulated at mRNA and protein levels using quantitative real-time PCR (figure 4A, p<0.001) and western blot analysis (figure 4B), respectively. Consistent with miR-212-3p overexpression, we further found NFIA knockdown significantly suppressed cell proliferation (figure 4C, p<0.001) and promoted apoptosis (figure 4D, p<0.001) in both T24 and J82 cells.
Downregulation of NFIA affected cell proliferation and apoptosis in BC cells. T24 and J82 cells were transfected with siNFIA or siNC, respectively. (A) Quantitative real-time PCR and (B) western blot were used to determine the expression of miR-212-3p in T24 and J82 cells. (C) Cell proliferation was determined using the MTT assay. (D) Flow cytometry with double staining with Annexin V/7-AAD was utilized to analyze cell apoptosis in T24 and J82 cells. Data are expressed as means ± SD of three independent experiments. **p<0.01 vs siNC.
4 Discussion
This study shows for the first time that, miR-212-3p is under-expressed in BC tissues and that miR-212-3p depresses BC cell (T24 and J82) proliferation and induces apoptosis. MiR-212-3p was reported to be downregulated in human prostate cancer (Ramalinga et al. 2015), glioblastoma (Liu et al. 2015), gastric cancer (Wada et al. 2010), hepatocellular carcinoma and derived cell lines (Tu et al. 2015). What’s more, previous studies have shown that miR-212-3p suppresses multiple pathways including cell proliferation, invasion, migration, autophagy, angiogenesis and senescence, as well as induces apoptosis (Ramalinga et al. 2015). Therefore, these findings appear very similar to previous reports that miR-212-3p is also down-regulated in BC tissues and inhibit BC carcinogenesis.
Accumulating evidence highlights a critical role for miRNAs in modulating their target mRNAs (Rupaimoole and Slack 2017). According to the statistical data by Lewis et al. (Lewis et al. 2005), an average of more than 200 mRNAs was predicted to be potential targets of each miRNA family members. Till now, several targets for miR-212-3p, including SRIT1 (Ramalinga et al. 2015), YAP1 (Tu et al. 2015), HMGB1 (Wada et al. 2010), SGK3 (Liu et al. 2015), Rab1a (Hou et al. 2018) and FOXA1 (Xie et al. 2018) have been identified. Moreover, miRNA–mRNA cross talk has been linked to the initiation and progression of human cancers (Lin and Gregory 2015). It has been reported that forced expression of miR-212-3p blocked proliferation and reduced survival of osteosarcoma cells and hepatocellular carcinoma cells via directly targeting FOXA1 (Xie et al. 2018). In glioblastoma, miR-212-3p functions as a tumor suppressor by targeting SGK3 (Liu et al. 2015). In prostate cancer, miRNA-132/212 targeting of SOX4 are thought to be responsible for suppression of TGF-β-mediated epithelial–mesenchymal transition (Fu et al. 2016).
NFIA is a key transcriptional modulator of brown fat genetic program (Hiraike et al. 2017). In recent years, much attention has been devoted to the role of NFIA in human malignancies, since it has been reported and implicated in several types of human cancers (Bernard et al. 2009; Zhao et al. 2017). Amplification and overexpression of NFIA generally occur in glioma and esophageal squamous carcinoma, and act as a novel promoter of tumor growth, proliferation and survival (Lee et al. 2014; Yang et al. 2018). In this study, using the publicly available databases TargetScan, we observed that NFIA contained a miR-212-3p seed match at the 3′UTR region. Further luciferase reporter assay revealed that the relative luciferase reporter intensity was remarkably reduced in HEK293T cells that co-transfected with WT NFIA and miR-212-3p compared with co-transfection with MUT NFIA and miR-212-3p, the result confirmed that the sequence 5′-CUGACAA-3′of miR-212-3p bound to NFIA mRNAs. We also observed that NFIA is highly expressed in BC tissues, and miR-212-3p expression levels are inversely correlated with NFIA levels. Moreover, the phenotype of BC cells with depletion of NFIA tends to imitate the BC cells that overexpressed miR-212-3p. These results indicate that miR-212-3p exerts its anti-tumor effect in BC cell through targeting NFIA. Interestingly, the members of NFIs could form homo- or heterodimers to active or inactive gene transcription according to cellular circumstances (Semenova et al. 2016). We suggest that NFIA is a regulator of BC proliferation and apoptosis by distinct modulation of pivotal genes that governs the biological behavior of BC.
In summary, the results of this study reveal that miR-212-3p suppresses the proliferation and induces apoptosis of BC cells by directly targeting NFIA. Our study may contribute to deep understanding of the role of miR-212-3p in human BC.
References
Al-Ahmadie HA, Iyer G, Lee BH, Scott SN, Mehra R, Bagrodia A, Jordan EJ, Gao SP, Ramirez R, Cha EK, Desai NB, Zabor EC, Ostrovnaya I, Gopalan A, Chen YB, Fine SW, Tickoo SK, Gandhi A, Hreiki J, Viale A, Arcila ME, Dalbagni G, Rosenberg JE, Bochner BH, Bajorin DF, Berger MF, Reuter VE, Taylor BS and Solit DB 2016 Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat. Genet. 48 356–358
Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A and Bray F 2017 Bladder cancer incidence and mortality: A global overview and recent trends. Eur. Urol. 71 96–108
Bernard F, Gelsi-Boyer V, Murati A, Giraudier S, Trouplin V, Adelaide J, Rey J, Olschwang S, Vainchenker W, Chaffanet M, Vey N, Mozziconacci MJ and Birnbaum D 2009 Alterations of NFIA in chronic malignant myeloid diseases. Leukemia 23 583–585
Chen Y, Gao DY and Huang L 2015 In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 81 128–141
Chiyomaru T, Seki N, Inoguchi S, Ishihara T, Mataki H, Matsushita R, Goto Y, Nishikawa R, Tatarano S, Itesako T, Nakagawa M and Enokida H 2015 Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int. J. Oncol. 46 487–496
Fu W, Tao T, Qi M, Wang L, Hu J, Li X, Xing N, Du R and Han B 2016 MicroRNA-132/212 upregulation inhibits TGF-beta-mediated epithelial-mesenchymal transition of prostate cancer cells by targeting SOX4. Prostate 76 1560–1570
Hansen KF, Sakamoto K, Aten S, Snider KH, Loeser J, Hesse AM, Page CE, Pelz C, Arthur JS, Impey S and Obrietan K 2016 Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn. Mem. 23 61–71
Hiraike Y, Waki H, Yu J, Nakamura M, Miyake K, Nagano G, Nakaki R, Suzuki K, Kobayashi H and Yamamoto S 2017 NFIA co-localizes with PPARγ and transcriptionally controls the brown fat gene program. Nat. Cell Biol. 19 1081–1092
Hou P, Kang Y and Luo J 2018 Hypoxia-mediated miR-212-3p downregulation enhances progression of intrahepatic cholangiocarcinoma through upregulation of Rab1a. Cancer Biol. Ther. 1–10
Jin H, Sun W, Zhang Y, Yan H, Liufu H, Wang S, Chen C, Gu J, Hua X, Zhou L, Jiang G, Rao D, Xie Q, Huang H and Huang C 2018 MicroRNA-411 downregulation enhances tumor growth by upregulating MLLT11 expression in human bladder cancer. Mol. Ther. Nucleic. Acids. 11 312–322
Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmström P-U, Choi W, Guo CC, Lotan Y and Kassouf W 2016 Bladder cancer. Lancet 388 2796–2810
Kang CM, Hu YW, Nie Y, Zhao JY, Li SF, Chu S, Li HX, Huang QS and Qiu YR 2016 Long non-coding RNA RP5-833A20.1 inhibits proliferation, metastasis and cell cycle progression by suppressing the expression of NFIA in U251 cells. Mol. Med. Rep. 14 5288–5296
Lee JS, Xiao J, Patel P, Schade J, Wang J, Deneen B, Erdreich-Epstein A and Song HR 2014 A novel tumor-promoting role for nuclear factor IA in glioblastomas is mediated through negative regulation of p53, p21, and PAI1. Neuro Oncol 16 191–203
Lee J, Hoxha E and Song HR 2017 A novel NFIA-NFkappaB feed-forward loop contributes to glioblastoma cell survival. Neuro Oncol 19 524–534
Lewis BP, Burge CB and Bartel DP 2005 Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15–20
Lin S and Gregory RI 2015 MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15 321–333
Liu H, Li C, Shen C, Yin F, Wang K, Liu Y, Zheng B, Zhang W, Hou X, Chen X, Wu J, Wang X, Zhong C, Zhang J, Shi H, Ai J and Zhao S 2015 MiR-212-3p inhibits glioblastoma cell proliferation by targeting SGK3. J. Neurooncol. 122 431–439
Livak KJ and Schmittgen TD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔ C T method. Methods–A Companion Method Enzymol. 25 402–408
Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S, Itesako T, Nakagawa M and Enokida H 2015 Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br. J. Cancer 113 282–289
Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M and Seki N 2016 Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget. 7 28460–28487
Messina G, Biressi S, Monteverde S, Magli A, Cassano M, Perani L, Roncaglia E, Tagliafico E, Starnes L, Campbell CE, Grossi M, Goldhamer DJ, Gronostajski RM and Cossu G 2010 Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell 140 554–566
Ramalinga M, Roy A, Srivastava A, Bhattarai A, Harish V, Suy S, Collins S and Kumar D 2015 MicroRNA-212 negatively regulates starvation induced autophagy in prostate cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis and cellular senescence. Oncotarget. 6 34446–34457
Rupaimoole R and Slack FJ 2017 MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discovery 16 203–222
Semenova EA, Kwon MC, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, Kuilman T, Peters D, Buikhuisen WA, Smit EF, Pritchard C, Cozijnsen M, van der Vliet J, Zevenhoven J, Lambooij JP, Proost N, van Montfort E, Velds A, Huijbers IJ and Berns A 2016 Transcription factor NFIB is a driver of small cell lung cancer progression in mice and marks metastatic disease in patients. Cell. Rep. 16 631–643
Tu H, Wei G, Cai Q, Chen X, Sun Z, Cheng C, Zhang L, Feng Y, Zhou H, Zhou B and Zeng T 2015 MicroRNA-212 inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting FOXA1. Onco. Targets Ther. 8 2227–2235
Wada R, Akiyama Y, Hashimoto Y, Fukamachi H and Yuasa Y 2010 miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int. J. Cancer 127 1106–1114
Wu R, Li F, Zhu J, Tang R, Qi Q, Zhou X, Li R, Wang W, Hua D and Chen W 2014 A functional variant at miR-132-3p, miR-212-3p, and miR-361-5p binding site in CD80 gene alters susceptibility to gastric cancer in a Chinese Han population. Med. Oncol. 31 60
Xie C, Chen B, Wu B, Guo J and Cao Y 2018 LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed. Pharmacother. 97 1645–1653
Yang B, Zhou ZH, Chen L, Cui X, Hou JY, Fan KJ, Han SH, Li P, Yi SQ and Liu Y 2018 Prognostic significance of NFIA and NFIB in esophageal squamous carcinoma and esophagogastric junction adenocarcinoma. Cancer. Med. 7 1756–1765
Zhao J, Qiao CR, Ding Z, Sheng YL, Li XN, Yang Y, Zhu DY, Zhang CY, Liu DL, Wu K and Zhao S 2017 A novel pathway in NSCLC cells: miR191, targeting NFIA, is induced by chronic hypoxia, and promotes cell proliferation and migration. Mol. Med. Rep. 15 1319–1325
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Rita Mulherkar
Rights and permissions
About this article
Cite this article
Wu, X., Chen, H., Zhang, G. et al. MiR-212-3p inhibits cell proliferation and promotes apoptosis by targeting nuclear factor IA in bladder cancer. J Biosci 44, 80 (2019). https://doi.org/10.1007/s12038-019-9903-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-019-9903-5