Abstract
Nicotine has a broad impact on both the central and peripheral nervous systems. Over the past decades, an increasing number of genes potentially involved in nicotine addiction have been identified by different technical approaches. However, the molecular mechanisms underlying nicotine addiction remain largely unknown. Under such situation, prioritizing the candidate genes for further investigation is becoming increasingly important. In this study, we presented a multi-source-based gene prioritization approach for nicotine addiction by utilizing the vast amounts of information generated from for nicotine addiction study during the past years. In this approach, we first collected and curated genes from studies in four categories, i.e., genetic association analysis, genetic linkage analysis, high-throughput gene/protein expression analysis, and literature search of single gene/protein-based studies. Based on these resources, the genes were scored and a weight value was determined for each category. Finally, the genes were ranked by their combined scores, and 220 genes were selected as the prioritized nicotine addiction-related genes. Evaluation suggested the prioritized genes were promising targets for further analysis and replication study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cigarette smoking is the most common form of tobacco use [1] and is one of the most significant sources of morbidity and death worldwide [2]. The World Health Organization estimates that there are 1.3 billion tobacco users worldwide, with more than 5 million dying from tobacco-related illness each year [3,4]. If the current patterns persist, annual tobacco-attributable deaths will rise to more than 10 million a few decades hence. Smoking presents key issues in public health in both developed and developing countries. For example, in the United States, more than 20 % of adults are current smokers [5], and cigarette smoking is responsible for approximately 438,000 premature deaths and an estimated economic cost of $167 billion annually [6]. In China, about 350 million people are smokers, and more than 50 % of the population is exposed to second-hand smoke, which results in 1.2 million annual deaths attributed to tobacco use [7]. Although a large fraction of smokers try to quit [5], available treatments are effective for only a fraction of them [8,9]. Thus, development of therapeutic approaches that can help smokers achieve and sustain abstinence from smoking, as well as methods that can prevent people from starting smoking, remains a huge challenge in public health.
Smoking is a complex behavior that involves the interplay of genetic and environmental factors [9–12]. As the main psychoactive ingredient responsible for smoking addiction, nicotine mainly evokes its physiological effects through interactions with nicotinic acetylcholine receptors (nAChRs) in the central nervous system. Nicotine exposure not only stimulates the mesocorticolimbic dopamine system in the outer shell of the nucleus accumbens and other brain regions [13–16], but also modulates the release of neurotransmitters such as norepinephrine, serotonin, and GABA [17–19]. Nicotine treatment can regulate the expression of genes/proteins involved in various functions including ERK1/2 and CREB [20], as well as their downstream targets such as c-FOS and FOSB [21–23]. Furthermore, biochemical pathways underlying various physiological processes, e.g., MAPK signaling, phosphatidylinositol phosphatase signaling, growth factor signaling, and ubiquitin–proteasome pathways, are modulated by nicotine [24–26]. Through its direct or indirect interactions with these genes and biological pathways, nicotine is involved in the regulation of various physiological processes, such as learning and memory, angiogenesis, energy metabolism, synaptic function, response to oxidative stress, and addiction [27–33].
Although nicotine exposure can evoke multiple effects in the neuronal system, the underlying molecular mechanism has not been completely understood. Studies have indicated that for complex behaviors like cigarette smoking, the individual differences can be attributed to hundreds of genes and their variants. Genes involved in different biological functions may act in concert to account for the risk of vulnerability to smoking behavior, with each gene having a moderate effect [34–36]. Rather than acting as sole factors, a large number of genes may cooperate in a synergistic manner in modifying the risk of smoking or responding to nicotine. Consistent with this belief, more and more genes have been found to be correlated with nicotine addiction over the past decades.
A lot of efforts have been devoted to identify the susceptibility genes and genetic markers underlying nicotine addiction via different approaches, among which include the techniques and experimental methods that focus on the function(s) or interaction(s) of one or a few genes/proteins, genetic association studies, linkage analysis, and high-throughput expression studies. Via these approaches, many genes potentially related to the physiological response to nicotine exposure or smoking behaviors have been identified, but none of these methods are powerful enough to identify all the molecular targets related to nicotine addiction. Although much of our knowledge of the molecular mechanisms underlying nicotine–neuron interaction has been accumulated through relatively traditional experiments or candidate gene-based genetic association analysis, these procedures usually focus on one or a small number of genes that may affect response to nicotine (e.g., nAChRs and nicotine metabolism) or the key neurotransmitter pathways (e.g., dopamine and serotonin) [37]. On the other hand, high-throughput technologies, such as microarray and proteomics approaches, and genome-wide association studies (GWASs) can provide information regarding genes’ functions and their interactions on a much larger scale without the requirement of preselecting target genes, and have been increasingly used to explore the genetic variants associated with nicotine addiction [33,38–43]. But these methods have their own limitations. For example, due to the complexities of the transcriptome and proteome of neuronal system, and the limitations of current technology, not all genes/proteins associated with brain disorder can be detected by microarray or proteomics approach reliably [43,44]; for GWAS, it has to overcome issues and limitations such as insufficient sample size, difficulty in control for multiple testing, and control for population stratification [45]. Moreover, for the many plausible candidate genes reported to be related to nicotine addiction, only a small number (e.g., nAChRs and dopamine signaling) have been partially replicated in different studies, the others have seldom been verified by independent analyses. This is especially true for high-throughput expression analysis and GWAS.
In such a situation, a systematic approach that is able to integrate information from different sources and to reveal the biochemical processes underlying the genes associated with nicotine exposure will not only help us to understand the relations of these genes, but also provide further evidence of the validity of these candidates. Till now, there are few studies devoted to collect those data together for the prioritization of genes related to nicotine addiction. This calls for an approach to integrate all the data sources to prioritize candidate genes for nicotine addiction in the further analysis.
In this study, we utilized a multi-source-based gene prioritization approach for nicotine addiction. In this approach, we collected and managed multiple genetic data sets of nicotine addiction or related phenotypes, including association studies, linkage analysis, gene expression studies, and single-gene/protein-based studies. By scoring the genes from different sources and assigning a weight to each source, we were able to rank the genes by their combined scores.
Materials and Methods
Identification of Nicotine Addiction-Related Genes
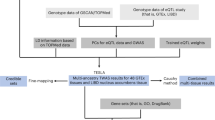
We utilized a comprehensive approach to prioritize candidate genes involved in biological response to nicotine. This approach included five steps, i.e., gene collection, gene scoring, weight optimization, gene prioritization, and evaluation. Genes were collected from the following four sources, i.e., association studies, linkage analysis, gene expression studies, and literature search of single-gene/protein-based studies. Second, we scored the candidate genes in the light of different categories. Third, we searched the optimal weight matrix by using simulated annealing (SA), with the values of different dimensions reflecting the importance of corresponding sources. Fourth, with the different matrix and scores of different categories, we got combined scores for the candidate genes and then prioritized them based on their combined scores. Fifth, we evaluated the top genes by gene set enrichment analysis. The framework of our study was shown in Fig. 1.
The flow chart for nicotine addiction-related genes prioritization. Genes are collected from four resources, i.e., association study, linkage analysis, gene expression analysis, and literature search of single-gene/protein-based studies. When a gene shows up in a certain category, a score of 1 point is assigned; otherwise, 0 is assigned. Each of the four categories has a weight value, which is determined by the optimization algorithm simulated annealing. The genes are ranked by their combined scores computed from scores corresponding to the four categories and their weights. Genes are ranked and prioritized by their combined scores, and further analysis is performed for the selected genes
Gene Collection
For association study, the list of candidate genes for smoking-related phenotypes was constructed by searching all human genetics association studies deposited in PUBMED (http://www.ncbi.nlm.nih.gov/pubmed/). Similar to earlier work [46,47], we queried the item “(Smoking [MeSH] OR Tobacco Use Disorder [MeSH]) AND (Polymorphism [MeSH] OR Genotype [MeSH] OR Alleles [MeSH]) NOT (Neoplasms [MeSH]),” and a total of 2,780 hits was retrieved by July 2013. The abstracts of these articles were reviewed, and the association studies of smoking-related behaviors, such as smoking initiation, smoking dependence, smoking cessation, or other neuronal disorders, were selected. From the selected publications, we narrowed our selection by focusing on those reporting a significant association of one or more genes with any of the phenotypes. To reduce the number of false-positive findings, the studies reporting negative or insignificant associations were not included, although it is likely that some genes analyzed in these studies might be associated with the phenotypes we were interested in. The full reports of the selected publications were reviewed to ensure the conclusions were supported by the content. From these studies, genes reported to be associated with each phenotype were selected for the current study. The results from several GWASs were also included [48–50]. For such studies, all the genes nominated to be nicotine addiction related by the original reports were included in our list. In another study on smoking cessation, Uhl et al. [40] performed a GWAS on three independent samples to identify genes facilitating smoking cessation success with bupropion hydrochloride versus nicotine replacement therapy. Multiple genes involved in cell adhesion, transcription regulation, transportation, and signaling transduction were suggested to contribute to successful smoking cessation. Among genes reported by Uhl et al., those showed significant association with smoking cessation in two or three samples were retrieved (63 genes). As a result, we retrieved 267 genes reported to be positive associated with nicotine addiction in the association studies.
Linkage analysis is useful in detecting genetic loci linked with susceptibility to nicotine addiction or smoking-related diseases. Multiple genome-wide linkage scans on smoking behavior have been performed using a variety of smoking behavior assessments. Numerous putative susceptibility loci have been identified. On the base of 15 genome-wide linkage scans of smoking behavior, Han et al. performed a comprehensive meta-analysis and identified the chromosome regions linked with smoking behavior with nominal significance [51]. From each of these chromosome regions, we retrieved all the genes within it. This resulted in a total of 5,701 genes, including both known genes with official gene symbols and the hypothetical genes.
The use of high-throughput expression profiling tools such as microarray and proteomics is becoming more integrated into research and their application to drug abuse studies is no exception. New insights gained through the use of microarray and proteomics technology have helped us to improve the understanding of the biological effects of drugs on animal or tissues of interest [25]. These techniques are also important to discover the genes/proteins that may be associated with nicotine addiction. Altogether, 33 datasets were collected from 31 publications reporting the effect of nicotine on cell lines or animal brains via microarray or proteomics; the genes or proteins reported to be significantly modulated by nicotine exposure were retrieved from these datasets. This resulted in a total of 1,938 genes.
A large fraction of our insight into the molecular mechanisms of nicotine addiction has been achieved via relatively traditional experimental approaches, which mainly focus on only one or a few genes or proteins for detailed analyses. Due to the large number of studies available, it is infeasible to collect all the publications to check the relations between genes/proteins and nicotine or smoking-related behaviors reported in them. Since co-occurrence of two items in a document can be utilized to identify their relationship [52], we searched the PUBMED for information on the potential correlation between genes and nicotine exposure. For this reason, this approach was referred to as literature search of single-gene/protein-based studies or simply as literature search in this study. Briefly, the human gene set was downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/gene/) and 26,811 known or predicted protein-coding genes extracted. Nicotine or tobacco smoking-related behaviors were evaluated with four terms, i.e., ‘smoking,’ ‘nicotine,’ ‘tobacco,’ and ‘nicotinic.’ For every gene, the combinations of the gene symbol and each of the four terms was used to query the related reports in PUBMED. For example, gene BDNF and term ‘nicotine’ formed a query item ‘BDNF and nicotine,’ and 68 hits were returned. If a gene has multiple aliases, then each alias was searched separately, and the result was then combined. If a gene had one or more hits with any of the four keywords, it was assigned 1 point, and if it did not co-occur with any of the keywords, 0 point was assigned. The total hit number of each gene was obtained by pooling all the hits of the combinations of its aliases and the four keywords searched. If a gene had a total hit number less than 5, then the abstracts of the corresponding articles were reviewed to make sure at least one study reporting the connection between gene and the keywords; otherwise, it was re-assigned 0 point. In total, 7,716 genes were collected via this approach.
Combined Scores
By these steps, we collected genes related to the effect of nicotine or tobacco smoking identified by genetic association analysis, genetic linkage analysis, high-throughput expression analysis, as well as single-gene/protein-based study approaches. When a gene has been identified by one approach, we assigned a score of 1 to it; otherwise, 0 was assigned. By this approach, we can evaluate the relation of a gene with nicotine addiction by analyzing the types of studies involved. However, the four types of evidences are not equal to each other. For example, when a gene is found to be significantly associated with smoking-related behavior in a genetic association study, then this study provides more specific evidence than another study reporting the inclusion of this gene in a chromosome region linked with the same behavior. Thus, different weight values should be assigned to different evidences when a gene has been analyzed by multiple types of studies.
The overall relation between a gene and nicotine addiction was measured by a combined score derived from its scores in the four categories, i.e.,
where n is the number of categories (n = 4), Score i is the score of a gene in the ith source, and w i is the corresponding weight value. When a gene has been identified in a source, Score i = 1; otherwise, Score i = 0.
To obtain the combined score S, a weight matrix is defined according to the relative importance of the four sources. The procedure used to prioritize the genes related to nicotine exposure is summarized in Fig. 1. Briefly, the genes collected from each of the four categories are assigned a category-specific score described above. For each gene, the scores multiply the corresponding weight values, and their sum is the combined score. Then, all the genes are ranked based on their combined scores, a gene with a higher rank in the list indicating a potential higher correlation with nicotine addiction.
Search for the Optimal Weight Matrix
The combined score of a gene depends on its scores from each category and the corresponding weight values. In order to prioritize the genes collected so that the genes more likely correlated with nicotine addiction can be ranked higher in the list, a suitable weight matrix needs to be determined. In this study, the following procedure was adopted:
-
1.
Randomly selecting weight value between 0 ~ 1.0 for each data category and normalizing the weight matrix to have a sum of 1;
-
2.
Calculating the combined score S for all genes by Eq. 1;
-
3.
Ranking all genes according to their combined scores;
-
4.
Calculating ratio R: the proportion of a set of genes known to be related to nicotine addiction in the top c percentage of all candidate genes accounting for all genes known to be related to nicotine addiction;
-
5
Making a small change to the weight matrix and normalizing the weight matrix to have a sum of 1;
-
6.
Repeating steps 2–5 until no larger R can be found, and then the weight matrix obtained is the optimal weight matrix.
In this procedure, in order to prioritize the genes collected, a set of genes known to be correlated with nicotine addiction or smoking-related phenotypes were utilized to set up the ranking criterion. Although multiple gene sets related to nicotine abuse have been reported [46,53], the genes suggested by Li and Burmeister [54] were selected in this study. Of the 62 candidate genes involved in the addiction of two or more drugs, such as nicotine, alcohol, heroin, cocaine, or amphetamine, 46 genes were suggested to be associated with the addiction of nicotine, and one or more other addictive drugs were retrieved. These genes were called base genes in this study (Supplemental Table 1).
In this study, 11,781 candidate genes potentially related to nicotine addiction were collected. The correlation between each gene and nicotine addiction was measured by its combined score. As mentioned above, a suitable weight matrix should assign higher ranks to the genes with higher correlation with nicotine addiction. Since it was unknown how many candidate genes should be selected, we used the proportion of known nicotine addiction-related genes (i.e., the base genes) included in the top c% candidate genes to evaluate the performance of the weight matrix. Obviously, for a larger c, more base genes would be included, but, at the same time, it had a larger chance to include genes less correlated with nicotine addiction. We tested different values for c, i.e., c = 2, 3, 4, or 5, and found that the selection of c did not affect the final weight matrix much. When c ≥ 3, the number of base genes included in the top c% candidate genes were same, and the algorithm converged to the optimal weight matrix quickly. Thus, c = 3 was selected to check how many members of the base genes were included in the selected candidate genes. Furthermore, the ratio R, i.e., the proportion of base genes in the top c% genes accounting for all base genes, was calculated. Since 46 base genes were collected, R = m/46, where m was the number of base genes included in the top c% candidate genes. According to this schema, a better weight matrix corresponded to a larger R value.
Instead of performing an exhaustive enumeration of all possible combinations of weight values, the weight matrix was optimized by SA algorithm [55]. SA is a generic probabilistic metaheuristic for both discrete and continuous global optimization problems [56,57]. Its inspiration comes from annealing in metallurgy, a technique involving heating and controlled cooling of a material to increase the size of its crystals and reduce their defects. SA is an iterative process that demands a variable T similar to temperature in annealing process in metallurgy. T starts initially with a high value and then gradually reduces toward zero at each step following an annealing schedule and acceptance criterion, with which the candidate solution is accepted or rejected. This process includes a means to escape the local optima by accepting worse solutions, but the chance of accepting a worse solution reduces as T decreases when solution space is searched. In this way, the system wanders initially towards a broad region of the search space containing good solutions, ignoring small features of the object function and then drifts towards regions including the optimal solutions. In our case, a random weight matrix was generated and used as the starting point for the algorithm. Then, the weight matrix was slightly modified to see whether more genes in the base gene set could be included in the top 3 % candidate genes. If yes, then the new weight matrix was used to replace the existing weight matrix; otherwise, a T-dependent probability was calculated to decide whether the existing weight matrix should be replaced or kept. Eventually, the search procedure converged to a weight matrix giving the highest R value.
With the optimal weight matrix, the combined scores of the candidate genes were calculated and used to rank the candidate genes.
Evaluation of the Prioritized Genes
The relation of the prioritized genes with nicotine addiction was evaluated by analyzing the Gene Ontology (GO) biological processes or biochemical pathways enriched in these genes. ToppGene (http://toppgene.cchmc.org) [58] was used for GO term enrichment analysis, in which the module ToppFun was able to detect functional enrichment of the input gene list based on transcriptome, proteome, regulome, GO, and so on. To simplify the analysis, only GO biological processes terms were selected. The exported terms were filtered by false discovery rate (FDR), only those with FDR value smaller than 0.05 were kept.
The biochemical pathways enriched in the prioritized genes were analyzed by Ingenuity Pathway Analysis (IPA; https://analysis.ingenuity.com) with the goal of revealing the enriched biochemical pathways. This pathway-based software is designed to identify global canonical pathways, dynamically generated biological networks, and global functions from a given list of genes. Basically, the genes with their symbol and/or corresponding GenBank Accession Numbers were uploaded into the IPA and compared with the genes included in each canonical pathway. All the pathways with one or more genes overlapping the candidate genes were extracted. In IPA, each of these pathways was assigned a p value, which denoted the probability of overlap between the pathway and input genes, via Fisher’s exact test. Because a relatively large number of pathways were examined, multiple comparison correction for the individually calculated p values was necessary in order to obtain reliable statistical inference. The pathways with FDR value less than 0.01, including three or more prioritized genes were considered to be significantly enriched. The corresponding FDR was calculated with the method of Benjamini and Hockberg [59].
Results
Genes Collected from the Four Sources
In this study, we collected the candidate genes mainly from four categories of resources, i.e., genetic association study, genetic linkage analysis, high-throughput gene expression study, and literature search of single-gene/protein-based studies. The numbers of genes collected from these categories were not equal. For association study, 267 genes were collected; for linkage analysis, 5,701 genes were included in human chromosome regions potentially linked with tobacco smoking; by microarray or proteomic expression analysis, 1,938 genes/proteins were identified to be differentially expressed under the treatment of nicotine while 7,716 genes were found to co-occur with ‘smoking,’ ‘nicotine,’ ‘tobacco,’ or ‘nicotinic’ in the abstracts of publications deposited in PUBMED (http://www.ncbi.nlm.nih.gov/pubmed). After removing the redundancy, a total of 11,781 candidate genes were left and were used as the candidate gene pool. The distribution of these genes among the four sources was shown in Table 1. Of the genes collected, 26 were identified in all of the four categories; 446 were identified in three of the four categories; 2,871 showed up in two of the four sources, and 8,438 genes were collected from only one resource.
Search for the Optimal Weight Matrix
As mentioned earlier, as long as a gene was identified by evidence from one category, it was assigned a score of 1 for this category; otherwise, 0 was assigned. On the other hand, information from each data resource could not be treated equally. For example, when the correlation between a gene and nicotine addiction is examined in a genetic association study, the gene usually is selected based on a priori information; for genes identified via GWASs, pre-selection of candidate genes is not necessary, but only a subset of genes significantly associated with the phenotype under investigation are identified [40,48–50]. In a typical GWAS, a gene is considered to be significantly associated with the phenotype under test, if one or more of single-nucleotide polymorphisms (SNPs) corresponding to the gene have p values smaller than a certain threshold. Due to the multiple testing issues, the p value threshold for significance should be corrected. If multiple SNPs are significantly associated with the phenotype, then the one with the smallest p value can be used to represent the relevance between this gene and the phenotype. Both of these two approaches provide more specific information than linkage analysis that identifies genomic regions co-segregating with a given phenotype. In this study, different weight values were assigned to the four categories. A larger weight for a category resulted in a higher score for the genes in this group. The final rank of each gene in the candidate gene list was based on its combined score derived from the weight values and the scores in the four categories.
The weight values for the four categories were searched by SA. According to our definition, a better weight matrix would assign higher ranks for the genes with larger correlations with nicotine addiction. We evaluated the weight matrix by measuring its performance in ranking the 46 base genes known to be involved in the addiction of nicotine, i.e., counting the numbers of the base genes included in the top 3 % (353 of the 11,781 genes) of all the candidate genes ranked by their combined scores given a weight matrix. The optimal weight matrix obtained by this procedure was [0.403 0.129 0.203 0.265], with the weight values corresponding to association study, linkage study, gene expression study, and literature search of single-gene/protein-based studies, respectively. With this weight matrix, 39 out of the 46 base genes (84.8 %) were included in the top 3 % of all the candidate genes ranked by combined scores. As a comparison, when the weights of the four categories were set to be equal, i.e., [1/4 1/4 1/4 1/4], 29 (63.0%) out of the 46 base genes were included in the top 3 % of all candidate genes, indicating the optimized weights led to higher ranks of the base genes in the overall candidate gene list.
Among these weights, the genetic association category had a higher value than the other three categories, which means association study may provide more reliable evidence in candidate gene search for complex diseases than the other approaches.
Identification of the Threshold
The correlation between the genes and nicotine addiction was measured by the combined scores. Compared with other genes, the base genes tended to have higher combined scores and thus were enriched in the top fraction of the gene list (Figs. 2 and 3). Most of the base genes had combined scores equal or higher than 0.665, while for the other genes only a small fraction had scores larger than this value. Based on such observation, two thresholds for the combined score were selected. For the first cutoff value (S 1 = 0.665), 220 candidate genes were selected (Supplemental Table 2), among which 38 base genes were included, and they accounted for 82.6 % of all the base genes. To make the prioritized gene list more comprehensive, another cutoff value was selected (S 2 = 0.598), with which 580 candidate genes were obtained. Among these genes, 39 base genes were included, which accounted for 84.8 % of all the base genes (Supplemental Table 2). When the threshold decreased from 0.665 to 0.598, more candidate genes were selected (580 vs. 220), but the number of base genes included remained stable (38 vs. 39). This was caused by the fact that most candidate genes had moderate or small combined scores, which probably indicated a higher false-positive rate among the prioritized genes as the combined scores became smaller. So, in the following analysis, we mainly focused on the 220 genes selected by the first threshold.
The distribution of the combined scores of the candidate genes. The genes are ranked by their combined scores. The x-axis is the order of the candidate genes. The y-axis on the left side is the combined score of the candidate genes, and the y-axis on the right side is the number of base genes. The solid line shows the distribution of the combined scores of candidate genes, and the dashed line shows the number of base genes included in the candidate genes. It can be seen that the score drops quickly from 1.0 to about 0.60 and then drops to about 0.47; after that, the combined scores decrease slowly. Such a distribution indicates that a relatively small number of genes have higher combined scores, while the majority genes have moderate or small scores
Distribution of the combined scores of all candidate genes and the base genes. The percentage of each histogram bin is measured by the genes with scores falling in the bin divided by the total number of candidate genes or the number of the base genes. Points marked by A and B are the two thresholds to select the genes
Biological Function of the Prioritized Genes
The biological function of these genes was analyzed by ToppGene Suite. The major enriched GO biological processes included cell–cell signaling, synaptic transmission, neurological system process, response to drug, and so on (Table 2). Most genes were associated with neurodevelopment-related processes and signal transduction. This outcome was consistent with the conclusion of Sun et al. based on the analysis of a group of addiction-related genes identified from genetic studies [60]. Also, it can be seen that some genes are associated with the transport of amine, which is consistent with the earlier reports [61,62]. So it may be concluded that our approach is reliable to prioritize candidate genes for complex diseases.
On the basis of the 220 genes potentially related to nicotine addiction, enriched biochemical pathways were identified by IPA and other bioinformatics tools (Table 3). Among these pathways, included are those signal transduction pathways related to neuronal function, e.g., cAMP-mediated signaling, calcium signaling, G-protein coupled receptor signaling, dopamine receptor signaling, serotonin receptor signaling, and glutamate receptor signaling. Pathways involved in drug or neurotransmitter metabolism were enriched in the genes, such as nicotine degradation II, dopamine degradation, xenobiotic metabolism signaling, and aryl hydrocarbon receptor signaling. Some immune response-related pathways were also enriched, e.g., role of cytokines in mediating communication between immune cells, T helper cell differentiation, IL-8 signaling, and ILK signaling.
Discussion
Over the past years, much has been learnt about the molecular mechanisms underlying nicotine addiction from studies on human subjects, animal, or cell models. Numerous genes and pathways have been found to play a role, either directly or indirectly, in smoking-related phenotypes. On the other hand, although more and more genes/proteins have been identified through various approaches, a detailed understanding of the biological processes related to the effects of nicotine treatment at the molecular level is still far from complete. Under such situation, integrating evidences obtained from different sources to prioritize the genes and the biochemical pathways associated with them will not only guide us to select the most likely vulnerable genes for further analysis, but also provide insight about the major biological mechanisms underlying nicotine addiction by reducing the potentially less important genes.
In this study, we utilized a multi-source-based approach to prioritize the genes involved in nicotine addiction. For the base genes known to be related to the addiction of nicotine and other addictive drugs, most of them were correctly included in the prioritized nicotine addiction-related gene list, indicating our method is reliable in identifying the potential targets by incorporating information from different sources.
Of the 220 genes in the prioritization list, genes having been studied extensively in nicotine addiction are include, such as the nicotinic receptors (e.g., CHRNA1, CHRNA4, CHRNA7, CHRNA10, and CHRNB2) and dopamine receptors (DRD1, DRD2, DRD3, DRD4 and DRD5). The GO biological processes enriched in these genes, e.g., cell–cell signaling, synaptic transmission, neurological system process, the transport of amine, and ion transport, are also among the major GO terms underlying a group of addiction-related genes identified from genetic studies [60]. Several essential biological processes, e.g., response to drug, response to alcohol, response to nicotine, response to cocaine, and response to organic nitrogen are also among the enriched GO terms. Thus, it is likely most of the prioritized genes may be involved in addiction-related biological functions, and our results, especially, provide further evidence that nicotine may share some biological mechanisms with other substances in addiction conditions.
Pathway analysis, which takes account of the biochemical relevance of genes, can not only be more robust to potential false-positives caused by various factors in different studies, but may also yield a more comprehensive view of the molecular mechanism underlying nicotine addiction. Thus, pathway analysis becomes more necessary to detect the main biological themes from the genes involved in different functions. Pathways enriched in the prioritized genes further reveal that the prioritized genes are involved in a wide range of biological processes. For example, we found that several signaling pathways related to neural activity are enriched in the prioritized genes, e.g., cAMP-mediated signaling, calcium signaling, dopamine receptor signaling, serotonin receptor signaling, glutamate receptor signaling, and synaptic long-term potentiation. In an earlier study, based on genes identified from genetic association analysis, we reported that these pathways were involved in different tobacco smoking-related behaviors, including smoking initiation/progression, nicotine addiction, and smoking cessation [46]. However, in this study, a larger gene set prioritized from multiple sources were used, and in each overrepresented pathway, more genes were included (Table 3). Moreover, some pathways not overrepresented in the earlier study were enriched in the current gene set, e.g., nicotine degradation II, dopamine degradation, serotonin degradation, noradrenaline and adrenaline degradation, and dopamine-DARPP32 feedback in cAMP signaling.
These results suggest that the mechanisms underlying nicotine addiction are complex and the pathways presented here, and the genes included may be potential targets for further investigation.
Several base genes were not among the 220 genes prioritized, which included CYP2E1, DDC, FAAH, GRIN1, HOMER1, HOMER2, OPRD1, and SLC6A2. CYP2E1 had a combined score of 0.606 and was included in the 580 prioritized genes. The other seven genes had values smaller than 0.598. The relation between these genes and nicotine addiction was examined by reviewing the available reports in PUBMED. For these genes, there are relatively few available reports on their roles in nicotine addiction, but some studies have focused on the association of these genes with the addiction of alcohol or other drugs. Thus, more investigation is needed to elucidate the roles of these genes in nicotine addiction. On the other hand, it means describing the correlation of a gene with nicotine addiction with a priori information used in this analysis is insufficient.
It should be noted that this study has some limitations. First, the prioritization procedure is based on the evidence from available association studies, linkage analyses, high-throughput expression analyses, and literature search of single-gene/protein-based studies, which means it cannot be used to predict novel genes related to nicotine addiction. Second, since the identification of susceptibility genes for nicotine addiction is still an ongoing process, the genes and pathways identified in this report are incomplete. At the same time, current available studies are unbalanced and incomprehensive. For example, for single-gene/protein-based studies, the candidates selected are usually biased toward better-studied targets; for high-throughout gene expression analysis, although no candidates are pre-selected, the results are still limited by available models and experiment conditions. It can be expected that, as more studies become available in each category, more genetic factors and pathways related to nicotine addiction will be determined.
There are several available studies devoted to the collection and prioritization of nicotine addiction related genes. By comparing genes located in chromosome regions implicated in nicotine addiction from a genome-wide linkage scan with a list of genes suggested by microarray studies of experimental nicotine exposure and candidate genes from the literature, Sullivan et al. [47] found that genes such as mitogen-activated protein kinase (MAPK), nuclear factor kappa B (NFKB), neuropeptide Y (NPY), nicotinic receptor subunit alpha 2 (CHRNA2), and related biochemical pathways were identified by all these approaches. They further developed a bioinformatic tool to provide a searchable archive of findings from genome-wide linkage, genome-wide association, and microarray studies for psychiatric disorders including nicotine addiction [63]. By integrating two gene resources (Entrez Gene and HomoloGene) and three pathway resources (KEGG, Reactome, and BioCyc), Sahoo et al. created a semantic web-based knowledge base for nicotine dependence, which could be used for gene query and pathway identification [64]. To balance the statistical evidence of genotype–phenotype correlation with the a priori evidence of biological relevance in a GWAS for complex disorders like nicotine addiction, Saccone at al. [65] developed a method to prioritize SNPs for further study after a GWAS. The method combined evidence from genotype–phenotype correlation, known pathways, SNP/gene functional properties, comparative genomics, prior evidence of genetic linkage, and linkage disequilibrium. In an earlier study [46], we collected genes associated with the risk of smoking initiation and progression, nicotine dependence, and smoking cessation by reviewing the related association studies. Further analysis revealed a number of common and unique pathways enriched in the genes associated with these phenotypes. These studies have demonstrated that integrating information from different sources to explore the molecular candidates related to nicotine addiction is not only feasible, but also necessary in order to obtain a comprehensive and unbiased understand about these genetic factors. These reports mainly focused on some specific types of study such as linkage analysis or association study, or a relatively small number of studies. The current study, on the other hand, has tried to provide a more comprehensive collection and analysis on the information from different sources.
By applying their method to a GWAS data [48,49], Saccone et al. [65] prioritized the SNPs associated with nicotine dependence. Of the top ten prioritized SNPs, nine SNPs could be mapped to eight candidate genes, i.e., rs16969968 (CHRNA5), rs1051730 (CHRNA3), rs6474413 (CHRNB3), rs578776 (CHRNA3), rs4142041 (CTNNA3), rs999 (PBX2), rs12623467 (NRXN1), rs12380218 (VPS13A), and rs2673931 (TRPC7). Among these eight genes, seven were included in the 220 prioritized genes in our study except PBX2 (this gene was not included in our candidate genes), indicating the high accuracy of our method. Similarly, Lewinger et al. [66] developed a hierarchical regression modeling approach to prioritize a subset of SNPs from a genome-wide association scan for further test. Rather than simply selecting a subset of most significant SNPs at certain cutoff, they utilized a prior model and included the prior information of the markers, such as their location relative to genes or evolutionary conserved regions, or prior linkage or association data. Then, the SNPs on the top ranked posterior expectations were selected for confirmation in following analysis. The methods developed by Saccone at al. and Lewinger et al., as well as the one described in this study, all take advantage of the prior knowledge. While those two methods are devoted for the prioritization of SNPs in a genome-wide association scan, our method is more suitable for the prioritization of genes related to complex diseases.
In conclusion, by incorporating information from multiple sources, we developed a gene prioritization approach to prioritize nicotine addiction-related genes. Evaluation suggested this approach was reliable for candidate gene prioritization for complex diseases, and the prioritized genes and the related pathways were potential targets for further analysis and replication study.
References
Smith SS, Fiore MC (1999) The epidemiology of tobacco use, dependence, and cessation in the United States. Prim Care 26(3):433–461
Murray S (2006) A smouldering epidemic. CMAJ 174(3):309–310
Jha P, Peto R (2014) Global effects of smoking, of quitting, and of taxing tobacco. N Engl J Med 370(1):60–68
Jha P (2009) Avoidable global cancer deaths and total deaths from smoking. Nat Rev Cancer 9(9):655–664
CDC (2007) Cigarette smoking among adults—United States, 2006. Morb Mortal Wkly Rep [serial online] 56(44):1157–1161
CDC (2005b) Annual smoking-attributable mortality, years of potential life lost, and economic costs—United States, 1997--2001. Morb Mortal Wkly Rep [serial online] 54:625–628
Zhang J, Ou JX, Bai CX (2011) Tobacco smoking in China: prevalence, disease burden, challenges and future strategies. Respirology 16(8):1165–1172
Hughes JR, Stead LF, Lancaster T (2007) Antidepressants for smoking cessation. Cochrane Database Syst Rev 1:CD000031
Lerman CE, Schnoll RA, Munafo MR (2007) Genetics and smoking cessation improving outcomes in smokers at risk. Am J Prev Med 33(6 Suppl):S398–S405
Lerman C, Berrettini W (2003) Elucidating the role of genetic factors in smoking behavior and nicotine dependence. Am J Med Genet B Neuropsychiatr Genet 118B(1):48–54
Goode EL et al (2003) Multiple genome-wide analyses of smoking behavior in the Framingham Heart Study. BMC Genet 4(Suppl 1):S102
Osler M et al (2001) Influence of genes and family environment on adult smoking behavior assessed in an adoption study. Genet Epidemiol 21(3):193–200
Tammimaki A et al (2006) Effect of quinpirole on striatal dopamine release and locomotor activity in nicotine-treated mice. Eur J Pharmacol 531(1–3):118–125
Gaddnas H et al (2001) Enhanced motor activity and brain dopamine turnover in mice during long-term nicotine administration in the drinking water. Pharmacol Biochem Behav 70(4):497–503
Benwell ME, Balfour DJ (1992) The effects of acute and repeated nicotine treatment on nucleus accumbens dopamine and locomotor activity. Br J Pharmacol 105(4):849–856
Benwell ME, Balfour DJ (1997) Regional variation in the effects of nicotine on catecholamine overflow in rat brain. Eur J Pharmacol 325(1):13–20
Barik J, Wonnacott S (2006) Indirect modulation by alpha7 nicotinic acetylcholine receptors of noradrenaline release in rat hippocampal slices: interaction with glutamate and GABA systems and effect of nicotine withdrawal. Mol Pharmacol 69(2):618–628
Barik J, Wonnacott S (2009) Molecular and cellular mechanisms of action of nicotine in the CNS. Handb Exp Pharmacol 192:173–207
Kenny PJ, File SE, Rattray M (2001) Nicotine regulates 5-HT(1A) receptor gene expression in the cerebral cortex and dorsal hippocampus. Eur J Neurosci 13(6):1267–1271
Brunzell DH, Russell DS, Picciotto MR (2003) In vivo nicotine treatment regulates mesocorticolimbic CREB and ERK signaling in C57Bl/6J mice. J Neurochem 84(6):1431–1441
Pagliusi SR et al (1996) The reinforcing properties of nicotine are associated with a specific patterning of c-fos expression in the rat brain. Eur J Neurosci 8(11):2247–2256
Nisell M et al (1997) Chronic nicotine enhances basal and nicotine-induced Fos immunoreactivity preferentially in the medial prefrontal cortex of the rat. Neuropsychopharmacology 17(3):151–161
Soderstrom K et al (2007) Nicotine increases FosB expression within a subset of reward- and memory-related brain regions during both peri- and post-adolescence. Psychopharmacology (Berl) 191(4):891–897
Tang K et al (1998) A crucial role for the mitogen-activated protein kinase pathway in nicotinic cholinergic signaling to secretory protein transcription in pheochromocytoma cells. Mol Pharmacol 54(1):59–69
Li MD et al (2004) Time-dependent changes in transcriptional profiles within five rat brain regions in response to nicotine treatment. Brain Res Mol Brain Res 132(2):168–180
Konu O et al (2001) Region-specific transcriptional response to chronic nicotine in rat brain. Brain Res 909(1–2):194–203
Dasgupta P, Chellappan SP (2006) Nicotine-mediated cell proliferation and angiogenesis: new twists to an old story. Cell Cycle 5(20):2324–2328
Dasgupta P et al (2006) Nicotine induces cell proliferation by beta-arrestin-mediated activation of Src and Rb-Raf-1 pathways. J Clin Invest 116(8):2208–2217
Robinson TE, Kolb B (2004) Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology 47(Suppl 1):33–46
Dajas-Bailador F, Wonnacott S (2004) Nicotinic acetylcholine receptors and the regulation of neuronal signalling. Trends Pharmacol Sci 25(6):317–324
Harkness PC, Millar NS (2002) Changes in conformation and subcellular distribution of alpha4beta2 nicotinic acetylcholine receptors revealed by chronic nicotine treatment and expression of subunit chimeras. J Neurosci 22(23):10172–10181
Kane JK et al (2004) Nicotine coregulates multiple pathways involved in protein modification/degradation in rat brain. Brain Res Mol Brain Res 132(2):181–191
Hwang YY, Li MD (2006) Proteins differentially expressed in response to nicotine in five rat brain regions: identification using a 2-DE/MS-based proteomics approach. Proteomics 6(10):3138–3153
Lessov CN et al (2004) Genetics and drug use as a complex phenotype. Subst Use Misuse 39(10–12):1515–1569
Tyndale RF (2003) Genetics of alcohol and tobacco use in humans. Ann Med 35(2):94–121
Hall W, Madden P, Lynskey M (2002) The genetics of tobacco use: methods, findings and policy implications. Tob Control 11(2):119–124
Ho MK, Tyndale RF (2007) Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J 7(2):81–98
Loukola A et al. (2013) Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol Psychiatry
Thorgeirsson TE et al (2010) Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 42(5):448–453
Uhl GR et al (2008) Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry 65(6):683–693
Wang J et al. (2007) Strain- and region-specific gene expression profiles in mouse brain in response to chronic nicotine treatment. Genes Brain Behav
Wang J et al (2009) Significant modulation of mitochondrial electron transport system by nicotine in various rat brain regions. Mitochondrion 9(3):186–195
Wang J, Yuan W, Li MD (2011) Genes and pathways co-associated with the exposure to multiple drugs of abuse, including alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine, and/or nicotine: a review of proteomics analyses. Mol Neurobiol 44(3):269–286
Blalock EM et al (2005) Harnessing the power of gene microarrays for the study of brain aging and Alzheimer's disease: statistical reliability and functional correlation. Ageing Res Rev 4(4):481–512
Pearson TA, Manolio TA (2008) How to interpret a genome-wide association study. JAMA 299(11):1335–1344
Wang J, Li MD (2010) Common and unique biological pathways associated with smoking initiation/progression, nicotine dependence, and smoking cessation. Neuropsychopharmacology 35(3):702–719
Sullivan PF et al (2004) Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am J Med Genet B Neuropsychiatr Genet 126B(1):23–36
Bierut LJ et al (2007) Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 16(1):24–35
Saccone SF et al (2007) Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 16(1):36–49
Uhl GR et al (2007) Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet 8:10
Han S et al (2010) Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biol Psychiatry 67(1):12–19
Roberts PM (2006) Mining literature for systems biology. Brief Bioinform 7(4):399–406
Li CY, Mao X, Wei L (2008) Genes and (common) pathways underlying drug addiction. PLoS Comput Biol 4(1):e2
Li MD, Burmeister M (2009) New insights into the genetics of addiction. Nat Rev Genet 10(4):225–231
Dekkers A, Arats E (1991) Global optimization and simulated annealing. Mathmatical Programing 50(1-3):367–393
Corana A et al (1987) Minimizing multimodal functions of continuous variables with the ‘simulated annealing’ algorithm. ACM Trans Math Softw 13(3):262–280
Locatelli M (2000) Simulated annealing algorithms for continuous global optimization. J Optim Theory Appl 104(1):121–133
Chen J et al (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37(Web Server issue):W305–W311
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57(1):289–300
Sun J, Zhao Z (2010) Functional features, biological pathways, and protein interaction networks of addiction-related genes. Chem Biodivers 7(5):1153–1162
Zhu J, Reith ME (2008) Role of the dopamine transporter in the action of psychostimulants, nicotine, and other drugs of abuse. CNS Neurol Disord Drug Targets 7(5):393–409
Sulzer D (2011) How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron 69(4):628–649
Konneker T et al (2008) A searchable database of genetic evidence for psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 147B(6):671–675
Sahoo SS et al (2008) An ontology-driven semantic mashup of gene and biological pathway information: application to the domain of nicotine dependence. J Biomed Inform 41(5):752–765
Saccone SF et al (2008) Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics 24(16):1805–1811
Lewinger JP et al (2007) Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet Epidemiol 31(8):871–882
Acknowledgments
This work was supported in part by grants from National Natural Science Foundation of China (Grant No. 31271411) and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry of China. We are grateful to Prof. Ming D Li of University of Virginia for his help on this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xinhua Liu and Meng Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, X., Liu, M., Li, X. et al. Prioritizing Genes Related to Nicotine Addiction Via a Multi-source-Based Approach. Mol Neurobiol 52, 442–455 (2015). https://doi.org/10.1007/s12035-014-8874-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-8874-7