Abstract
Intracytoplasmic inclusions of protein aggregates in dopaminergic cells (Lewy bodies) are the pathological hallmark of Parkinson’s disease (PD). Ubiquitin (Ub), alpha (α)-synuclein, p62/sequestosome 1, and oxidized proteins are the major components of Lewy bodies. However, the mechanisms involved in the impairment of misfolded/oxidized protein degradation pathways in PD are still unclear. PD is linked to mitochondrial dysfunction and environmental pesticide exposure. In this work, we evaluated the effects of the pesticide paraquat (PQ) and the mitochondrial toxin 1-methyl-4-phenylpyridinium (MPP+) on Ub-dependent protein degradation pathways. No increase in the accumulation of Ub-bound proteins or aggregates was observed in dopaminergic cells (SK-N-SH) treated with PQ or MPP+, or in mice chronically exposed to PQ. PQ decreased Ub protein content, but not its mRNA transcription. Protein synthesis inhibition with cycloheximide depleted Ub levels and potentiated PQ-induced cell death. The inhibition of proteasomal activity by PQ was found to be a late event in cell death progression and had neither effect on the toxicity of either MPP+ or PQ, nor on the accumulation of oxidized sulfenylated, sulfonylated (DJ-1/PARK7 and peroxiredoxins), and carbonylated proteins induced by PQ. PQ- and MPP+-induced Ub protein depletion prompted the dimerization/inactivation of the Ub-binding protein p62 that regulates the clearance of ubiquitinated proteins by autophagy. We confirmed that PQ and MPP+ impaired autophagy flux and that the blockage of autophagy by the overexpression of a dominant-negative form of the autophagy protein 5 (dnAtg5) stimulated their toxicity, but there was no additional effect upon inhibition of the proteasome. PQ induced an increase in the accumulation of α-synuclein in dopaminergic cells and membrane-associated foci in yeast cells. Our results demonstrate that the inhibition of protein ubiquitination by PQ and MPP+ is involved in the dysfunction of Ub-dependent protein degradation pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteins are continually at risk of damage, misfolding, and aggregation. If not properly degraded, misfolded protein aggregates can cause cellular toxicity and disease. Cells have evolved an elaborated network of protein quality control mechanisms to maintain the integrity of the proteome. Protein homeostasis (proteostasis) involves specific processes that guard protein synthesis, folding, and trafficking. In addition, protein degradation pathways such as the ubiquitin (Ub)-proteasome system (UPS) and autophagy degrade misfolded or aggregated proteins to avoid proteotoxic stress [1]. A dysfunction in protein quality control mechanisms is a hallmark in neurodegenerative diseases [1, 2]. Lewy bodies (LBs) found in Parkinson’s disease (PD) brains are composed of misfolded protein aggregates. A number of proteins have been identified as major components of LBs including α-synuclein, Ub, and p62 [3–5]. Inhibition of proteasomal activity has been proposed to lead to the accumulation of Ub-bound proteins including α-synuclein [2]. Interestingly, other reports have demonstrated the presence of Ub-negative protein inclusions in PD brains [6, 7], suggesting that different mechanisms other than impaired proteasomal activity can be involved in the accumulation of misfolded protein aggregates.
A disruption in autophagic pathways has also been linked to PD pathogenesis [1, 2, 8, 9]. It is now well established that protein ubiquitination (or ubiquitylation) directs the recognition of selective cargo for degradation via the autophagosome-lysosome system [10, 11]. Conditional disruption of autophagy in dopaminergic cells leads to the accumulation of ubiquitinated protein aggregates in vivo [12, 13]. Recognition of ubiquitinated proteins for their degradation by autophagy is mediated by the adapter protein p62/sequestosome 1 (SQSTM1), and the neighbor of BRCA1 gene 1 (NBR1). p62 binds ubiquitinated proteins via its Ub-associated (UBA) C-terminal domain, while its binding to autophagosomal LC3/GABARAP proteins involves a short linear sequence known as LIR (LC3-interacting region) [11, 14]. Interestingly, p62 also mediates the autophagic clearance of non-ubiquitinated proteins [15, 16], and it may mediate the degradation of some polyubiquitinated proteins by the proteasome [17, 18].
A large variety of oxidative protein modifications can be induced by reactive oxygen/nitrogen species, or by-products of oxidative stress. Oxidized proteins can form oligomeric complexes resulting in the formation of protein aggregates. Irreversibly oxidized proteins such as protein carbonyls have to be degraded in order to maintain proper cellular homeostasis. Degradation of oxidized proteins by the 26S or 20S proteasome in a Ub-dependent and independent manner has been reported. However, covalent crosslinks, disulfide bonds, hydrophobic interactions, and heavily oxidized stable protein aggregates are not suitable for proteasomal degradation. Recent evidence suggests that autophagy plays a major role in the removal of oxidized protein aggregates by their incomplete degradation within the lysosomal compartment that results in the formation of polymerized lipofuscin-like aggregates consisting of oxidized polypeptides [19, 20]. Interestingly, p62 silencing enhances the accumulation of oxidized proteins [21], supporting a role for protein ubiquitination in the clearance of oxidized proteins by autophagy [22].
Mitochondrial dysfunction and oxidative stress are causative factors for dopaminergic cell loss in PD. Sporadic (non-hereditary) PD accounts for >80 % of reported cases, while genetic mutations only account for 5 % of sporadic PD occurrence [23]. Exposures to environmental toxicants, including pesticides (paraquat (PQ) and rotenone), are recognized as risk factors for an increased susceptibility to develop PD [24–29]. Thus, mitochondrial toxins such as inhibitors of complex I (1-methyl-4-phenylpyridinium (MPP+) and rotenone) and pesticides (PQ and rotenone as well) are used as toxicological models to dissect the molecular mechanisms by which mitochondrial dysfunction and oxidative stress mediate dopaminergic cell death. It has been reported that PQ and MPP+ induce the accumulation of Ub-bound protein aggregates by impairment of the proteasomal activity [30–32]. We and the others have reported that the impairment of autophagy facilitates dopaminergic cell death induced by PQ and MPP+ [33, 34]. Both autophagy and the UPS are complementary protein degradation pathways where inhibition of the UPS triggers the clearance of Ub-bound proteins or aggregates by autophagy [1, 2, 35, 36]. However, their exact and complementary contribution to dopaminergic cell death and the clearance of misfolded/oxidized protein aggregates induced by environmental/mitochondrial toxins has not been clarified.
In this work, we demonstrate that the environmental toxicant PQ and the mitochondrial complex I inhibitor MPP+ decrease protein ubiquitination in dopaminergic cells. Inhibition of the proteasome activity was found a late stage during cell death progression and did not modulate the toxicity of either PQ or MPP+. Depletion of Ub was shown to parallel p62 dimerization/inactivation, and the accumulation of oxidized proteins and α-synuclein. Inhibition of autophagy stimulated PQ and MPP+ toxicity. Our results demonstrate that early impairment in Ub protein synthesis by environmental and mitochondrial insults inactivates p62 and Ub-dependent degradation pathways.
Materials and Methods
Reagents
Chloroquine (CQ), 6-hydroxydopamine (6-OHDA), 1-methyl-4-phenylpyridinium iodide (MPP+), and rotenone were obtained from Sigma-Aldrich. 6-OHDA was prepared as described previously [37]. PQ (1,1′-dimethyl-4,4′-bipyridinium dichloride) and cycloheximide (CHX) were purchased from Acros Organics. (S)-MG132 (carbobenzoxy-l-leucyl-l-leucyl-l-leucinal, Z-LLL-CHO) was obtained from Cayman Chemical. Pyr-41 (4-[4-[(5-nitro-2-furanyl)methylene]-3,5-dioxo-1-pyrazolidinyl]benzoic acid ethyl ester) was purchased from Tocris. Stock solutions for MG132, Pyr-41, and rotenone were prepared in DMSO (vehicle). All other chemicals were from Sigma-Aldrich, Thermo Fisher Scientific, or Acros Organics.
Cell Culture and Treatments
The dopaminergic properties of the neuroblastoma cell line SK-N-SH and their cell culture have been detailed before [33]. The Lund human mesencephalic (LUHMES) neuronal precursor cell line, a subclone of the tetracycline-controlled v-myc-overexpressing human mesencephalic-derived cell line MESC2.10, was purchased from the American Type Culture Collection (ATCC, Biosource Center). Culture of LUHMES cells was done according to Scholz et al. [38]. Briefly, culture ware was pre-coated with 50 μg/ml poly-L-ornithine (Sigma-Aldrich) and 1 μg/ml fibronectin (BD Biosciences). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 nutrient mixture (GIBCO or Hyclone) supplemented with Neuroplex N-2 (Gemini or Life Technologies), 2 M l-glutamine (GIBCO or Hyclone), and 40 ng/ml recombinant basic fibroblast growth factor (bFGF, Peprotech or StemRD). Cells were grown at 37 °C in a 5 % CO2 humidified atmosphere and were treated as indicated in the figures. Control conditions included the appropriate vehicle, which never exceed >0.01 % (v/v).
Protein Extraction and Western Immunoblot
Protein extraction and quantification were done as explained before [39]. For the detection of sulfenic acid-modified proteins (PSOH), the cells were incubated with the cell-permeable nucleophilic reagent dimedone prior to harvesting. Dimedone selectively reacts with the electrophilic sulfur atom in sulfenic acid to form a stable thioether that can be detected using the anti-sulfenic acid-modified 2-thiodimedone-specific antibody [40]. The protein carbonyl content was determined by the reaction of carbonyl groups in protein side chains with 2,4-dinitrophenylhydrazine (DNPH, Sigma-Aldrich) to form 2,4-dinitrophenylhydrazone (DNP), which is detected using anti-DNP antibodies (Sigma-Aldrich). The cells were lysed in the presence of 1 mM DTPA (Sigma-Aldrich). Ten micrograms of protein were dissolved with a final concentration of 6 % sodium dodecyl sulfate (SDS) w/v and were derivatized by the addition of 20 mM DNPH in 10 % trifluoroacetic acid (TFA, v/v) [41, 42]. After derivatization, the samples were neutralized with 2 M Tris, 30 % glycerol (v/v) and 10.2 % β-mercaptoethanol (v/v) to obtain a final concentration of 2.8 % (v/v).
Polyacrylamide gel electrophoresis (PAGE) was performed using Bis-Tris (with 3-(N-morpholino) propansulfonic acid (MOPS) + 5 mM sodium bisulfite-based running buffer), or Tris-glycine gels. Proteins were transferred to nitrocellulose (GE Healthcare Life Sciences) or PVDF membranes (Maine Manufacturing). Blots were blocked and incubated with the corresponding primary antibodies as recommended by the manufacturers: anti-Ub P4D1 (Cat no. 3936) and α-synuclein (Cat no. 2642, carboxy-terminal sequence) were from Cell Signaling; anti-green fluorescent protein (GFP, Cat no. 1020) was from Aves Labs; anti-SQSTM1/p62 (Cat no. Ab109012), anti-sulfonylated peroxiredoxins (Prx-SO3H, Cat no. ab16830), and anti-sulfonylated DJ-1 (DJ-1-SO3H, Cat no. ab169520) were from Abcam; anti-PSOH-modified 2-thiodimedone-specific antibody (Cat no. ABS30) was from Millipore; and anti-DNP (Cat no. D9656) and anti-microtubule-associated protein 1B-light chain (3LC3B, Cat no. L7543) were from Sigma-Aldrich. The blots were probed with β-actin (Cat no. A2228, Sigma-Aldrich) to verify equal protein loading. Peroxidase-conjugated secondary anti-rabbit, anti-mouse, or anti-chicken antibodies (Thermo Scientific or Cell Signaling Technology) were used, and bands were detected using enhanced chemiluminescence (ECL) Western blotting substrate (Thermo Fisher Scientific/Pierce or Amersham/GE Healthcare Life Sciences) in a C‑DiGit chemiluminescence Western blot scanner (LI‑COR Biosciences) or a VersaDoc Gel Imaging System (Bio-Rad).
Analysis of high molecular weight aggregates (HMW) of α-synuclein from soluble and insoluble fractions (SDS-PAGE) and native α-synuclein conformation by Blue Native-polyacrylamide gel electrophoresis (BN-PAGE) from Triton (X-100) insoluble fractions were performed as explained before [43].
Filter Trap Assay for Ubiquitinated Protein Aggregates
Cells were harvested as explained before, and proteins were denatured in lithium dodecyl sulfate (LDS)-sample buffer. Ten micrograms to 50 μg of protein was filtered in a nitrocellulose membrane previously equilibrated in transfer buffer using a dot blotter (Scie-Plas). Membranes were washed twice with 2 % SDS (w/v); 10 mM Tris-EDTA, pH 7.5 buffer, and Ub-bound protein aggregates were detected by immunoblotting [44].
Cell Death Determination (Loss of Plasma Membrane Integrity) and Oxidative Stress
Loss of cell viability was determined using flow cytometry by measuring propidium iodide uptake (PI, 1 μg/ml) (Life Technologies or Sigma-Aldrich) as a marker of plasma membrane integrity loss. Flow cytometry was performed as explained before [33, 37, 45].
Evaluation of Ubiquitin-Dependent Protein Degradation
The plasmid encoding the Ub‐dependent (GFPμ) fluorescence reporter was kindly provided by Dr. Ron Kopito (Stanford University) [46]. GFPμ consists of the fusion of a 16-amino acid CL1 degron (a degradation signal identified in yeast) with the carboxyl terminus of GFP. CL1 targets GFP for ubiquitination, aggregation, and degradation by the proteasome [46]. The GFPμ plasmid was linearized with Nde I and transfected into SK-N-SH cells using FuGENE HD reagent (Promega). Cells stably overexpressing GFPμ were selected in medium containing 0.3 mg/ml geneticin (G418, Acros Organics), and GFPμ positive cells were sorted in a FACSAria cell sorter (BD Biosciences). After treatment, cells were harvested in phosphate-buffered saline (PBS) and analyzed by flow cytometry. GFPμ was excited with a 488-nm laser, and the emission was detected through a 530/30 emission filter in a FACSort (BD Biosciences / Cytek-DxP-10 upgrade) flow cytometer. The geometric mean of GFPμ fluorescence intensity was assessed in viable cells (PI negative (−)).
Determination of Proteasomal Activity in Total Cell Lysates
Total cell lysates were prepared on ice by homogenization in radioimmunoprecipitation assay buffer (RIPA, 50 mM Tris-HCl pH 8.0, 150 mM NaCl2, 1.0 % Igepal (NP-40, v/v), 0.5 % sodium deoxycholate (w/v), 0.1 % SDS (w/v)). Lysates were cleared by centrifugation and protein content was determined by the bicinchoninic acid assay (BCA) method. The chymotrypsin-like activity was measured using the proteasome activity assay kit (Abcam 107921) that utilizes a 7-amino-4-methylcoumarin (AMC)-tagged peptide substrate (Suc-LLVY-AMC), which, upon cleavage, releases free fluorescent AMC. Proteasomal-mediated AMC release was measured kinetically for 120 min using a microplate reader (Tecan, excitation/emission of 350/440 nm) in the presence or absence of MG132. Results were expressed as U/ml/mg of protein, where 1 unit (U) of proteasome activity is defined as the amount of proteasome that generates 1.0 nmol of AMC per minute (nmol/min).
Evaluation of Cellular Proteasomal Activity
After treatment, cells were incubated with the boron-dipyrromethene (Bodipy)-tagged cell-permeable proteasome activity probe BodipyFL-Ahx3L3VS (200 nM) for 2 h, which was synthesized as explained before [47, 48]. Cells were harvested in PBS. BodipyFL-Ahx3L3VS was excited with a 488-nm laser, and the emission was detected through a 530/30 emission filter in a BDFACSort (Cytek-DxP-10 upgrade). The geometric mean of BodipyFL-Ahx3L3VS fluorescence intensity was assessed to evaluate changes in fluorescence that directly relate to proteasome activity.
Evaluation of Ubiquitin B mRNA Levels
RNA was extracted with Trizol (Life Technologies) following the manufacturer’s instructions and quantified in a Nanodrop 2000 (Thermo Fisher Scientific). cDNA strands were synthesized using 5 μg of RNA and Moloney murine leukemia virus reverse transcriptase M-MLV (200 U/μl, Life Technologies), Oligo(dT) primer (0.5 μg/μl, Life Technologies), and dNTPs (2.5 mM each, Applied Biosystems) during 1 cycle of amplification under the following conditions: (1) 65 °C/5 min, (2) 37 °C/50 min, and (3) 70 °C/15 min (Applied Biosystems 2720 Thermal Cycler). The ubiquitin B (UBB) mRNA expression levels were determined by real-time PCR (RT-PCR) using 100 ng of cDNA as template, TaqMan Universal PCR Master Mix (Applied Biosystems), and specific TaqMan probes for the human polyubiquitin gene UBB (Hs00430290_m1 FAM, Applied Biosystems). The probe used to detect UBB mRNA amplifies six different variants (NM_018955 and NM_001281716-9). RT-PCR was performed in an ABI Prism 7500 (Applied Biosystems) under the following conditions: step (1) 50 °C/2 min, step (2) 95 °C/10 min, and step (3) 40 cycles of 95 °C/15 s, followed by 60 °C/1 min. Data were normalized to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs02758991_g1 VIC) as endogenous housekeeping gene by the relative standard curve method (http://www.uic.edu/depts/rrc/cgf/realtime/stdcurve), and the results were expressed as relative expression levels with respect to the control group.
Recombinant Adenoviral Vectors
The replication-deficient recombinant adenovirus (Ad5CMV) encoding a dominant-negative form (dn) of the autophagy protein 5 (ATG5) was kindly provided by Dr. Gökhan S. Hotamisligil (Harvard School of Public Health, Boston, MA) [49]. Adenoviruses encoding wild-type (WT) or mutant A53T α-synuclein were provided by Dr. Jean-Christophe Rochet (Purdue University) and have been described elsewhere [43, 50]. Adenovirus containing only the CMV promoter (Ad-Empty) was used as a negative control. Viruses were amplified and tittered in HEK293T cells as previously described [39, 51]. The cells were infected with viral particles at the indicated multiplicity of infection (MOI), and 24 h post-infection, they were washed and treated under the specified experimental conditions.
In Vivo Mouse Model of Paraquat Toxicity
C57BL/6 mice (8–10 weeks old) (Jackson Labs) were administered an intraperitoneal injection of 10 mg/kg PQ or PBS twice per week for 9 consecutive weeks [52]. Animals were analyzed 1 week after the last injection. Mice were decapitated and the midbrains were removed for Western immunoblot (WB) analysis. For immunohistochemistry, mice were perfused intracardially with 4 % paraformaldehyde (PFA) in 0.1 M sodium phosphate buffer (pH 7.4). The brains were removed, post-fixed for 24 h in 4 % PFA, and cryoprotected with 30 % sucrose. Frozen brains were cut into 30-μm coronal sections using a H/I sliding microtome (Hacker Instruments & Industries Inc.) at −16 °C and stored in PBS at 4 °C until the immunohistochemical analysis. Endogenous peroxidase activity was inactivated. Sections were blocked with normal horse serum (Life Technologies) and incubated 48 h with anti-tyrosine hydroxylase antibody (TH, Calbiochem, EMD/Millipore Cat no. AB1542) or anti-Ub at 4 °C. After rinsing, sections were incubated in secondary Alexa 647-anti-mouse or Alexa 568-anti-sheep (Jackson ImmunoResearch) for 1 h at RT. Sections were mounted with Fluoro-Gel (Electron Microscopy Sciences) containing 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei. Images were collected on an Olympus IX 81 inverted confocal scanning fluorescent microscope (×10 or ×60 oil lens) (Olympus America) using Fluoview 500 Software. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC ) of the University of Nebraska-Lincoln (Project 1025).
Yeast Experiments
Saccharomyces cerevisiae W303-1A strain (MATa can1-100 ade2-1his3-1,15 leu2-3,112 trp1-1 ura3-1) harboring chromosomally integrated human α-synuclein-GFP expression cassette under the control of the inducible GAL1 promoter was generated and handled as previously described [43]. For confocal microscopy, live cells were visualized with ×100 oil lens. For survival assays, aliquots of the yeast culture were diluted to 300 cells and plated onto YP (yeast peptone) plates containing 2 % glucose (YPD, w/v) or 2 % galactose (YPGal, w/v) as the sole carbon source. The colony-forming units or degenerative colonies were scored following 2 (YPD plates) or 4 (YPGal plates) days of incubation at 28 °C.
Statistical Analysis
Experimental replicas were independent and performed on separate days. Collected data were analyzed by using one-way, two-way or three-way ANOVA, and the appropriate post hoc test using SigmaPlot/Stat package. When ANOVA assumptions were not met (normality (Shapiro–Wilk test) or equal variance), Kruskal-Wallis one-way ANOVA on ranks or data transformation (two-way ANOVA) was performed on the collected data. Data were plotted as mean ± standard error (SE) using the same package for statistical analysis. Flow cytometry plots and immunoblots presented show the results of representative experiments. Relative densitometry analysis of WBs and dot blots was made using the ImageJ Program (National Institutes of Health, http://rsb.info.nih.gov/ij).
Results
Effect of PD-Related Toxicants on the Accumulation of Ub-Bound Proteins
PD is linked to mitochondrial dysfunction and environmental pesticides exposure [53, 54]. Previous reports have demonstrated that PD-related toxicants impair the activity of the proteasome leading to the accumulation of Ub-bound protein aggregates [30–32, 55, 56]. We found that exposure of dopaminergic neuroblastoma cells (SK-N-SH) to PQ induces a dose-dependent decrease in ubiquitinated protein levels (Suppl. Fig. 1a). Lower non-toxic concentrations of PQ (≤0.2 mM) induced no changes in the levels of ubiquitin-bound proteins (Suppl. Fig. 1a). Higher toxic concentrations of PQ (≥0.5–1 mM) induced a slight decrease in ubiquitinated proteins (Suppl. Fig. 1a). Similarly, PQ induced a dose-dependent decrease in the accumulation of Ub-bound proteins in the mesencephalic neuronal precursor cell line LUHMES (Suppl. Fig. 1a), whose sensitivity to PQ is significantly higher (~100 to 200 μM, not shown). These results demonstrate that the decrease in ubiquitinated protein levels induced by PQ is not cell type specific.
A decrease in ubiquitinated proteins can be ascribed to different phenomena including an enhanced proteasomal activity, a decrease in protein ubiquitination, and/or a reduced availability of free Ub monomers/chains. To test this possibility, cells were treated with PQ and incubated with the cell-permeable proteasome inhibitor MG132 (0.2 μM) 24 h prior to analysis (see Fig. 1a upper box). As previously reported [43], at this concentration and time of incubation, MG132 efficiently inhibits the proteasome activity leading to the accumulation of Ub-bound proteins without triggering significant cell death by itself (Suppl. Fig. 1b and Fig. 2a, b). PQ induced a dose-dependent decrease in the accumulation of Ub-bound proteins in the presence of MG132 (Fig. 1a, quantified in Fig. 1 (with respect to control) and in Suppl. Fig. 1c (with respect to each treatment in the absence of MG132)), suggesting that the decreased accumulation of ubiquitinated proteins is ascribed to a reduction in protein ubiquitination. Similarly, treatment with the mitochondrial toxin MPP+, but not rotenone or 6-OHDA, also reduced protein ubiquitination (Fig. 1c).
PQ and MPP+ impair protein ubiquitination. a Ub-bound proteins were evaluated by WB in SK-N-SH cells treated with PQ for 48 h. Cells were treated with the proteasome inhibitor MG132 (0.2 μM) 24 h prior to analysis. b Relative quantification (densitometry) of Ub-bound proteins in the presence of MG132 (●). Cells were treated as explained before (a). Data was normalized to β-actin and represented as fold change with respect to control (no PQ treatment). Cell death (○) was determined by the loss of plasma membrane integrity (PI uptake) and represented as percentage of cells with high PI fluorescence. Data are means ± SE of at least n = 3. One-way ANOVA, Holm-Sidak post hoc test a, p < 0.05; Kruskal-Wallis one-way ANOVA on ranks, Dunn’s post hoc test b, p < 0.05 compared to the corresponding control (no drug treatment). c Changes in the levels of Ub-bound proteins were determined by WB in cells treated for 48 h with the complex I inhibitors MPP+ (2.5 mM) and rotenone (Rot, 4 μM) and the hydroxylated dopamine analog 6-OHDA (6-OH, 50 μM). d Cells were treated as explained above (a and c), and ubiquitinated proteins and aggregates were evaluated by dot blot. Relative quantification of ubiquitinated proteins (numbers in italics) was normalized to β-actin and represented with respect to the indicated control. e Immunohistochemistry detection of ubiquitinated proteins in the substantia nigra of C57Bl/6 mice treated for 9 weeks with PBS (e1–e5) or PQ (e6–e10). Blue squares in e1 or e6 depict the area of magnification for e2–e5 (PBS) or e7–e10 (PQ) panels. Scale bars, e1 and e6 200 μm; e2–e5 and e7–e10: 20 μm. TH+ tyrosine hydroxylase-positive neurons
PQ induces a transient accumulation of GFPμ, a reporter for the UPS. In a and c, the simultaneous analysis of cell death and changes in the levels of GFPμ induced by MG132 (24 h) or PQ treatment (48 h) was done by flow cytometry. Data are represented in two-dimensional 5 % probability contour plots of changes in PI uptake (y axis) vs changes in GFPμ fluorescence (x axis). Broken squares depict how viable cells were selected. Percentages in contour plots represent the number of cells per quadrant. Using this type of analysis (gating), the changes in GFPμ induced by MG132 or PQ were quantified in b, d, and e. The geometric mean of GFPμ fluorescence intensity (●) was assessed in viable cells (PI negative (–)) and represented as fold change with respect to control. Cell death (○) was represented as percentage of cells with high PI fluorescence. Data in graphs are means ± SE of at least n = 3. Kruskal-Wallis one-way ANOVA on ranks, Student-Newman-Keuls (SNK) post hoc test a, p < 0.05 compared to the corresponding control (no drug treatment)
A dysfunction in the UPS has been shown to lead to the formation of SDS-resistant aggregates (aggresomes) [57, 58]. Protein complexes and aggregates are not well resolved by WB due to their high molecular weight (MW). Thus, we evaluated the changes in both ubiquitinated proteins and aggregates by filter trap (retardation) dot blot assay. Figure 1d corroborates that both PQ and MPP+ induce a decrease in protein ubiquitination, evaluated in the presence of MG132. Finally, to evaluate the effect of PQ exposure on the levels of Ub-bound proteins in dopaminergic cells in vivo, C57Bl/6 mice were exposed chronically to PQ (9 weeks). Immuno histochemistry analysis of TH+ neurons and Ub shows that no major increase in Ub staining was induced by PQ in dopaminergic (TH+) cells (Fig. 1e).
We have previously demonstrated that cell death induced by PQ is a stochastic process [33, 39, 45], which means that it occurs at different rates within the same cell population. As such, WB analysis does not allow us to determine if the changes in Ub-bound protein levels occur before or after cell death (i.e., samples are composed by a mixed population of cells at different stages during the cell death process). Thus, we next evaluated the accumulation of GFPμ, a substrate for the UPS, in live cells (as depicted in broken square regions in Fig. 2a, c). Treatment with MG132 induces the accumulation of GFPμ (Fig. 2a, b). Lower non-toxic concentrations of PQ (≤0.2 mM) induced a significant accumulation of GFPμ (Fig. 2c, d), which agrees with the overall decrease in protein ubiquitination previously seen (Fig. 1a, b, PQ + MG132 data). Surprisingly, while higher toxic concentrations of PQ decreased protein ubiquitination further (Fig. 1a, b), this was not translated in added accumulation of GFPμ (Fig. 2c, d). A time-course analysis of changes in GFPμ levels induced by a toxic concentration of PQ (0.5 mM, 48 h, Suppl. Fig. 2a), or a sub-toxic PQ concentration (0.1 mM, 96 h, Fig. 2e), also evidenced an early accumulation of GFPμ followed by a reduction in its levels in live cells irrespective to the dose of PQ used or the length of exposure. Similarly, MPP+, which also impairs protein ubiquitination (Fig. 1c), also induced a reduction in GFPμ content (Suppl. Fig. 2b-c). Polyubiquitin-tagged proteins have a half-life (t 1/2) of ≤30 min [59]. The GFPμ reporter has a reported short-term t 1/2 = 30 min [60, 61], while GFP itself has a t 1/2 = 24 h [62]. Thus, our results suggest that while the transient accumulation of UPS substrates (GFPμ) induced by PQ is linked to an impairment of protein-ubiquitination (Fig. 2d, e), the subsequent Ub-independent decrease in GFPμ induced by both PQ and MPP+ is likely linked to impaired protein synthesis.
Impairment of Proteasomal Activity by PQ Is a Late Step in the Cell Death Process
Previous studies have demonstrated that PD-related toxicants including PQ and MPP+ impair proteasomal activity [30, 31, 56, 63, 64]. We found that the activity of the proteasome decreased in lysates from cells treated with toxic PQ concentrations (≥0.5 mM) (Fig. 3a). However, due to the stochastic nature of cell death progression, this assay does not allow us to evaluate if changes in the activity of the proteasome occur prior to cell death or if they are only an epiphenomenon associated with the loss of cellular viability. Thus, we evaluated the changes in the activity of the proteasome in intact cells using the cell-permeable proteasome activity probe BodipyFL-Ahx3L3VS [47, 48]. MG132 inhibits the processing of BodipyFL-Ahx3L3VS by the proteasome (Fig. 3c and Suppl. Fig. 3a). PQ (Fig. 3b, c) and MPP+ (Suppl. Fig. 3b) induced a dose-dependent increase in proteasome activity. Accordingly, previous studies have reported that PQ and MPP+ induce an early increase in proteasomal activity [65–67]. Thus, our results suggest that the impairment in proteasomal activity induced by PQ and other PD-related insults is a late event associated with the loss of cell viability. Accordingly, MG132 had no effect on PQ-, MPP+-, rotenone-, or 6-OHDA-induced toxicity (Fig. 3d).
Effects of PQ and MPP+ on the activity of the proteasome. Cells were treated with the indicated toxicants for 48 h. a The proteasomal 20S chymotripsyn-like activity was evaluated in total cell lysates by evaluation of the changes in AMC fluorescence released from the hydrolysis of Suc-LLVY-AMC. The proteasome inhibitor MG132 was used as a positive control (5 μM for 6 h). Data are expressed as (U/ml/mg) protein. b Changes in the activity of the proteasome were evaluated in live cells using the cell-permeable proteasome substrate BodipyFL-Ahx3L3VS. Histograms represent the distribution of cells with different levels of BodipyFL-Ahx3L3VS fluorescence. c The geometric mean of BodipyFL-Ahx3L3VS fluorescence intensity was quantified and represented as fold change with respect to control. MG132 was used as a positive control (0.2 μM for 16 h prior to the incubation with BodipyFL-Ahx3L3VS). Background fluorescence (Control- in (b)) was subtracted. d Cell death was determined in the presence or absence of MG132 (0.2 μM) by evaluating the loss of plasma membrane integrity (PI uptake) and represented as percentage of cells with high PI fluorescence. Data in graphs are means ± SE of n = 3–5. Kruskal Wallis one-way ANOVA on ranks, Student-Newman-Keuls post hoc test a, p < 0.05; Mann-Whitney rank sum t-test *p < 0.05 compared to the corresponding control (no drug treatment)
PQ Reduces Ub Protein Content But Not Ub mRNA Transcription
The decrease in protein ubiquitination induced by PQ and MPP+ might be mediated by impairment in the activity of Ub-activating (E1s) and Ub-conjugating enzymes (E2s), and/or Ub ligases (E3s). A decrease in the activity of these enzymes is translated in the accumulation of free Ub monomers/chains [68]. However, we found that treatment of cells with PQ induced a decrease in Ub monomers as well (Fig. 4a and Suppl. Fig. 3c), suggesting that a decrease in the Ub protein pool, rather than an impairment in the E1-E2-E3 system, is linked to the reduced levels of protein ubiquitination. Ub is encoded in mammals by four genes. UBA52 and RPS27A genes code for a single copy of Ub fused to ribosomal proteins, while the UBB and UBC genes code for poly-Ub precursor proteins. We evaluated if the reduction in Ub protein levels induced by PQ is mediated by a decrease in gene transcription. Surprisingly, a significant increase in UBB mRNA transcription/stability was observed at non-toxic concentrations of PQ (≤0.2 mM) (Fig. 4b). These results demonstrate that Ub protein synthesis or stability, but not Ub-gene transcription, is impaired by PQ.
PQ impairs Ub protein synthesis. a Changes in the levels of Ub monomers and free chains were evaluated by WB (18 % Tris-glycine gels) in lysates from SK-N-SH cells treated with PQ for 48 h. Cells were treated with the proteasome inhibitor MG132 (0.2 μM) 24 h prior to analysis. Relative quantification of ubiquitin monomers (numbers in italics) was normalized to β-actin and represented with respect to control. b RT-PCR analysis of the changes in polyubiquitin gene UBB mRNA levels in cells treated with PQ for 48 h. Data were normalized using GAPDH as endogenous housekeeping gene. Results are expressed as changes in the relative expression levels compared to the control (no PQ treatment). c Ub-bound proteins were evaluated by WB in cells treated with CHX (μM) for 48 h in the presence or absence of the proteasome inhibitor MG132 (as explained in (a)). d Cell death was determined in cells treated with PQ, in the presence or absence of CHX (100 μM), by evaluating the loss of plasma membrane integrity (PI uptake) and represented as percentage of cells with high PI fluorescence. Data in graphs are means ± SE of n = 3–5. One-way ANOVA, Holm-Sidak post hoc test a, p < 0.05 compared to the corresponding control (no drug treatment). Two-way ANOVA, Holm-Sidak post hoc test b, p < 0.05 compared to the corresponding PQ concentration without CHX treatment
We next evaluated the role of Ub protein synthesis inhibition/depletion in PQ toxicity. Because Ub is encoded by four genes, its knockdown is experimentally cumbersome as reported in previous studies demonstrating that while UBB knockdown reduces Ub monomers by 70 %, it only decreases Ub-bound proteins by 30 % [69]. A previous study demonstrated that the protein synthesis inhibitor CHX depletes cellular Ub resulting in a decrease in steady-state polyubiquitinated proteins [70]. Depletion of Ub-bound proteins was induced by CHX treatment (48 h, Fig. 4c), which only resulted in a slight increase in cell death (Fig. 4d). CHX significantly stimulated PQ toxicity (Fig. 4d), suggesting that Ub depletion, but not inactivation of the proteasome, contributes to PQ toxicity. Ub depletion by itself does not induce cell death, suggesting that additional events linked to PQ or MPP+ exposure (oxidative damage or mitochondrial dysfunction) in addition to impaired protein ubiquitination are required for cell death progression.
The Accumulation of Oxidized Proteins Induced by PQ Is Not Regulated by the Proteasome
Clearance of oxidized proteins has been shown to be mediated by both Ub-dependent and Ub-independent proteasomal degradation pathways [71, 72]. PQ induced a dose-dependent accumulation of sulfenylated protein cysteine (PSOH), irreversibly oxidized (sulfonylated PSO3H) DJ-1 and peroxiredoxins (Prxs), as well as protein carbonyls (Fig. 5a–c and Suppl. Fig. 4a-b). However, inhibition of the proteasome with MG132 did not increase further the accumulation of oxidized protein byproducts (Fig. 5a–c and Suppl. Fig. 4a-b), suggesting that the increased load in oxidized proteins is ascribed to impaired protein ubiquitination but not to a decrease in proteasomal activity. We have previously demonstrated that oxidative stress induced by MPP+ is primarily restricted to the mitochondria matrix [45]. Accordingly, no accumulation of oxidized DJ-1 or Prxs was observed upon exposure to MPP+(Suppl. Fig. 4c).
The accumulation of oxidized proteins induced by PQ is not modulated by inhibition of the proteasome. Cells were treated with PQ for 48 h in the presence or absence of 0.2 μM MG132 (as exemplified in Fig. 1a). a PSOHs were determined in cells incubated with the PSOH selective probe dimedone prior to the analysis. PSOHs were visualized using the anti-PSOH-modified cysteine 2-thiodimedone-specific antibody. b Levels of irreversibly oxidized DJ-1 and Prxs (PSO3H) were detected by WB. Relative quantification of oxidized proteins (numbers in italics) was normalized to β-actin and represented with respect to the indicated control. c Protein carbonyls were detected by WB in total cell lysates derivatized with DNPH. Carbonylated proteins were detected using anti-DNP antibody. Graphs indicate the densitometry analysis of changes in the levels of PSOHs or protein carbonyls normalized to β-actin and expressed as fold change with respect to control. Data are means ± SE of five independent experiments. Kruskal-Wallis one-way ANOVA on ranks, Student-Newman-Keuls (SNK) post hoc test a, p < 0.05 compared to the corresponding control (no drug treatment)
Dimerization of p62 Parallels the Impairment in Protein Ubiquitination and Autophagy Flux
Selective degradation of ubiquitinated protein aggregates is also mediated by autophagy via the Ub-binding receptor p62 [10]. p62 binds to ubiquitinated proteins via its UBA C-terminal domain. Interestingly, the UBA domain, which has a low affinity for Ub, also mediates the formation of highly stable symmetrically inactive dimers. p62 dimerization and Ub-binding are mutually exclusive [73]. Thus, we considered that Ub depletion induced by PQ might dimerize/inactivate p62. Inhibition of Ub-activating enzymes (E1s) with Pyr 41 induced p62 dimerization (Fig. 6a and Suppl. Fig. 5a) [74]. PQ and, to a lesser extent, MPP+ and 6-OHDA but not rotenone also induced a dose-dependent dimerization of p62 (Fig. 6b and Suppl. Fig. 5b-c). Overexpression of a dominant-negative form of ATG5 (dnATG5), which together with ATG12 and ATG16 are essential for autophagosome formation [75], enhanced p62 dimerization (Fig. 6b and Suppl. Fig. 5b). Overexpression of dnATG5 did not impair the decrease in ubiquitinated proteins/aggregates induced by PQ, (Fig. 6c) These results demonstrate that Ub protein depletion parallels p62 dimerization.
PQ treatment induces the dimerization/inactivation of p62 and impairs autophagy. a Cells were treated with the Ub-conjugating enzyme (E1) inhibitor Pyr-41 for 24 h. p62 dimerization (inactivation) was evaluated by WB. Dimerization of p62 is evidenced by an increase in p62 dimers with the concomitant decrease p62 monomers. b p62 dimerization was also evaluated in cells treated with PQ for 48 h. When indicated, cells were transduced with dnATG5 or Empty viruses (-dnATG5) 24 h before PQ treatment. Relative quantification of p62 dimerization was represented as p62 dimers/p62 monomers ratio, normalized to β-actin and expressed with respect to the indicated control (numbers in italics in (a) and (b)). c Cells were treated as explained above (b), and ubiquitinated proteins and aggregates were evaluated by dot blot. Relative densitometry quantification of ubiquitinated proteins was normalized to β-actin and represented with respect to the indicated control (numbers in italics). d Alterations in autophagy flux induced by PQ were determined by changes in the levels of the autophagosome marker LC3-II in the presence of CQ (40 μM, incubated 4 h prior to analysis), an inhibitor of lysosomal cargo degradation. Relative densitometry quantification of LC3-II (numbers in italics) was normalized to β-actin and represented with respect to the indicated control. e Cell death was determined in cells treated with PQ, in the presence or absence of MG132 (0.2 μM). When indicated, cells were transduced with dnATG5 or Empty (-dnATG5) viruses 24 h before PQ treatment. Cell death was evaluated by the loss of plasma membrane integrity (PI uptake) and represented as percentage of cells with high PI fluorescence. Data are means ± SE of n = 3–5. Two-way ANOVA, Holm-Sidak post hoc test analysis of data without MG132 a, p < 0.05 dnATG5 vs Empty within the corresponding PQ category
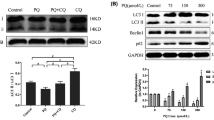
The accumulation of autophagosomes is evidenced by an increase in the levels of the microtubule-associated protein light chain (LC3–I) protein in its lipidated form (LC3–II). As previously reported [36, 76, 77], inhibition of the proteasome with MG132 induces autophagy (Suppl. Fig. 5d). We and the others have previously demonstrated that PQ and the complex I inhibitors MPP+ and rotenone impair autophagy flux [33, 34, 78], defined as the complete process beginning with the formation of the phagophore and ending after the fusion of the autophagosomes with the lysosome for the degradation of lysosomal cargo [9, 33]. Autophagy flux was inferred by WB analysis of LC3–II turnover in the presence of cloroquine (CQ), the inhibitor of lysosomal cargo degradation that specifically blocks the acid-dependent breakdown of autolysosome content without affecting autophagosome–lysosome fusion, resulting in the accumulation of autophagolysosomes that cannot be cleared [33]. Figure 6d and Suppl. Fig. 5e-f corroborate that PQ induces a dose- and time-dependent impairment in autophagy flux. We have previously demonstrated that overexpression of dnATG5 inhibits autophagy and potentiates PQ and MPP+ toxicity [33]. Protein degradation mechanisms are complementary, and dysregulation of either the UPS or autophagy has been reported to be mutually compensated, particularly in the clearance of aggregated proteins linked to neurodegenerative disorders [35, 36, 79–84]. While inhibition of autophagy with dnATG5 overexpression stimulated PQ toxicity, MG132 exerted no additional toxicity when combined with dnATG5 overexpression Fig. 6e. These results demonstrate that Ub protein depletion induced by PQ and MPP+ is linked to the inactivation of p62 that precedes the decrease in autophagy flux. Autophagy but not the proteasome regulates the progression of PQ- and MPP+-induced dopaminergic cell death.
Paraquat Increases the Pathological Accumulation of α-Synuclein in Dopaminergic Cells and Membrane-Associated Foci in Yeast
The clearance of misfolded/aggregated α-synuclein has been shown to be mediated by both autophagy and the ubiquitin/proteasome pathways [83–85]. Previous studies have reported that PQ upregulates the levels of α-synuclein [34, 86–89]. We observed no effect of PQ on the total endogenous levels of α-synuclein in SK-N-SH dopaminergic cells (data not shown) or in PQ treated C57Bl/6 (Fig. 7a). Thus, we evaluated if PQ could alter the pathological accumulation of α-synuclein when overexpressed (as a PD model of SNCA multiplication) or when mutated. Overexpression of α-synuclein (WT or A53T mutant) in SK-N-SH cells for 72 h, in the presence or absence of PQ, did not induce the accumulation of HMW aggregates of α-synuclein (SDS-PAGE analysis of whole cell lysates containing soluble and insoluble fractions), but it increased the accumulation α-synuclein in its monomeric form (Fig. 7b). Under native conditions, α-synuclein is reported to exist predominantly as stable unfolded monomers that migrate as 57–60 kDa proteins (Fig. 7c). It is unclear whether the larger than expected size of the bands is a result from the monomers adopting an unfolded extended conformation, which results in a larger than expected hydrodynamic radius [90–92], or if it represents a fraction of α-synuclein existing as a stable tetramer [93, 94]. We have previously demonstrated that the inhibition of the proteasome promotes the accumulation of α-synuclein in this unfolded state (or tetramer) and the appearance of a band with enhanced lower MW [43]. Similarly, PQ induced the accumulation of unlfoded α-synuclein (Fig. 7c) and the accumulation of a low MW band immunoreactive for α-synuclein (depicted with asterisk in Fig. 7c).
Effect of PQ on α-synuclein accumulation and distribution. a WB analysis of α-synuclein levels in the midbrain of C57Bl/6 mice treated for 9 weeks with PQ or PBS. b, c Cells were transduced with Ad-Empty, Ad-α-synuclein, or Ad-A53T for 24 h (3 MOI), washed, and then treated with or without PQ (48 h). Whole cell lysates or TX-100 insoluble fractions were analyzed by SDS (b) or BN-PAGE (c), respectively, and α-synuclein was visualized by WB. Numbers (italics) represent the densitometry analysis normalized to β-actin with respect to the corresponding control. d Wild-type S. cerevisiae cells containing genome-integrated human α-synuclein-GFP expression cassette under the control of GAL1 promoter were cultured in the medium containing 2 % glucose or 2 % galactose. The galactose-grown cells were then treated with the indicated amounts of PQ for 1 h (acute treatment) and 48 h (chronic treatment). Subcellular distribution of α-synuclein-GFP was visualized by confocal microscopy. Shown are representative images of cells that have been acutely treated with PQ. Scale bars are 5 μm. Arrows indicate membrane-associated foci of α-synuclein-GFP. Bar graphs show quantitation of α-synuclein-GFP foci. At least 300 cells per condition per sample were analyzed. e Yeast cultures described above were diluted to 300 cells and plated for survival on glucose- or galactose-containing plates with or without PQ. Following 4 days incubation at 28 °C, the plates were inspected for colony-forming units and the presence of small, degenerative colonies. The left panel shows representative images of cell growth on galactose plates with and without 1 mM PQ. Arrows indicate degenerative colonies. The panels on the right show quantitation of cell survival and percentage of degenerative colonies on glucose and galactose-containing plates containing the indicated amounts of PQ. Bar graphs are means ± SD (n = 4); *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired t-test
To further interrogate the effect of PQ on α-synuclein distribution, we used the budding yeast S. cerevisiae genetic model overexpressing an inducible promoter-driven fusion of α-synuclein-GFP. Yeast has been extensively used as a valid experimental platform to elucidate the fundamental mechanisms associated with neurodegenerative diseases [95]. The inducible expression of α-synuclein can result in no toxicity, intermediate toxicity, or high toxicity in relation to the levels of α-synuclein being expressed. α-Synuclein-GFP overexpressed in yeast at non-toxic levels localizes at the plasma membrane, consistent with its known affinity to phospholipids (Fig. 7d). This is the expected localization of a protein that localizes at synaptic vesicles in neurons, when considering that yeast has constitutive vesicular secretion [95]. Acute or chronic treatment of yeast cells with PQ induced the accumulation of membrane-associated foci (Fig. 7d). While α-synuclein overexpression had no effect in SK-N-SH (not shown) or yeast cell death induced by PQ (Fig. 7e), in yeast it induced the formation of degenerative colonies (Fig. 7e) (smaller in size and impeded in their ability to propagate normally). These results suggest that the accumulation of monomeric α-synuclein and its localization at the plasma membrane is regulated by PQ.
Discussion
The etiology of PD involves the convergence of aging, genetic, and environmental risk factors [53, 96]. A disruption in protein quality control mechanisms is linked to the accumulation of protein inclusions and neuronal cell loss observed in neurodegenerative disorders including PD [2, 97]. Ub selectively targets cargo to the three major protein degradation pathways, the proteasome, the lysosome, and the autophagosome [98]; and environmental/mitochondrial toxicants inhibit the activity of the proteasome and impair autophagy flux [33, 56]. However, the effects of environmental/mitochondrial toxicants on other components of Ub-dependent protein degradation pathways have not been studied in detail, and inhibition of the proteasome is still considered the major mechanism involved in the impairment of the Ub-dependent degradation of misfolded/damage proteins. In this work, we demonstrated that impaired protein ubiquitination is an early and central step in the impairment of protein degradation pathways induced by PQ and MPP+. The depletion of the Ub protein pool induced by both agents was observed at both sub-toxic and toxic (subchronic) exposures, and was not cell type specific. Furthermore, Ub protein depletion was paralleled by the inactivation of p62 and in the case of PQ, the accumulation of oxidized protein byproducts and alterations in the levels of monomeric α-synuclein and its distribution at the plasma membrane (Fig. 8). These results might explain the heterogeneity of protein inclusions in PD brains (LBs), particularly, the presence of Ub-negative protein inclusions [6, 7].
PQ- and MPP+-induced Ub protein depletion and p62 inactivation are early steps in the impairment of Ub-dependent protein degradation pathways (UPS and autophagy). Our data suggests that PD-related toxicants PQ and MPP+ deplete the Ub protein pool (red arrow) by a mechanism that might involve oxidative mRNA damage, energy failure, or altered Ub protein stability. Ub depletion leads to the inactivation (dimerization) of the ubiquitin binding receptor p62 that directs ubiquitinated cargo for degradation via the autophagosome-lysosome pathway. Ub protein depletion and p62 inactivation impair Ub-dependent protein degradation pathways and parallel the accumulation of oxidized/misfolded proteins (yellow arrows) and alterations in the levels/distribution of α-synuclein (not exemplified). Severe/chronic oxidative stress and/or energy failure induced by environmental and mitochondrial toxicants would eventually lead to a decrease in the activity of the proteasome and impaired autophagy flux (red crosses)
A decrease in the activity of the proteasome has been found in PD brains [99–102]. Previous studies have also demonstrated that environmental and mitochondrial toxins including PQ and MPP+ impair the activity of the proteasome by (1) direct inhibition of the proteasome, (2) mitochondrial dysfunction and energy depletion, or (3) oxidative stress [30, 31, 56, 63, 103]. Similar to previous reports [30, 31, 34, 104], we observed that high and toxic PQ concentrations induced a decrease in the chymotrypsin-like activity of the proteasome. However, biochemical assays of proteasomal activity are usually done in lysates from cell samples (or PD tissues), where the decrease in proteasome activity might be confounded by the loss of cells (viability). Using a novel cell-permeable proteasomal substrate (BodipyFL-Ahx3L3VS), we found that both sub-toxic and toxic concentrations of PQ and MPP+ increase the activity of the proteasome prior to cell death. Interestingly, previous studies have demonstrated that MPP+-induced dopaminergic cell death actually requires an increase in the activity of the proteasome [65, 66]. Our findings do not argue against previous studies demonstrating that impairment in proteasome activity is induced by PD-related toxicants [30, 31, 56, 63, 103]. However, our results suggest that inhibition of the proteasome by environmental/mitochondrial toxicants might be a late event only ascribed to the loss of cell viability. As such, inhibition of the proteasome had no effect on PQ- or MPP+-induced toxicity. Accordingly, previous in vivo studies using the MPP+ precursor 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) found no stimulatory effect of proteasome inhibition on dopaminergic cell death [105].
Previous reports have demonstrated that PD-related toxicants induce the accumulation of Ub-bound protein aggregates, which has been ascribed primarily to proteasome inhibition by these agents [30–32, 55, 56, 103]. In vivo, MPTP has been shown to induce both an increase and a decrease in the levels of Ub-bound proteins using different experimental paradigms [32, 106]. While we did observe a slight increase in the accumulation of the UPS fluorescent substrate GFPμ; this transient increase seems to precede the late decrease in proteasomal activity. Proteasomal activity is very robust and GFPμ accumulation induced by proteasome inhibitors has been reported to require about 70 % inhibition of proteasomal activity [46]. Thus, our results are more consistent with the notion that the transient increase in GFPμ accumulation is primarily linked to impaired ubiquitination and not to the inhibition of the proteasome.
We demonstrated here that the primary effect of acute or prolonged treatment with PQ and MPP+ at either high (toxic) or low (sub-toxic) doses is a decrease in protein ubiquitination (Fig. 8). We and the others have previously shown that chronic inhibition of the proteasome depletes cells from Ub [43, 107, 108]. Bence et al. found that the GFPμ fluorescence rapidly declined in cells after the exposure to a protein synthesis inhibitor due to the impaired synthesis of short-lived proteins such as GFPμ [60, 61]. However, PQ-induced Ub protein depletion seems to precede the inhibition of the proteasome, discarding a possible negative feedback loop from the proteasome to Ub protein synthesis. Interestingly, while MPP+ reduced protein ubiquitination, rotenone (another complex I inhibitor) had no such effect, which adds to the cumulative evidence that the toxicity induced by MPP+ and rotenone might actually involve different mechanisms [33, 45, 109–114].
Levels of endogenous Ub conjugates depend on the balance between (1) the rate of Ub conjugation determined by the availability of Ub, the activity of the E1-E2-E3 system, and ATP levels, and (2) the rate of the turnover of Ub conjugates (degradation/deubiquitination), which depends on the activity of the proteasome, autophagy, and deubiquitinating enzymes [98]. Several components of the UPS can present different sensitivities to oxidative damage/modulation [115–122]. Deubiquitinating enzymes are inhibited by oxidation of their catalytic cysteine residue [123, 124]. The proteasome has been reported to be more susceptible to oxidative inhibition than Ub-conjugating enzymes [115, 125, 126]. Accordingly, mild to moderate oxidative stress upregulates Ub and the Ub-conjugating system promoting the formation of Ub conjugates and reducing the activity of the proteasome. In contrast, extensive but not lethal oxidative stress reduces the formation of Ub conjugates by inactivating Ub-conjugating enzymes promoting the accumulation/aggregation of damaged/abnormal proteins [125]. While it is possible that PQ might interfere with the activity of the Ub-conjugating system (E1, E2, and E3s), this should have been translated into the accumulation of Ub monomers/free chains. In contrast, PQ clearly depleted cells from Ub even at sub-toxic concentrations. Thus, our results imply that PQ-induced depletion of Ub-bound proteins is associated with a decrease in the Ub protein pool.
While PQ clearly reduced the Ub protein pool, Ub mRNA levels were shown to increase in response to PQ, which is consistent with the notion that Ub is a stress-inducible protein [69, 127]. These findings suggest that PQ or MPP+ might impair the synthesis of Ub at the post-transcriptional level or modify Ub protein stability. Protein synthesis has been shown to be more sensitive to oxidative stress than DNA/RNA synthesis [128]. Ub protein depletion induced by PQ and MPP+ was also paralleled by a decrease in GFPμ fluorescence in stable cells. Thus, our results indicate that PQ and MPP+ are likely impairing overall protein synthesis, which should initially affect short-lived proteins such as Ub and GFPμ (Fig. 8). MPP+ has been previously shown to inhibit protein synthesis [129], but the mechanisms involved remain unknown. PQ and MPP+ induce oxidative stress and energy failure, which can affect overall protein synthesis. The correct attachment of amino acids to each tRNA species that is required for protein synthesis is an energy-dependent process carried out by aminoacyl-tRNA synthetases. Additionally, aminoacyl-tRNA synthetases also hydrolyze (edit) an incorrectly attached amino acid, and oxidative stress induces protein mistranslation and degradation by impairment of aminoacyl-tRNA synthetase editing [130]. Oxidative stress also diverts tRNA synthetases to the nucleus to protect against oxidative damage [131]. Moreover, oxidized mRNA, protein mistranslation, and subsequent degradation have been recently recognized as important contributors to neurodegeneration [132–137]. To determine whether energy failure, oxidative stress, or both impair Ub protein synthesis will require further investigation. However, we have demonstrated here and in previous studies that oxidative stress induced by MPP+ and low PQ concentrations is primarily ascribed to mitochondria [45], suggesting that energy failure might be the primary mechanism involved in impaired protein (Ub) synthesis.
An increased accumulation of oxidized protein byproducts is found in PD brains. Elevated levels of carbonylated proteins [138] and cysteine-oxidized proteins including the hydrogen peroxide scavengers Prxs and the early onset PD-related protein-gene DJ-1/PARK7 [139–141] are important oxidative biomarkers detected in PD brains. We observed that PQ induced an increase in the accumulation of PSOH, precursors of irreversibly oxidative protein sulfinic (PSO2H) and PSO3H acid modifications. Accordingly, PQ also induced a dose-dependent accumulation of irreversibly oxidized DJ-1-SO3H and Prxs-SO3H, as well as protein carbonyls. Turnover of both DJ-1 and Prx has been proposed to be mediated by the UPS [142–144]. Cysteine sulfenylation at the N-terminus of proteins is an important step in the endoproteolytic cleavage and formation of N-degrons recognized by the UPS [145]. Both Ub-dependent and Ub-independent degradation of oxidized proteins has been reported [22, 71, 72, 125, 146]. Accumulation of oxidized proteins in the absence of Ub-bound protein aggregates suggests that ubiquitination might be required for the degradation of oxidized proteins induced by PQ. Aging is the main risk factor in the development of PD [147]. Similar to our results, it was observed that in an aging yeast model polyubiquitinated proteins are significantly reduced even in the presence of a decrease in proteasomal activity leading to an increase/accumulation of oxidized proteins [148]. Moreover, a decrease in Ub conjugates as well as in de novo Ub conjugation activity was reported in lenses from aged rats, and these effects were associated with the accumulation of damaged proteins [149].
It has been recently recognized the important role that autophagy plays in the degradation of ubiquitinated cargo. Both the UPS and autophagy play complementary roles in the degradation of misfolded protein aggregates such as α-synuclein [2, 83, 85, 150, 151]. Heavily oxidized stable protein aggregates are not suitable for proteasomal degradation, and autophagy is thought to also play a major role in the degradation of oxidized protein aggregates [19, 20]. The UPS and autophagy are complementary pathways, where alterations in the rate of one system are reported to modify those of the other one. In particular, impairment of the UPS system triggers autophagy [81, 82, 152]. Interestingly, while contradicting results exist regarding the ability of proteasome inhibitors to induce neurodegeneration [153–158], impairment of autophagy seems to selectively induce dopaminergic cell loss in vivo [12, 13, 159]. Indeed, we observed that MG132 induces autophagy. However, whereas inhibition of ATG5-dependent autophagy stimulated PQ and MPP+ toxicity, no additional effect was induced by proteasome inhibition. The fact that inhibition of autophagy stimulates PQ and MPP+ toxicity without exerting an effect on the accumulation of Ub-bound proteins (this work and [33]) suggests that the protective effects of autophagy are independent from its role in protein degradation.
Ubiquitinated proteins are selectively targeted to the autophagosome-lysosome system via Ub-binding proteins, primarily p62/SQSTM [10, 98]. p62 has been found in LBs from PD and dementia with LBs (DLB) brains [160–163], neurofibrillary tangles from Alzheimer’s disease brains, and in Huntingtin aggregates [4, 164, 165]. Likewise, it has been shown that α-synuclein inclusions and oxidized proteins can be degraded through the p62-dependent autophagy clearance [21, 163]. A recent report demonstrated that the UBA domain, which has a low affinity for Ub, also mediates the formation of inactive p62 dimers and that p62 dimerization and Ub-binding are mutually exclusives [73]. In our study, we found that the inhibition of Ub-activating enzymes (E1s) and the depletion of the Ub protein pool induced by PQ and MPP+ are paralleled by p62 inactivation/dimerization (Fig. 8). As we reported before [33], PQ and MPP+ induce a dose- and time-dependent impairment in autophagy flux. Thus, our results suggest that p62 inactivation by Ub protein depletion might be a mechanism by which PQ and MPP+ also impair autophagy (Fig. 8). p62 has been reported to regulate mitophagy and stress response signaling via the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and nuclear factor (erythroid-derived 2)-like 2 (Nrf2) among others [14, 73, 166–168]. It is likely that p62 dimerization might be regulating the stress response of cells treated with PQ and MPP+, and we expect to address this in the future.
Clearance of misfolded/aggregated α-synuclein has been shown to be mediated by both autophagy and the ubiquitin/proteasome pathways [83–85]. Ubiquitination of α-synuclein by E3 Ub ligases (seven in absentia homologue-1 (SIAH1), E6-associated protein (E6-AP), neural precursor cell expressed developmentally downregulated protein 4 (Nedd4), tumor necrosis factor-receptor associated factor 6 (TRAF6)) has been proposed to regulate its degradation via the proteasome or endosomal-lysosomal degradation pathways [169–173]. Interestingly, Ub-independent α-synuclein degradation via the 20S proteasome has also been reported [174]. Recent studies have demonstrated that ubiquitination of α-synuclein in different lysine residues mediates diverse effects including the formation of protein inclusions or Lewy bodies [175–177], which is now considered a protective mechanism against the accumulation of toxic protofibrillar intermediates, as aggregation of α-synuclein might be required for its detoxification [178]. PQ alone had no effect on the endogenous α-synuclein levels, but when WT or mutant A53T α-synuclein was overexpressed, PQ induced an increase accumulation of α-synuclein in its monomeric form. Indeed, ubiquitination of monomeric and fibrillar α-synuclein has been reported previously [179]. Furthermore, we observed that in yeast cells, where α-synuclein is localized at the plasma membrane, PQ treatment induced the accumulation of membrane foci, suggesting that alterations in protein ubiquitination might alter membrane sorting or turnover of α-synuclein. In fact it has been demonstrated that in yeast, localization of α-synuclein to the plasma membrane requires the secretory pathway [180], and α-synuclein was also reported to be ubiquitinated by Nedd4 [169], homologous to the E6-AP carboxyl terminus (HECT) domain E3 that functions at the plasma membrane in the turnover/sorting of a number of membrane-associated proteins [181].
Overall, our results uncover a new mechanism by which environmental (PQ) and mitochondrial toxicants (MPP+) impair Ub-dependent proteostatic mechanisms. Post-transcriptional depletion of the Ub protein pool impairs both proteasome- and p62-autophagy-mediated protein degradation pathways. Depletion of the Ub protein pool seems to be associated with the accumulation of oxidized proteins, alterations in the levels/distribution of α-synuclein, and precede the impairment in proteasome activity and autophagy flux (Fig. 8).
References
Tanaka K, Matsuda N (2014) Proteostasis and neurodegeneration: the roles of proteasomal degradation and autophagy. Biochim Biophys Acta 1843(1):197–204. doi:10.1016/j.bbamcr.2013.03.012
Cook C, Stetler C, Petrucelli L (2012) Disruption of protein quality control in Parkinson’s disease. Cold Spring Harb Perspect Med 2(5):a009423. doi:10.1101/cshperspect.a009423
Wakabayashi K, Tanji K, Odagiri S, Miki Y, Mori F, Takahashi H (2013) The Lewy body in Parkinson’s disease and related neurodegenerative disorders. Mol Neurobiol 47(2):495–508. doi:10.1007/s12035-012-8280-y
Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M et al (2002) p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol 160(1):255–263. doi:10.1016/S0002-9440(10)64369-6
Kuzuhara S, Mori H, Izumiyama N, Yoshimura M, Ihara Y (1988) Lewy bodies are ubiquitinated. A light and electron microscopic immunocytochemical study. Acta Neuropathol 75(4):345–353
Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M (1998) alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci U S A 95(11):6469–6473
Gomez-Tortosa E, Newell K, Irizarry MC, Sanders JL, Hyman BT (2000) alpha-Synuclein immunoreactivity in dementia with Lewy bodies: morphological staging and comparison with ubiquitin immunostaining. Acta Neuropathol 99(4):352–357
Choi AM, Ryter SW, Levine B (2013) Autophagy in human health and disease. N Engl J Med 368(7):651–662. doi:10.1056/NEJMra1205406
Navarro-Yepes J, Burns M, Anandhan A, Khalimonchuk O, del Razo LM, Quintanilla-Vega B, Pappa A, Panayiotidis MI et al (2014) Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxid Redox Signal 21(1):66–85. doi:10.1089/ars.2014.5837
Shaid S, Brandts CH, Serve H, Dikic I (2013) Ubiquitination and selective autophagy. Cell Death Differ 20(1):21–30. doi:10.1038/cdd.2012.72
Rogov V, Dotsch V, Johansen T, Kirkin V (2014) Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 53(2):167–178. doi:10.1016/j.molcel.2013.12.014
Ahmed I, Liang Y, Schools S, Dawson VL, Dawson TM, Savitt JM (2012) Development and characterization of a new Parkinson’s disease model resulting from impaired autophagy. J Neurosci 32(46):16503–16509. doi:10.1523/JNEUROSCI.0209-12.2012
Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z (2012) Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J Neurosci 32(22):7585–7593. doi:10.1523/JNEUROSCI.5809-11.2012
Komatsu M, Kageyama S, Ichimura Y (2012) p62/SQSTM1/A170: physiology and pathology. Pharmacol Res 66(6):457–462. doi:10.1016/j.phrs.2012.07.004
Watanabe Y, Tanaka M (2011) p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J Cell Sci 124(Pt 16):2692–2701. doi:10.1242/jcs.081232
Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW et al (2009) Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem 111(4):1062–1073. doi:10.1111/j.1471-4159.2009.06388.x
Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24(18):8055–8068. doi:10.1128/MCB.24.18.8055-8068.2004
Babu JR, Geetha T, Wooten MW (2005) Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem 94(1):192–203. doi:10.1111/j.1471-4159.2005.03181.x
Dunlop RA, Brunk UT, Rodgers KJ (2009) Oxidized proteins: mechanisms of removal and consequences of accumulation. IUBMB Life 61(5):522–527. doi:10.1002/iub.189
Chondrogianni N, Petropoulos I, Grimm S, Georgila K, Catalgol B, Friguet B, Grune T, Gonos ES (2012) Protein damage, repair and proteolysis. Mol Asp Med. doi:10.1016/j.mam.2012.09.001
Wang L, Cano M, Handa JT (2014) p62 Provides dual cytoprotection against oxidative stress in the retinal pigment epithelium. Biochim Biophys Acta 1843(7):1248–1258. doi:10.1016/j.bbamcr.2014.03.016
Marques C, Pereira P, Taylor A, Liang JN, Reddy VN, Szweda LI, Shang F (2004) Ubiquitin-dependent lysosomal degradation of the HNE-modified proteins in lens epithelial cells. FASEB J 18(12):1424–1426. doi:10.1096/fj.04-1743fje
Klein C, Westenberger A (2012) Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med 2(1):a008888. doi:10.1101/cshperspect.a008888
Tanner CM, Kamel F, Ross GW, Hoppin JA, Goldman SM, Korell M, Marras C, Bhudhikanok GS et al (2011) Rotenone, paraquat, and Parkinson’s disease. Environ Health Perspect 119(6):866–872. doi:10.1289/ehp.1002839
Kamel F (2013) Epidemiology. Paths from pesticides to Parkinson’s. Science 341(6147):722–723. doi:10.1126/science.1243619
van der Mark M, Brouwer M, Kromhout H, Nijssen P, Huss A, Vermeulen R (2012) Is pesticide use related to Parkinson disease? Some clues to heterogeneity in study results. Environ Health Perspect 120(3):340–347. doi:10.1289/ehp.1103881
Caudle WM, Guillot TS, Lazo CR, Miller GW (2012) Industrial toxicants and Parkinson’s disease. Neurotoxicology 33(2):178–188. doi:10.1016/j.neuro.2012.01.010
Jang H, Boltz DA, Webster RG, Smeyne RJ (2009) Viral parkinsonism. Biochim Biophys Acta 1792(7):714–721. doi:10.1016/j.bbadis.2008.08.001
Gorell JM, Rybicki BA, Cole Johnson C, Peterson EL (1999) Occupational metal exposures and the risk of Parkinson’s disease. Neuroepidemiology 18(6):303–308
Caneda-Ferron B, De Girolamo LA, Costa T, Beck KE, Layfield R, Billett EE (2008) Assessment of the direct and indirect effects of MPP+ and dopamine on the human proteasome: implications for Parkinson’s disease aetiology. J Neurochem 105(1):225–238. doi:10.1111/j.1471-4159.2007.05130.x
Yang W, Tiffany-Castiglioni E (2007) The bipyridyl herbicide paraquat induces proteasome dysfunction in human neuroblastoma SH-SY5Y cells. J Toxic Environ Health A 70(21):1849–1857. doi:10.1080/15287390701459262
Fornai F, Schluter OM, Lenzi P, Gesi M, Ruffoli R, Ferrucci M, Lazzeri G, Busceti CL et al (2005) Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and alpha-synuclein. Proc Natl Acad Sci U S A 102(9):3413–3418. doi:10.1073/pnas.0409713102
Garcia-Garcia A, Anandhan A, Burns M, Chen H, Zhou Y, Franco R (2013) Impairment of Atg5-dependent autophagic flux promotes paraquat- and MPP+-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol Sci 136(1):166–182. doi:10.1093/toxsci/kft188
Wills J, Credle J, Oaks AW, Duka V, Lee JH, Jones J, Sidhu A (2012) Paraquat, but not maneb, induces synucleinopathy and tauopathy in striata of mice through inhibition of proteasomal and autophagic pathways. PLoS One 7(1):e30745. doi:10.1371/journal.pone.0030745
Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA et al (2007) HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447(7146):859–863. doi:10.1038/nature05853
Janen SB, Chaachouay H, Richter-Landsberg C (2010) Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia 58(14):1766–1774. doi:10.1002/glia.21047
Lei S, Zavala-Flores L, Garcia-Garcia A, Nandakumar R, Huang Y, Madayiputhiya N, Stanton RC, Dodds ED et al (2014) Alterations in energy/redox metabolism induced by mitochondrial and environmental toxins: a specific role for glucose-6-phosphate-dehydrogenase and the pentose phosphate pathway in paraquat toxicity. ACS Chem Biol. doi:10.1021/cb400894a
Scholz D, Poltl D, Genewsky A, Weng M, Waldmann T, Schildknecht S, Leist M (2011) Rapid, complete and large-scale generation of post-mitotic neurons from the human LUHMES cell line. J Neurochem 119(5):957–971. doi:10.1111/j.1471-4159.2011.07255.x
Rodriguez-Rocha H, Garcia-Garcia A, Zavala-Flores L, Li S, Madayiputhiya N, Franco R (2012) Glutaredoxin 1 protects dopaminergic cells by increased protein glutathionylation in experimental Parkinson’s disease. Antioxid Redox Signal. doi:10.1089/ars.2011.4474
Seo YH, Carroll KS (2009) Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A 106(38):16163–16168. doi:10.1073/pnas.0903015106
Luo S, Wehr NB (2009) Protein carbonylation: avoiding pitfalls in the 2,4-dinitrophenylhydrazine assay. Redox Rep 14(4):159–166. doi:10.1179/135100009X392601
Levine RL, Williams JA, Stadtman ER, Shacter E (1994) Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol 233:346–357
Anandhan A, Rodriguez-Rocha H, Bohovych I, Griggs AM, Zavala-Flores L, Reyes-Reyes EM, Seravalli J, Stanciu LA et al (2014) Overexpression of alpha-synuclein at non-toxic levels increases dopaminergic cell death induced by copper exposure via modulation of protein degradation pathways. Neurobiol Dis. doi:10.1016/j.nbd.2014.11.018
Myeku N, Metcalfe MJ, Huang Q, Figueiredo-Pereira M (2011) Assessment of proteasome impairment and accumulation/aggregation of ubiquitinated proteins in neuronal cultures. Methods Mol Biol 793:273–296. doi:10.1007/978-1-61779-328-8_18
Rodriguez-Rocha H, Garcia-Garcia A, Pickett C, Li S, Jones J, Chen H, Webb B, Choi J et al (2013) Compartmentalized oxidative stress in dopaminergic cell death induced by pesticides and complex I inhibitors: distinct roles of superoxide anion and superoxide dismutases. Free Radic Biol Med 61C:370–383. doi:10.1016/j.freeradbiomed.2013.04.021
Bence NF, Bennett EJ, Kopito RR (2005) Application and analysis of the GFPu family of ubiquitin-proteasome system reporters. Methods Enzymol 399:481–490. doi:10.1016/S0076-6879(05)99033-2
Berkers CR, van Leeuwen FW, Groothuis TA, Peperzak V, van Tilburg EW, Borst J, Neefjes JJ, Ovaa H (2007) Profiling proteasome activity in tissue with fluorescent probes. Mol Pharm 4(5):739–748. doi:10.1021/mp0700256
de Jong A, Schuurman KG, Rodenko B, Ovaa H, Berkers CR (2012) Fluorescence-based proteasome activity profiling. Methods Mol Biol 803:183–204. doi:10.1007/978-1-61779-364-6_13
Yang L, Li P, Fu S, Calay ES, Hotamisligil GS (2010) Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 11(6):467–478. doi:10.1016/j.cmet.2010.04.005
Liu F, Hindupur J, Nguyen JL, Ruf KJ, Zhu J, Schieler JL, Bonham CC, Wood KV et al (2008) Methionine sulfoxide reductase A protects dopaminergic cells from Parkinson’s disease-related insults. Free Radic Biol Med 45(3):242–255. doi:10.1016/j.freeradbiomed.2008.03.022
Barde I, Salmon P, Trono D (2010) Production and titration of lentiviral vectors. Curr Protoc Neurosci Chapter 4:Unit 4 21. doi:10.1002/0471142301.ns0421s53
Srivastava G, Dixit A, Yadav S, Patel DK, Prakash O, Singh MP (2012) Resveratrol potentiates cytochrome P450 2 d22-mediated neuroprotection in maneb- and paraquat-induced parkinsonism in the mouse. Free Radic Biol Med 52(8):1294–1306. doi:10.1016/j.freeradbiomed.2012.02.005
Goldman SM (2014) Environmental toxins and Parkinson’s disease. Annu Rev Pharmacol Toxicol 54:141–164. doi:10.1146/annurev-pharmtox-011613-135937
Subramaniam SR, Chesselet MF (2013) Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog Neurobiol 106–107:17–32. doi:10.1016/j.pneurobio.2013.04.004
Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT (2000) Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci 3(12):1301–1306. doi:10.1038/81834
Sun F, Kanthasamy A, Anantharam V, Kanthasamy AG (2007) Environmental neurotoxic chemicals-induced ubiquitin proteasome system dysfunction in the pathogenesis and progression of Parkinson’s disease. Pharmacol Ther 114(3):327–344. doi:10.1016/j.pharmthera.2007.04.001
Corcoran LJ, Mitchison TJ, Liu Q (2004) A novel action of histone deacetylase inhibitors in a protein aggresome disease model. Curr Biol 14(6):488–492. doi:10.1016/j.cub.2004.03.003
Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE (2001) Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 12(5):1393–1407
Stack JH, Whitney M, Rodems SM, Pollok BA (2000) A ubiquitin-based tagging system for controlled modulation of protein stability. Nat Biotechnol 18(12):1298–1302. doi:10.1038/82422
Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292(5521):1552–1555. doi:10.1126/science.292.5521.1552
Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273(52):34970–34975
Corish P, Tyler-Smith C (1999) Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12(12):1035–1040
Shamoto-Nagai M, Maruyama W, Kato Y, Isobe K, Tanaka M, Naoi M, Osawa T (2003) An inhibitor of mitochondrial complex I, rotenone, inactivates proteasome by oxidative modification and induces aggregation of oxidized proteins in SH-SY5Y cells. J Neurosci Res 74(4):589–597. doi:10.1002/jnr.10777
Yamamuro A, Yoshioka Y, Ogita K, Maeda S (2006) Involvement of endoplasmic reticulum stress on the cell death induced by 6-hydroxydopamine in human neuroblastoma SH-SY5Y cells. Neurochem Res 31(5):657–664. doi:10.1007/s11064-006-9062-6
Endo R, Saito T, Asada A, Kawahara H, Ohshima T, Hisanaga S (2009) Commitment of 1-methyl-4-phenylpyrinidinium ion-induced neuronal cell death by proteasome-mediated degradation of p35 cyclin-dependent kinase 5 activator. J Biol Chem 284(38):26029–26039. doi:10.1074/jbc.M109.026443
Sawada H, Kohno R, Kihara T, Izumi Y, Sakka N, Ibi M, Nakanishi M, Nakamizo T et al (2004) Proteasome mediates dopaminergic neuronal degeneration, and its inhibition causes alpha-synuclein inclusions. J Biol Chem 279(11):10710–10719. doi:10.1074/jbc.M308434200
Prasad K, Winnik B, Thiruchelvam MJ, Buckley B, Mirochnitchenko O, Richfield EK (2007) Prolonged toxicokinetics and toxicodynamics of paraquat in mouse brain. Environ Health Perspect 115(10):1448–1453. doi:10.1289/ehp.9932
Seufert W, Jentsch S (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9(2):543–550
Oh C, Park S, Lee EK, Yoo YJ (2013) Downregulation of ubiquitin level via knockdown of polyubiquitin gene Ubb as potential cancer therapeutic intervention. Sci Rep 3:2623. doi:10.1038/srep02623
Hanna J, Leggett DS, Finley D (2003) Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol 23(24):9251–9261
Shringarpure R, Grune T, Mehlhase J, Davies KJ (2003) Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem 278(1):311–318. doi:10.1074/jbc.M206279200
Kastle M, Reeg S, Rogowska-Wrzesinska A, Grune T (2012) Chaperones, but not oxidized proteins, are ubiquitinated after oxidative stress. Free Radic Biol Med 53(7):1468–1477. doi:10.1016/j.freeradbiomed.2012.05.039
Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B, Williamson MP, Layfield R et al (2010) Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J Mol Biol 396(1):178–194. doi:10.1016/j.jmb.2009.11.032
Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP et al (2007) Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res 67(19):9472–9481. doi:10.1158/0008-5472.CAN-07-0568
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8(4):445–544
Bang Y, Kang BY, Choi HJ (2014) Preconditioning stimulus of proteasome inhibitor enhances aggresome formation and autophagy in differentiated SH-SY5Y cells. Neurosci Lett 566:263–268. doi:10.1016/j.neulet.2014.02.056
Lan D, Wang W, Zhuang J, Zhao Z (2015) Proteasome inhibitor-induced autophagy in PC12 cells overexpressing A53T mutant alpha-synuclein. Mol Med Rep 11(3):1655–1660. doi:10.3892/mmr.2014.3011
Lim J, Lee Y, Jung S, Youdim MB, Oh YJ (2014) Impaired autophagic flux is critically involved in drug-induced dopaminergic neuronal death. Parkinsonism Relat Disord 20(Suppl 1):S162–S166. doi:10.1016/S1353-8020(13)70039-7
Myeku N, Figueiredo-Pereira ME (2011) Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. J Biol Chem 286(25):22426–22440. doi:10.1074/jbc.M110.149252
Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L et al (2014) Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J Biol Chem 289(36):24944–24955. doi:10.1074/jbc.M114.580357
Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC (2009) Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 33(4):517–527. doi:10.1016/j.molcel.2009.01.021
Rubinsztein DC (2007) Autophagy induction rescues toxicity mediated by proteasome inhibition. Neuron 54(6):854–856. doi:10.1016/j.neuron.2007.06.005
Ebrahimi-Fakhari D, Cantuti-Castelvetri I, Fan Z, Rockenstein E, Masliah E, Hyman BT, McLean PJ, Unni VK (2011) Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of alpha-synuclein. J Neurosci 31(41):14508–14520. doi:10.1523/JNEUROSCI.1560-11.2011
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC (2003) Alpha-synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278(27):25009–25013. doi:10.1074/jbc.M300227200
Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D (2004) Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305(5688):1292–1295. doi:10.1126/science.1101738
Chorfa A, Betemps D, Morignat E, Lazizzera C, Hogeveen K, Andrieu T, Baron T (2013) Specific pesticide-dependent increases in alpha-synuclein levels in human neuroblastoma (SH-SY5Y) and melanoma (SK-MEL-2) cell lines. Toxicol Sci 133(2):289–297. doi:10.1093/toxsci/kft076
Chorfa A, Lazizzera C, Betemps D, Morignat E, Dussurgey S, Andrieu T, Baron T (2014) A variety of pesticides trigger in vitro alpha-synuclein accumulation, a key event in Parkinson’s disease. Arch Toxicol. doi:10.1007/s00204-014-1388-2
Manning-Bog AB, McCormack AL, Li J, Uversky VN, Fink AL, Di Monte DA (2002) The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: paraquat and alpha-synuclein. J Biol Chem 277(3):1641–1644. doi:10.1074/jbc.C100560200
Mitra S, Chakrabarti N, Bhattacharyya A (2011) Differential regional expression patterns of alpha-synuclein, TNF-alpha, and IL-1beta; and variable status of dopaminergic neurotoxicity in mouse brain after Paraquat treatment. J Neuroinflammation 8:163. doi:10.1186/1742-2094-8-163
Fauvet B, Mbefo MK, Fares MB, Desobry C, Michael S, Ardah MT, Tsika E, Coune P et al (2012) Alpha-synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J Biol Chem 287(19):15345–15364. doi:10.1074/jbc.M111.318949
Lashuel HA, Overk CR, Oueslati A, Masliah E (2013) The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nat Rev Neurosci 14(1):38–48. doi:10.1038/nrn3406
Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT Jr (1996) NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 35(43):13709–13715. doi:10.1021/bi961799n
Dettmer U, Newman AJ, Luth ES, Bartels T, Selkoe D (2013) In vivo cross-linking reveals principally oligomeric forms of alpha-synuclein and beta-synuclein in neurons and non-neural cells. J Biol Chem 288(9):6371–6385. doi:10.1074/jbc.M112.403311
Bartels T, Choi JG, Selkoe DJ (2011) Alpha-synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477(7362):107–110. doi:10.1038/nature10324
Khurana V, Lindquist S (2010) Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci 11(6):436–449. doi:10.1038/nrn2809
Trinh J, Farrer M (2013) Advances in the genetics of Parkinson disease. Nat Rev Neurol 9(8):445–454. doi:10.1038/nrneurol.2013.132
Chung KK, Dawson VL, Dawson TM (2001) The role of the ubiquitin-proteasomal pathway in Parkinson’s disease and other neurodegenerative disorders. Trends Neurosci 24(11 Suppl):S7–S14
Clague MJ, Urbe S (2010) Ubiquitin: same molecule, different degradation pathways. Cell 143(5):682–685. doi:10.1016/j.cell.2010.11.012
McNaught KS, Jenner P (2001) Proteasomal function is impaired in substantia nigra in Parkinson’s disease. Neurosci Lett 297(3):191–194
McNaught KS, Belizaire R, Jenner P, Olanow CW, Isacson O (2002) Selective loss of 20S proteasome alpha-subunits in the substantia nigra pars compacta in Parkinson’s disease. Neurosci Lett 326(3):155–158
McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW (2003) Altered proteasomal function in sporadic Parkinson’s disease. Exp Neurol 179(1):38–46
Martins-Branco D, Esteves AR, Santos D, Arduino DM, Swerdlow RH, Oliveira CR, Januario C, Cardoso SM (2012) Ubiquitin proteasome system in Parkinson’s disease: a keeper or a witness? Exp Neurol 238(2):89–99. doi:10.1016/j.expneurol.2012.08.008
Wang XF, Li S, Chou AP, Bronstein JM (2006) Inhibitory effects of pesticides on proteasome activity: implication in Parkinson’s disease. Neurobiol Dis 23(1):198–205. doi:10.1016/j.nbd.2006.02.012
Zeng BY, Iravani MM, Lin ST, Irifune M, Kuoppamaki M, Al-Barghouthy G, Smith L, Jackson MJ et al (2006) MPTP treatment of common marmosets impairs proteasomal enzyme activity and decreases expression of structural and regulatory elements of the 26S proteasome. Eur J Neurosci 23(7):1766–1774. doi:10.1111/j.1460-9568.2006.04718.x
Kadoguchi N, Umeda M, Kato H, Araki T (2008) Proteasome inhibitor does not enhance MPTP neurotoxicity in mice. Cell Mol Neurobiol 28(7):971–979. doi:10.1007/s10571-008-9271-4
Carvalho AN, Marques C, Rodrigues E, Henderson CJ, Wolf CR, Pereira P, Gama MJ (2013) Ubiquitin-proteasome system impairment and MPTP-induced oxidative stress in the brain of C57BL/6 wild-type and GSTP knockout mice. Mol Neurobiol 47(2):662–672. doi:10.1007/s12035-012-8368-4
Xu Q, Farah M, Webster JM, Wojcikiewicz RJ (2004) Bortezomib rapidly suppresses ubiquitin thiolesterification to ubiquitin-conjugating enzymes and inhibits ubiquitination of histones and type I inositol 1,4,5-trisphosphate receptor. Mol Cancer Ther 3(10):1263–1269
Ding Q, Dimayuga E, Markesbery WR, Keller JN (2006) Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J 20(8):1055–1063. doi:10.1096/fj.05-5495com
Giordano S, Lee J, Darley-Usmar VM, Zhang J (2012) Distinct effects of rotenone, 1-methyl-4-phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS One 7(9):e44610. doi:10.1371/journal.pone.0044610
Yacoubian TA, Slone SR, Harrington AJ, Hamamichi S, Schieltz JM, Caldwell KA, Caldwell GA, Standaert DG (2010) Differential neuroprotective effects of 14-3-3 proteins in models of Parkinson’s disease. Cell Death Dis 1:e2. doi:10.1038/cddis.2009.4
Song JX, Shaw PC, Wong NS, Sze CW, Yao XS, Tang CW, Tong Y, Zhang YB (2012) Chrysotoxine, a novel bibenzyl compound selectively antagonizes MPP(+), but not rotenone, neurotoxicity in dopaminergic SH-SY5Y cells. Neurosci Lett 521(1):76–81. doi:10.1016/j.neulet.2012.05.063
Huang Y, Xu J, Liang M, Hong X, Suo H, Liu J, Yu M, Huang F (2013) RESP18 is involved in the cytotoxicity of dopaminergic neurotoxins in MN9D cells. Neurotox Res 24(2):164–175. doi:10.1007/s12640-013-9375-6
Martins JB, Bastos Mde L, Carvalho F, Capela JP (2013) Differential effects of methyl-4-phenylpyridinium ion, rotenone, and paraquat on differentiated SH-SY5Y cells. J Toxicol 2013:347312. doi:10.1155/2013/347312
Zhu M, Li WW, Lu CZ (2014) Histone decacetylase inhibitors prevent mitochondrial fragmentation and elicit early neuroprotection against MPP+. CNS Neurosci Ther 20(4):308–316. doi:10.1111/cns.12217
Zhang X, Zhou J, Fernandes AF, Sparrow JR, Pereira P, Taylor A, Shang F (2008) The proteasome: a target of oxidative damage in cultured human retina pigment epithelial cells. Invest Ophthalmol Vis Sci 49(8):3622–3630. doi:10.1167/iovs.07-1559
Jahngen-Hodge J, Obin MS, Gong X, Shang F, Nowell TR Jr, Gong J, Abasi H, Blumberg J et al (1997) Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J Biol Chem 272(45):28218–28226
Obin M, Shang F, Gong X, Handelman G, Blumberg J, Taylor A (1998) Redox regulation of ubiquitin-conjugating enzymes: mechanistic insights using the thiol-specific oxidant diamide. FASEB J 12(7):561–569
Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM et al (2004) Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci U S A 101(29):10810–10814. doi:10.1073/pnas.0404161101
Ishii T, Sakurai T, Usami H, Uchida K (2005) Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry 44(42):13893–13901. doi:10.1021/bi051336u
Caballero M, Liton PB, Epstein DL, Gonzalez P (2003) Proteasome inhibition by chronic oxidative stress in human trabecular meshwork cells. Biochem Biophys Res Commun 308(2):346–352
Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K (1999) 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J Biol Chem 274(34):23787–23793
Ding Q, Keller JN (2001) Proteasome inhibition in oxidative stress neurotoxicity: implications for heat shock proteins. J Neurochem 77(4):1010–1017
Cotto-Rios XM, Bekes M, Chapman J, Ueberheide B, Huang TT (2012) Deubiquitinases as a signaling target of oxidative stress. Cell Rep 2(6):1475–1484. doi:10.1016/j.celrep.2012.11.011
Lee JG, Baek K, Soetandyo N, Ye Y (2013) Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nat Commun 4:1568. doi:10.1038/ncomms2532
Shang F, Taylor A (2011) Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med 51(1):5–16. doi:10.1016/j.freeradbiomed.2011.03.031
Shang F, Gong X, Taylor A (1997) Activity of ubiquitin-dependent pathway in response to oxidative stress. Ubiquitin-activating enzyme is transiently up-regulated. J Biol Chem 272(37):23086–23093
Monia BP, Ecker DJ, Crooke ST (1990) New perspectives on the structure and function of ubiquitin. Nat Biotechnol 8(3):209–215
Buttgereit F, Brand MD (1995) A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312(Pt 1):163–167
Notter MF, Irwin I, Langston JW, Gash DM (1988) Neurotoxicity of MPTP and MPP+ in vitro: characterization using specific cell lines. Brain Res 456(2):254–262
Ling J, Soll D (2010) Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci U S A 107(9):4028–4033. doi:10.1073/pnas.1000315107
Wei N, Shi Y, Truong LN, Fisch KM, Xu T, Gardiner E, Fu G, Hsu YS et al (2014) Oxidative stress diverts tRNA synthetase to nucleus for protection against DNA damage. Mol Cell 56(2):323–332. doi:10.1016/j.molcel.2014.09.006
Tanaka M, Chock PB, Stadtman ER (2007) Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci U S A 104(1):66–71. doi:10.1073/pnas.0609737104
Shan X, Tashiro H, Lin CL (2003) The identification and characterization of oxidized RNAs in Alzheimer’s disease. J Neurosci 23(12):4913–4921
Shan X, Chang Y, Lin CL (2007) Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J 21(11):2753–2764. doi:10.1096/fj.07-8200com
Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR et al (2008) Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One 3(8):e2849. doi:10.1371/journal.pone.0002849
Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS (2014) An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep 9(4):1256–1264. doi:10.1016/j.celrep.2014.10.042
Ding Q, Dimayuga E, Keller JN (2007) Oxidative stress alters neuronal RNA- and protein-synthesis: implications for neural viability. Free Radic Res 41(8):903–910. doi:10.1080/10715760701416996
Alam ZI, Daniel SE, Lees AJ, Marsden DC, Jenner P, Halliwell B (1997) A generalised increase in protein carbonyls in the brain in Parkinson’s but not incidental Lewy body disease. J Neurochem 69(3):1326–1329
Saito Y, Hamakubo T, Yoshida Y, Ogawa Y, Hara Y, Fujimura H, Imai Y, Iwanari H et al (2009) Preparation and application of monoclonal antibodies against oxidized DJ-1. Significant elevation of oxidized DJ-1 in erythrocytes of early-stage Parkinson disease patients. Neurosci Lett 465(1):1–5. doi:10.1016/j.neulet.2009.08.074
Choi J, Sullards MC, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI et al (2006) Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem 281(16):10816–10824. doi:10.1074/jbc.M509079200
Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA (2007) S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A 104(47):18742–18747. doi:10.1073/pnas.0705904104
Cho CS, Yoon HJ, Kim JY, Woo HA, Rhee SG (2014) Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci U S A 111(33):12043–12048. doi:10.1073/pnas.1401100111
Li X, Lu D, He F, Zhou H, Liu Q, Wang Y, Shao C, Gong Y (2011) Cullin 4B protein ubiquitin ligase targets peroxiredoxin III for degradation. J Biol Chem 286(37):32344–32354. doi:10.1074/jbc.M111.249003
Miller DW, Ahmad R, Hague S, Baptista MJ, Canet-Aviles R, McLendon C, Carter DM, Zhu PP et al (2003) L166P mutant DJ-1, causative for recessive Parkinson’s disease, is degraded through the ubiquitin-proteasome system. J Biol Chem 278(38):36588–36595. doi:10.1074/jbc.M304272200
Ravid T, Hochstrasser M (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9(9):679–690. doi:10.1038/nrm2468
Asher G, Reuven N, Shaul Y (2006) 20S proteasomes and protein degradation “by default”. BioEssays 28(8):844–849. doi:10.1002/bies.20447
Sohal RS, Mockett RJ, Orr WC (2002) Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic Biol Med 33(5):575–586
da Cunha FM, Demasi M, Kowaltowski AJ (2011) Aging and calorie restriction modulate yeast redox state, oxidized protein removal, and the ubiquitin-proteasome system. Free Radic Biol Med 51(3):664–670. doi:10.1016/j.freeradbiomed.2011.05.035
Shang F, Gong X, Palmer HJ, Nowell TR Jr, Taylor A (1997) Age-related decline in ubiquitin conjugation in response to oxidative stress in the lens. Exp Eye Res 64(1):21–30. doi:10.1006/exer.1996.0176
Yang F, Yang YP, Mao CJ, Liu L, Zheng HF, Hu LF, Liu CF (2013) Crosstalk between the proteasome system and autophagy in the clearance of alpha-synuclein. Acta Pharmacol Sin 34(5):674–680. doi:10.1038/aps.2013.29
Petroi D, Popova B, Taheri-Talesh N, Irniger S, Shahpasandzadeh H, Zweckstetter M, Outeiro TF, Braus GH (2012) Aggregate clearance of alpha-synuclein in Saccharomyces cerevisiae depends more on autophagosome and vacuole function than on the proteasome. J Biol Chem 287(33):27567–27579. doi:10.1074/jbc.M112.361865
Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584(7):1393–1398. doi:10.1016/j.febslet.2009.12.047
Bove J, Zhou C, Jackson-Lewis V, Taylor J, Chu Y, Rideout HJ, Wu DC, Kordower JH et al (2006) Proteasome inhibition and Parkinson’s disease modeling. Ann Neurol 60(2):260–264. doi:10.1002/ana.20937
Kim YM, Jang WH, Quezado MM, Oh Y, Chung KC, Junn E, Mouradian MM (2011) Proteasome inhibition induces alpha-synuclein SUMOylation and aggregate formation. J Neurol Sci 307(1–2):157–161. doi:10.1016/j.jns.2011.04.015
Kordower JH, Kanaan NM, Chu Y, Suresh Babu R, Stansell J 3rd, Terpstra BT, Sortwell CE, Steece-Collier K et al (2006) Failure of proteasome inhibitor administration to provide a model of Parkinson’s disease in rats and monkeys. Ann Neurol 60(2):264–268. doi:10.1002/ana.20935
Manning-Bog AB, Reaney SH, Chou VP, Johnston LC, McCormack AL, Johnston J, Langston JW, Di Monte DA (2006) Lack of nigrostriatal pathology in a rat model of proteasome inhibition. Ann Neurol 60(2):256–260. doi:10.1002/ana.20938
Sun F, Anantharam V, Zhang D, Latchoumycandane C, Kanthasamy A, Kanthasamy AG (2006) Proteasome inhibitor MG-132 induces dopaminergic degeneration in cell culture and animal models. Neurotoxicology 27(5):807–815. doi:10.1016/j.neuro.2006.06.006
Xie W, Li X, Li C, Zhu W, Jankovic J, Le W (2010) Proteasome inhibition modeling nigral neuron degeneration in Parkinson’s disease. J Neurochem 115(1):188–199. doi:10.1111/j.1471-4159.2010.06914.x
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K et al (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441(7095):885–889. doi:10.1038/nature04724
Kuusisto E, Salminen A, Alafuzoff I (2001) Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport 12(10):2085–2090
Odagiri S, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K (2012) Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in alpha-synucleinopathy. Acta Neuropathol 124(2):173–186. doi:10.1007/s00401-012-0975-7
Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K (2011) Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol Dis 43(3):690–697. doi:10.1016/j.nbd.2011.05.022
Watanabe Y, Tatebe H, Taguchi K, Endo Y, Tokuda T, Mizuno T, Nakagawa M, Tanaka M (2012) p62/SQSTM1-dependent autophagy of Lewy body-like alpha-synuclein inclusions. PLoS One 7(12):e52868. doi:10.1371/journal.pone.0052868
Kuusisto E, Salminen A, Alafuzoff I (2002) Early accumulation of p62 in neurofibrillary tangles in Alzheimer’s disease: possible role in tangle formation. Neuropathol Appl Neurobiol 28(3):228–237
Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N (2004) Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem 91(1):57–68. doi:10.1111/j.1471-4159.2004.02692.x
Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J (2005) The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J Biol Chem 280(42):35625–35629. doi:10.1074/jbc.C500237200
Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT (2013) K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell 51(3):283–296. doi:10.1016/j.molcel.2013.06.020
Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E et al (2010) A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30(13):3275–3285. doi:10.1128/MCB.00248-10
Tofaris GK, Kim HT, Hourez R, Jung JW, Kim KP, Goldberg AL (2011) Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A 108(41):17004–17009. doi:10.1073/pnas.1109356108
Zucchelli S, Codrich M, Marcuzzi F, Pinto M, Vilotti S, Biagioli M, Ferrer I, Gustincich S (2010) TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is localized to Lewy bodies in sporadic Parkinson’s disease brains. Hum Mol Genet 19(19):3759–3770. doi:10.1093/hmg/ddq290
Mulherkar SA, Sharma J, Jana NR (2009) The ubiquitin ligase E6-AP promotes degradation of alpha-synuclein. J Neurochem 110(6):1955–1964. doi:10.1111/j.1471-4159.2009.06293.x
Lee FK, Wong AK, Lee YW, Wan OW, Chan HY, Chung KK (2009) The role of ubiquitin linkages on alpha-synuclein induced-toxicity in a Drosophila model of Parkinson’s disease. J Neurochem 110(1):208–219. doi:10.1111/j.1471-4159.2009.06124.x
Lee JT, Wheeler TC, Li L, Chin LS (2008) Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet 17(6):906–917. doi:10.1093/hmg/ddm363
Tofaris GK, Layfield R, Spillantini MG (2001) Alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett 509(1):22–26
Meier F, Abeywardana T, Dhall A, Marotta NP, Varkey J, Langen R, Chatterjee C, Pratt MR (2012) Semisynthetic, site-specific ubiquitin modification of alpha-synuclein reveals differential effects on aggregation. J Am Chem Soc 134(12):5468–5471. doi:10.1021/ja300094r
Haj-Yahya M, Fauvet B, Herman-Bachinsky Y, Hejjaoui M, Bavikar SN, Karthikeyan SV, Ciechanover A, Lashuel HA et al (2013) Synthetic polyubiquitinated alpha-synuclein reveals important insights into the roles of the ubiquitin chain in regulating its pathophysiology. Proc Natl Acad Sci U S A 110(44):17726–17731. doi:10.1073/pnas.1315654110
Liu C, Fei E, Jia N, Wang H, Tao R, Iwata A, Nukina N, Zhou J et al (2007) Assembly of lysine 63-linked ubiquitin conjugates by phosphorylated alpha-synuclein implies Lewy body biogenesis. J Biol Chem 282(19):14558–14566. doi:10.1074/jbc.M700422200
Klucken J, Poehler AM, Ebrahimi-Fakhari D, Schneider J, Nuber S, Rockenstein E, Schlotzer-Schrehardt U, Hyman BT et al (2012) Alpha-synuclein aggregation involves a bafilomycin A 1-sensitive autophagy pathway. Autophagy 8(5):754–766. doi:10.4161/auto.19371
Sampathu DM, Giasson BI, Pawlyk AC, Trojanowski JQ, Lee VM (2003) Ubiquitination of alpha-synuclein is not required for formation of pathological inclusions in alpha-synucleinopathies. Am J Pathol 163(1):91–100
Dixon C, Mathias N, Zweig RM, Davis DA, Gross DS (2005) Alpha-synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics 170(1):47–59. doi:10.1534/genetics.104.035493
Goel P, Manning JA, Kumar S (2015) NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene 557(1):1–10. doi:10.1016/j.gene.2014.11.051
Acknowledgments
This work was supported by the National Institutes of Health Grants P20RR17675, Centers of Biomedical Research Excellence (COBRE), GM108975 (OK), the Scientist Development Grant of the American Heart Association (12SDG12090015), the Office of Research of the University of Nebraska-Lincoln, and by the National Council of Science and Technology of Mexico (CONACYT) Grant #104316. JNY was supported by a CONACYT scholarship #219185. We would like to thank the Flow Cytometry Core Facility from the Nebraska Center for Virology for the access to the flow cytometry instrumentation (NIGMS grant number P30 GM103509) and the personnel at the Life Science Annex at UNL. EB was supported by the Iowa State University College of Veterinary Medicine (CVM) Summer Scholar Research Program (NIH 2T35OD012199-11A1).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Juliana Navarro-Yepes and Annadurai Anandhan contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 283 kb)
Rights and permissions
About this article
Cite this article
Navarro-Yepes, J., Anandhan, A., Bradley, E. et al. Inhibition of Protein Ubiquitination by Paraquat and 1-Methyl-4-Phenylpyridinium Impairs Ubiquitin-Dependent Protein Degradation Pathways. Mol Neurobiol 53, 5229–5251 (2016). https://doi.org/10.1007/s12035-015-9414-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9414-9