Abstract
Streptomyces lividans uses mainly two pathways to target secretory proteins to the cytoplasmic membrane. The major pathway (Sec pathway) transports pre-proteins using the signal recognition particle, and the minor Tat pathway is responsible for the secretion using a folded conformation of a relatively low number of proteins. The signal peptides of the Sec-dependent alpha-amylase and the Tat-dependent agarase were interchanged and fused in-frame to the corresponding mature part of the other enzyme. Alpha-amylase was unable to use the Tat route when fused to the agarase signal peptide, while agarase used the Sec route when it was targeted by the alpha-amylase signal peptide. In addition to the signal peptide some yet unidentified parts of the secreted proteins may play a role in selecting the secretory route. Structure predictions for the Tat- and Sec-dependent proteins suggest that less structured proteins are more likely to be candidates for the Tat route.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomycetes are Gram-positive soil bacteria that have been used as a host for the secretory production of homologous and heterologous proteins employing Streptomyces lividans in particular as a host, a strain that comprises a relatively less active restriction modification system and a low endogenous protease activity in comparison to many other streptomycetes [1].
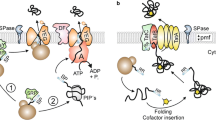
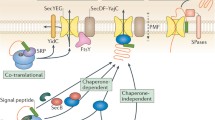
Bacteria generally make use of two pathways to target secretory proteins to the cytoplasmic membrane. The major pathway (Sec pathway), which transports pre-proteins using the signal recognition particle (SRP) [2] to interact with SecA that subsequently interacts dynamically with the channel formed by the heterotrimeric complex, SecYEG, to export the protein in an unfolded conformation. The second pathway, the Tat pathway, was initially discovered in the thylakoid membranes of plant plastids, and is shown to be a minor pathway responsible for exporting relatively few Tat proteins in Escherichia coli and Bacillus subtilis [3, 4] in a folded conformation. Up to 27 proteins have been found to be substrates of the S. coelicolor Tat system [5, 6]. TatA, TatB and TatC constitute the S. lividans Tat system [7]. In S. lividans soluble TatA and TatB form a heterocomplex that recognises the secretory pre-protein. The TatAB complex brings the substrate directly to TatC, which mediates the insertion of the precursor-Tat complex into the membrane. Subsequently, a functional TatABC translocation system is formed where TatA oligomerizes, enabling the pore to export a folded protein [8].
Secretory proteins have an amino-terminal sequence, the signal peptide, which is required for the correct targeting to the translocation pathway and must be cleaved by a signal peptidase type I during the course of translocation or shortly after translocation. Signal peptides have a similar overall architecture with a tripartite structure consisting of an N-, H- and C-region. Despite the fact that Sec- and Tat-targeting signal peptides are similar, there are differences that are crucial to facilitating the means to target the correct export pathway. Tat signal peptides contain a highly conserved twin-arginine motif, SRRXFLK, where the consecutive arginine residues are almost invariant, the Tat signal peptide H-regions are less hydrophobic than the equivalent Sec ones and the Tat signal peptide C-regions contain basic residues, which are currently not found in the C-region of the Sec signal peptides [9].
In the past, co-immunoprecipitation experiments performed in our laboratory using polyclonal antibodies against agarase, a Tat protein, or against the SRP components, indicated that pre-agarase was able to co-immunoprecipitate with the SRP components, suggesting that this protein may potentially be secreted via the Sec route in S. lividans [2].
To further investigate this, we constructed chimera protein coding sequences containing the signal peptide of a Sec protein (alpha-amylase) fused in-frame to the mature agarase coding sequence, or the mature alpha-amylase fused to the agarase signal peptide. The chimeras were propagated in the wild-type S. lividans strain and in the defective strains in the translocase complex (∆secG) or in the Tat route (∆tatC) in order to assess the capacity of the corresponding signal peptides to target the respective linked protein to the secretory route, which is not generally used by that protein. The obtained results confirmed that agarase could be secreted via the Sec route in S. lividans while there was no alpha-amylase secreted via the Tat route.
Materials and Methods
Bacterial Strains, Plasmids and Media
Streptomyces lividans TK21 [10], S.lividans ∆secG [11] and S. lividans ∆tatC are the wild type, and the secG and tatC are the deficient strains, respectively.
Plasmid pAMI11 [11] is a pIJ486 [12] derivative and plasmid pAGAs5 [13] is a pAGAs1 [14] derivative, carrying the S. lividans gene amlB and the S. coelicolor agarase gene (dagA), respectively, as well as a frame-shift-mutated thiostrepton resistance gene.
Plasmid pAMI2003 is a pAGAs20 [15] derivative carrying a chimerical agarase-alpha-amylase gene, and plasmid pAGA502 is a pAGA500 derivative carrying a chimerical alpha-amylase-agarase gene.
Plasmid pAMI2003 contains a sequence comprising the agarase gene expression control elements, and the sequence coding for the signal peptide and the first seven amino acids of the mature agarase protein fused in-frame to the mature alpha-amylase coding sequence. Plasmid pAGA502 contains a sequence comprising the agarase gene expression control elements, and the sequence coding for the signal peptide and the first three amino acids of the mature alpha-amylase fused in-frame to the mature agarase coding sequence.
Both plasmids were propagated in S. lividans TK21, S. lividans ∆secG and S.lividans ∆tatC.
Streptomyces strains were cultured in liquid NMMP medium at 30 °C in a rotary shaker at 250 rpm in the presence of the antibiotic required for the maintenance of the different plasmids (apramycin, kanamycin and/or thiostrepton, 10 μg/ml each).
When propagating the different recombinant strains in solid medium, apramycin (25 μg/ml), thiostrepton (50 μg/ml) and kanamycin (50 μg/ml) were added to the solid media, when required.
Construction of the tatC Mutant
A 7499-bp fragment containing tatC was amplified from the S. lividans TK21 chromosome using oligonucleotide primers TatC.e.FR (5′ GGTACGGTCATCCGTCGTAA3′) and TatC.e.RE (5′CCATGTAGGCCCTCTGAAAA 3′). The DNA fragment was cloned in pCR-XL-TOPO (Invitrogen, Life Technologies) to generate the plasmid pCR-XL-TatC. Subsequently, the ReDirect methodology was used, as previously described [16]. The disruption cassette encompassing oriT and the apramycin resistance gene (AmR) contained in plasmid pIJ773 [16] was generated by PCR amplification using oligonucleotide primers TatC.APRA.FR2
(5′TGATCTCCGGCCCGCCGCACGAGATGGGAACGTGGGTTGATTCCGGGGATCCGTCGACC 3′) and TatC.APRA.RE2 (5′GGGCGGCGACGCGTGCCGTCACCGCCCCGGCGGTCCTCATGTAGGCTGGAGCTGCTTC 3′), both of which carried DNA extensions around the start and stop codons of the Tat coding sequence.
The resulting amplified DNA fragment was gel purified and electroporated into E. coli BW25113 carrying the plasmid pCR-XL-TatC and the compatible plasmid pIJ779, which harbour the RED functions necessary for linear recombination events [ 16]. The selected recombinant was propagated in E. coli ET12567 (pUZ8002) and then introduced into S. lividans TK21 by conjugation, as previously described [16]. Double-crossover mutants were selected for their resistance to apramycin and sensitivity to kanamycin. The selected ∆tatC mutant strain (S. lividans ∆tatC) was further analysed by PCR amplification using oligonucleotide primers TatC.FR (5′ CTCAAGAGCGAGGCCAAG 3′) and TatC.RE (5′TCTGTCATCGTGCTTTCGAG 3′).
Protein Analysis and Western Blot Experiments
Standard extracellular protein analyses were essentially carried out as described [17]. For cell-associated protein analysis, cells grown in NMMP medium at different times of growth (16, 24, 36, 48 and 60 h) were harvested by centrifugation at 1400×g for 10 min, washed with P buffer [18] and sonicated on ice for three bursts of 5 s and resuspended in P buffer containing the EDTA-free protease inhibitor cocktail (Roche). For extracellular protein analyses, TCA was added at 10 % final concentration to the supernatants and the mixture was incubated at −20 °C for 1 h to precipitate the extracellular proteins. The proteins were then separated by centrifugation at 15,000×g for 20 min at 4 °C. Protein pellets were washed twice with ice-cold acetone and the residual acetone was removed by air-drying. Protein pellets were resuspended in 10 mM Tris-HCl (pH 7.5).
For Western blot analysis, cell-associated and extracellular proteins were fractionated by SDS-PAGE in 10 and 12 % (w/v) acrylamide gel [19]. For alpha-amylase and agarase detection, the fractionated protein was transferred onto immobilon polyvinylidene difluoride membranes (Millipore), as described [ 20]. The equivalent amount of protein loaded onto the SDS-PAGE acrylamide gel was corrected by the bacterial dry weight in each case. A ten-fold excess of extracellular protein was loaded onto the SDS gels in all cases [21].
The transferred material was incubated with rabbit polyclonal antibodies raised against S. lividans TK21 alpha-amylase (AmlB; a gift from C. Isiegas) and S. coelicolor agarase (DagA; [ 22] ) followed by incubation with HRP-conjugated protein A (Invitrogen Laboratories) diluted 1:10,000 in PBS containing 5 % (w/v) skimmed milk for 40 min at room temperature [23]. Peptides reacting with the antibodies were revealed using the ECL enhanced chemiluminescence system from Amersham after one min incubation and different exposures to X-ray film ranging from 20 s to 3 min.
The protein concentration in the different samples was determined using the BCA protein assay kit (Pierce) as indicated by the supplier. An equivalent amount of protein from each sample was analysed by SDS-PAGE.
Enzyme Activities
To determine extracellular alpha-amylase and agarase activity, the supernatants from the aliquots of bacterial cell cultures were concentrated at the indicated phases of growth by precipitation with ammonium sulphate brought to 80 % saturation; the precipitated protein was collected by centrifugation at 13,000×g for 30 min and dissolved in 20 mM phosphate buffer (pH 7) for alpha-amylase and in 50 mM imidazole–HCl (pH 6.5) for agarase. Alpha-amylase and agarase activities were determined as previously described [21, 22].
One unit of enzyme activity is the amount of enzyme that increased absorbance at 540 nm (alpha-amylase) or at 450 nm (agarase) by 0.001 per minute of incubation under the assay conditions. The specific activity was expressed as units per mg of dry weight. The enzyme activities used for representations are the average values of three independent experiments. The protein concentration in the different samples was determined using the BCA protein assay kit (Pierce), as indicated by the supplier.
Protein Structure
Secondary structure predictions were obtained using EMBOSS software [24] and online web servers JPred [25 ], Findex [26], SymPred [27] and Raptor-X [28]. The presence of disulphide bonds in the proteins was predicted using DIANNA [29].
Results and Discussion
A classic approach to engineering protein secretion in Streptomyces strains involves transferring the signal peptide from efficiently secreted proteins to others, so that the chimerical pre-protein can ideally be targeted more efficiently to the bacterial translocation machinery, something which has been effective for a small set of proteins to be exported via the major Sec route [1]. The discovery of a secondary secretion route in bacteria, the Tat route, where the secreted proteins appeared correctly folded in the cell culture supernatants, paved the way for a possible exploitation of this system to engineer the production of extracellular proteins whose activity no longer depended on folding enzymes. Hence, the Streptomyces Tat system deserves to be studied further as regards its possible exploitation for secretory protein production. Our previous experiments indicated that the pre-agarase Tat protein was able to co-immunoprecipitate with the SRP components [2], thus prompting us to further explore the feasibility of the potential use of the Sec system for agarase secretion in S. lividans, and of a Sec protein such as alpha-amylase to be targeted to the Tat route as well.
Alpha-amylase and agarase were overproduced when their respective genes were propagated in S. lividans TK21 harbouring multicopy plasmids pAMI11 and pAGAs5, respectively. These plasmids were also propagated in S. lividans TK21 strains deficient either in the Sec route translocase complex component SecG [11] or in the Tat secretory route component TatC. Deletion of tatC in S. lividans TK21 resulted in a deficient sporulation phenotype and a tendency to aggregate the mycelium in clumps when grown in liquid medium; the mycelium mass remained smaller with respect to that of the overproducer wild-type strains when grown in liquid medium, as described (Figs. 1, 2 and 3; [30]). The mycelium of the S. lividans TatC-deficient strain appears to be more fragile than that of the wild-type one and prone to lysis on shaking as previously described [5].
Growth curves of the S. lividans TK21, S. lividans ΔsecG and S. lividans ΔtatC. Time course at different times of growth (16, 24, 36, 48 and 60 h) of the S. lividans TK21 (black circles), S. lividans ΔsecG (white circles) or S. lividans ΔtatC (black squares) harbouring pIJ486. The data are the average of at least three independent determinations. Bars show standard error
Growth curves, alpha-amylase secretion and extracellular activity in S. lividans TK21, S. lividans ΔsecG and S. lividans ΔtatC harbouring plasmids pAMI11 or pAMI2003. Time course at different times of growth (16, 24, 36, 48 and 60 h) of the strains harbouring alpha-amylase synthesised under the control of its own promoter and exported by its own signal peptide in S. lividans TK21 (black circles), S. lividans ΔsecG (white circles) or S. lividans ΔtatC (black squares) (a) compared with the strains harbouring alpha-amylase synthesised under the control of the dagA promoter and exported by the DagA signal peptide in the three different bacterial hosts (b). The data are the average of at least three independent determinations. Bars show standard error. Cell-associated and extracellular amylase at different times of growth present in different strains containing the pAMI11 plasmid (a) and the pAMI2003 plasmid (b) at different times of growth were analysed by Western blotting with antibodies raised against AmlB. The amount of protein loaded onto the gels was corrected by the dry weight of the bacterial cell cultures. Arrows indicate the relative mobility of the mature AmlB. The corresponding alpha-amylase activities are shown. Alpha-amylase (c) activity was determined. The specific extracellular activity was expressed as units per mg of dry weight. The data are the average of at least three independent determinations. Bars show the standard errors
Growth curves, agarase secretion and extracellular activity in S. lividans TK21, S. lividans ΔsecG and S. lividans ΔtatC harbouring plasmids pAGAs5 or pAGA502. Time course at different times of growth (16, 24, 36, 48 and 60 h) of the strains harbouring agarase synthesised under the control of its own promoter and exported by its own signal peptide in S. lividans TK21 (black circles), S. lividans ΔsecG (white circles) or S. lividans ΔtatC (black squares) (a) is compared with that synthesised under the control of the dagA promoter and exported by the AmlB signal peptide in the three different bacterial hosts (b). The data are the average of at least three independent determinations. Bars show standard error. Cell-associated and extracellular amylase present in different strains containing the pAGAs5 plasmid (a) and the pAGA502 plasmid (b) at different times of growth were analysed by Western blotting with antibodies raised against DagA. The amount of protein loaded onto the gels was corrected by the dry weight of the bacterial cell cultures. Arrows indicate the relative mobility of the pre-mature DagA and mature DagA. Agarase activity was determined (c). The specific extracellular activity was expressed as units per mg of dry weight. The data are the average of at least three independent determinations. Bars show the standard errors
No precursor or mature forms of the proteins were revealed by the anti-AmlB or anti-DagA serum to be cell-associated with the alpha-amylase or agarase overproducers strains carrying plasmids pAMI11 or pAGAs5, respectively, suggesting that synthesis, transport and secretion of alpha-amylase and agarase appears to take place very efficiently in these strains (Figs. 2a, 3a).
Alpha-amylase (AmlB) is a secretory protein that is transported and secreted outside the S. lividans cell by the Sec pathway, hence when alpha-amylase was oversynthesised in the SecG-deficient strain it appears later in the supernatant, as expected (Fig. 2a; [11]), which is not the case when the protein is overproduced in the Tat-deficient strain, where SecG is fully functional (Fig. 2a).
The amount of extracellular alpha-amylase in the Tat-deficient strain is much smaller than in the case of the wild type, as expected, due to the less efficient growth of the TatC-defective strain in the liquid medium [5].
The maximum level of mature and active AmlB appeared at the exponential phase of growth and decreased when the culture reached the stationary phase (Fig. 2a; [21 ]), while in the case of the Tat protein, agarase (DagA) [5], the mature protein accumulates extracellularly at the stationary phase (Fig. 3a). The SecG deficiency apparently does not affect agarase production (Fig. 3a), as expected for a Tat-dependent protein.
TatC deficiency will render the Tat route non-operational and impede any processing of the intracellular pre-agarase, thus, no extracellular agarase was expected to be found in the TatC-deficient strain. Nevertheless, a small amount of mature agarase appeared at the TatC-deficient strain supernatants (Fig. 3a). This protein could result from pre-agarase being targeted by its own signal peptide to the functional Sec route, as described [2]. The TatC-deficient strain is prone to lysis [5], and should the appearance of this protein be caused by cellular lysis, one would expect to visualise the precursor form of the agarase rather than the detected mature enzyme, which turned out to be active (Fig. 3a, c).
Detection of extracellular enzymatic activity correlated well with the extracellular appearance of each enzyme in the different strains tested (Figs. 2c, 3c).
To determine the extent to which the two model proteins, alpha-amylase and agarase, could use the Tat or the Sec route respectively, their signal peptides were interchanged and fused in-frame to the corresponding mature part of the other enzyme, and the capacity of the chimerical proteins to choose either route was determined by Western blot assays using antibodies against agarase or alpha-amylase.
Propagation of the multicopy plasmid pAMI2003 encoding the mature alpha-amylase fused to the agarase signal peptide resulted in neither cell-associated nor extracellular detection of the enzyme, or extracellular activity of alpha-amylase in the wild-type strain, as if the secretion of alpha-amylase would not have been possible via the Tat route, perhaps due to its relative structural complexity. Consequently, neither protein nor activity was detected in the SecG- or TatC-deficient strains (Fig. 2b, c). Hence, the absence of cell-associated pre-alpha-amylase suggests a possible degradation by intracellular proteases.
Propagation in S. lividans TK21 of the multicopy plasmid pAGA502, encoding the mature agarase fused to the alpha-amylase signal peptide, resulted in the detection of pre-agarase associated with the bacterial cell and extracellular mature agarase. The presence of cell-associated chimerical pre-agarase seems to indicate that the pre-protein was retained in the cell, which is not the case when agarase is fused to its own signal peptide (Fig. 3b).
Agarase appeared later in the supernatant when plasmid pAGA502 was propagated in the SecG-deficient strain, as expected for a pre-protein containing a Sec signal peptide targeted to the Sec route (Fig. 3b). Consequently, the amount of cell-associated pre-agarase was larger than that of the wild-type strain carrying the same plasmid.
In the TatC-deficient strain carrying plasmid pAGA502, mature agarase accumulated at late phases of growth, while pre-agarase remained associated with the bacterial cell (Fig. 3b). The Tat route is not functional in the TatC-deficient strain, therefore, the appearance of agarase in the supernatants is a likely result if the alpha-amylase Sec signal peptide triggers agarase secretion via the Sec route.
The agarase activity measured in the wild-type and Sec-deficient strains carrying plasmid pAGA502 was lower mainly at later phases of growth than that of the corresponding strains harbouring plasmid pAGAs5, probably indicating the requirement of some Sec-specific protein-folding elements, absent at late phases of growth, that may be necessary to correctly fold the otherwise unfolded protein secreted via the Sec route. The agarase activity observed in the Tat-deficient strain harbouring plasmid pAGA502 is due to the agarase targeted by the Sec route and its low level correlated well with the low amount of secreted agarase observed in that strain (Fig. 3c).
These results, together with the extracellular absence of alpha-amylase targeted by the agarase signal peptide, strongly suggest that apart from the signal peptide it is likely that other parts of the mature proteins may play a role in the effective secretion of the active proteins in S. lividans, as previously described in E. coli [31], however, one would have expected the secondary or eventually tertiary structure of the proteins to evolve in such a way that the cell could determine the selection of a particular secretory route for each type of protein. Thus, most of the S. coelicolor unequivocally assessed Tat proteins [5] did not to have predicted disulphide bonds when the secondary structure was analysed, while the set of major Sec secretory proteins [32] had at least one predicted disulphide bond (data not shown). Therefore, the potentially less complex structure of a secretory protein, such as agarase, could facilitate its exportation via either the Sec or Tat routes. Sec proteins with a potentially more complex structure, such as alpha-amylase, are unlikely to use the Tat route, when being targeted by a Tat signal peptide.
The obtained results seem to indicate that the Sec route could be more effective when engineering extracellular secretory protein production in Streptomyces, while the Tat route could exclusively be used by a selected minor subset of proteins.
These features could be of particular importance when designing and engineering overproduction and secretion of homologous or heterologous proteins in Streptomyces strains.
References
Anné, J., Maldonado, B., Van Impe, J., Van Mellaert, L., & Bernaerts, K. (2012). Recombinant protein production and streptomycetes. Journal of Biotechnology, 158, 159–167.
Palacín, A., de la Fuente, R., Valle, I., Rivas, L. A., & Mellado, R. P. (2003). Streptomyces lividans contains a minimal functional signal recognition particle that is involved in protein secretion. Microbiology, 149, 2435–2442.
Berks, B. C., Palmer, T., & Sargent, F. (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Current Opinion in Microbiology, 8, 174–181.
Jongbloed, J. D., Antelmann, H., Hecker, M., Nijland, R., Bron, S., Airaksinen, U., et al. (2002). Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. Journal of Biological Chemistry, 277, 44068–44078.
Widdick, D. A., Dilks, K., Chandra, G., Bottrill, A., Naldrett, M., Pohlschroder, M., & Palmer, T. (2006). The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proceedings of the National Academy of Sciences of the United States of America, 103, 17927–17932.
Mellado, R. P. (2011). Summing up particular features of protein secretion in Streptomyces lividans. World Journal of Microbiology & Biotechnology, 27, 2231–2237.
Schaerlaekens, K., Schierová, M., Lammertyn, E., Geukens, N., Anné, J., & Van Mellaert, L. (2001). Twin-arginine translocation pathway in Streptomyces lividans. Journal of Bacteriology, 183, 6727–6732.
De Keersmaeker, S., Vrancken, K., Van Mellaert, L., Anné, J., & Geukens, N. (2007). The Tat pathway in Streptomyces lividans: Interaction of Tat subunits and their role in translocation. Microbiology, 153, 1087–1094.
Palmer, T., & Berks, B. C. (2012). The twin-arginine translocation (Tat) protein export pathway. Nature Reviews Microbiology, 10, 483–496.
Hopwood, D. A., Bibb, M. J., & Chater, K. F. (1985). Genetic manipulation of Streptomyces. A laboratory manual. Norwich: John Innes Foundation.
Palomino, C., & Mellado, R. P. (2008). Influence of a Streptomyces lividans SecG functional analogue on protein secretion. International Microbiology, 11, 25–31.
Ward, J. M., Janssen, G. R., Kieser, T., Bibb, M. J., Buttner, M. J., & Bibb, M. J. (1986). Construction and characterisation of a series of multi-copy promoter-probe vectors Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Molecular and General Genetics, 203, 468–475.
Palacín, A., Parro, V., Geukens, N., Anné, J., & Mellado, R. P. (2002). SipY is the Streptomyces lividans type I signal peptidase exerting a major effect on protein secretion. Journal of Bacteriology, 184, 4875–4880.
Parro, V., & Mellado, R. P. (1993). Heterologous recognition in vivo of promoter sequences from the Streptomyces coelicolor dagA gene. FEMS Microbiology Letters, 106, 347–356.
Isiegas, C., Parro, V., & Mellado, R. P. (1999). Streptomyces lividans as a host for the production and secretion of Escherichia coli TEM α-lactamase. Letters in Applied Microbiology, 28, 321–326.
Gust, B., Challis, G. L., Fowler, K., Kieser, T., & Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterne soil odor geosmin. Proceedings of the National Academy of Sciences of the United States of America, 100, 1541–1546.
Escutia, M. R., Val, G., Palacín, A., Geukens, N., Anné, J., & Mellado, R. P. (2006). Compensatory effect of the minor Streptomyces lividans type I signal peptidases on the SipY major signal peptidase deficiency as determined by extracellular proteome analysis. Proteomics, 6, 4137–4146.
Thompson, C. J., Kieser, T., Ward, J. M., & Hopwood, D. A. (1982). Physical analysis of antibiotic-resistant genes from Streptomyces and their use in vector construction. Gene, 20, 51–62.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Timmons, T. M., & Dunbar, B. S. (1990). Protein blotting and immunodetection. Methods in Enzymology, 182, 679–688.
Gullón, S., Vicente, L. R., & Mellado, R. P. (2012). A novel two-component system involved in secretion stress response in Streptomyces lividans. PLoS ONE, 7(11), e48987. doi:10.1371/journal.pone.0048987.
Parro, V., & Mellado, R. P. (1994). Effect of glucose on agarase overproduction by Streptomyces. Gene, 145, 49–55.
Rabindran, S. K., Haroun, R. I., Wisniewski, J., & Wu, C. (1993). Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science, 259, 230–234.
Rice, P., Longden, I., & Bleasby, A. (2000). EMBOSS: The European molecular biology open software suite. Trends in Genetics, 16, 276–277. doi:10.1016/S0168-9525(00)02024-2.
Cole, C., Barber, J. D., & Barton, G. J. (2008). The Jpred 3 secondary structure prediction server. Nucleics Acids Research, 36(suppl 2), W197–W201.
Prilusky, J., Felder, C. E., Zeev-Ben-Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., et al. (2005). FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics, 21, 3435–3438. doi:10.1093/bioinformatics/bti537.
Lin, H. N., Sung, T. Y., Ho, S. Y., & Hsu, W. L. (2010). Improving protein secondary structure prediction based on short subsequences with local structure similarity. BMC Genomics, 11(Suppl 4), S4. doi:10.1186/1471-2164-11-S4-S4.
Källberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., & Xu, J. (2012). Template-based protein structure modelling using the RaptorX web server. Nature Protocols, 7, 1511–1522. doi:10.1038/nprot.2012.085.
Ferrè, F., & Clote, P. (2006). DIANNA 1.1: An extension of the DIANNA web server for ternary cysteine classification. Nucleic Acids Research, 34(suppl 2), W182–W185.
Schaerlaekens, K., Van Mellaert, L., Lammertyn, E., Geukens, N., & Anné, J. (2004). The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome analysis. Microbiology, 150, 21–31.
Cristóbal, S., de Gier, J. W., Nielsen, H., & von Heijne, G. (1999). Competition between Sec-and TAT-dependent protein translocation in Escherichia coli. EMBO Journal, 18, 2982–2990.
Gullón, S., Palomino, C., Navajas, R., Paradela, A., & Mellado, R. P. (2012). Translocase and major signal peptidase malfunctions affect aerial mycelium formation in Streptomyces lividans. Journal of Biotechnology, 160, 112–122.
Acknowledgments
The Spanish Ministry of the Environment and Rural and Marine affairs has commissioned and supported this research (Grant No. EGO22008). Grant PIE201220E003 from the CSIC also supported this research. We wish to thank C. Palomino for her contribution to obtaining S. lividans ∆tatC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gullón, S., Vicente, R.L., Valverde, J.R. et al. Exploring the Feasibility of the Sec Route to Secrete Proteins Using the Tat Route in Streptomyces lividans . Mol Biotechnol 57, 931–938 (2015). https://doi.org/10.1007/s12033-015-9883-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9883-0